Cells that register logical relationships among proteins - PubMed (original) (raw)
Cells that register logical relationships among proteins
C W Xu et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Two-hybrid methods have augmented the classical genetic techniques biologists use to assign function to genes. Here, we describe construction of a two-bait interaction trap that uses yeast cells to register more complex protein relationships than those detected in existing two-hybrid systems. We show that such cells can identify bridge or connecting proteins and peptide aptamers that discriminate between closely related allelic variants. The protein relationships detected by these cells are analogous to classical genetic relationships, but lend themselves to systematic application to the products of entire genomes and combinatorial libraries. We show that, by performing logical operations on the phenotypic outputs of these complex cells and existing two-hybrid cells, we can make inferences about the topology and order of protein interactions. Finally, we show that cells that register such relationships can perform logical operations on protein inputs. Thus these cells will be useful for analysis of gene and allele function, and may also define a path for construction of biological computational devices.
Figures
Figure 1
The two-bait interaction trap. Cells contain bait1, a protein fusion of Tet repressor and protein X, and bait2, a fusion of LexA and protein Y. In cell 1, a prey, a fusion of protein Z1 and transcriptional activator B42, does not interact with either X or Y. The reporters are not activated, the cell grows on 5-FOA, does not grow on Ura− medium, and is white on X-Gal. In cell 2, another prey, a fusion of Z2 and B42 interacts with X but not with Y, the TetOp-URA3 reporter is selectively expressed, and the cell grows on Ura− medium but is white on X-Gal.
Figure 2
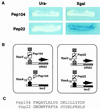
Peptide aptamers that discriminate between Ras alleles. (A) Reporter phenotypes of cells on Ura− or X-Gal medium. These cells contain TetR-RasV12 and LexA-RasA15, and also peptide aptamer Pep104 or Pep22. The Pep104 containing cell grows on Ura− medium but is white on X-Gal medium. The Pep22 cell grows on Ura− medium and is blue on X-Gal. (B) Protein relationships in these two-bait cells. Pep104 interacts only with RasV12 but not with RasA15, and thus selectively activates the TetOp-URA3 reporter. Pep22 interacts with both RasV12 and RasA15, and thus activates both the TetOp-URA3 and LexOp-lacZ reporters. (C) Sequences of the variable (recognition) regions of peptide aptamers Pep104 and Pep22.
Figure 3
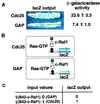
Cells that perform the logical And operation on input proteins. (A) Expression of the LexOp-lacZ reporter in cells containing TetR-Cdc25 (907–1,589) or GAP as measured by the intensity of blue color on X-Gal medium and by β-galactosidase activity in Miller units. (B) Inferred protein relationships in these cells. In the cell expressing TetR-Cdc25, Cdc25 loads LexA-Ras with GTP, increasing the amount of Ras that is in the GTP bound conformation (black outline) with which B42-c-Raf1 can interact to stimulate transcription of the lacZ reporter, and decreasing the amount of Ras in the GDP-bound conformation (grey outline) with which B42-c-Raf-1 cannot interact. In the cell expressing TetR-GAP, this switch is reversed. In this cell, TetR-GAP stimulates Ras GTPase activity, increasing the amount of Ras in the GDP-bound form (black outline), with which B42-c-Raf1 cannot interact, and decreasing the amount of Ras in the GTP-bound form (grey outline), thus resulting in lowered lacZ output. (C) Table depicting results of these operations on protein inputs. In the table, the 1’s in the first column indicate the presence of B42-Raf, the 0 and 1 in the second column denote, respectively, the presence of TetR-Gap and TetR-Cdc25, and the 0 and 1 in the third column denote respectively low and high outputs of β-galactosidase.
Similar articles
- "Mutagenesis" by peptide aptamers identifies genetic network members and pathway connections.
Geyer CR, Colman-Lerner A, Brent R. Geyer CR, et al. Proc Natl Acad Sci U S A. 1999 Jul 20;96(15):8567-72. doi: 10.1073/pnas.96.15.8567. Proc Natl Acad Sci U S A. 1999. PMID: 10411916 Free PMC article. - Understanding gene and allele function with two-hybrid methods.
Brent R, Finley RL Jr. Brent R, et al. Annu Rev Genet. 1997;31:663-704. doi: 10.1146/annurev.genet.31.1.663. Annu Rev Genet. 1997. PMID: 9442911 Review. - Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens.
Fromont-Racine M, Rain JC, Legrain P. Fromont-Racine M, et al. Nat Genet. 1997 Jul;16(3):277-82. doi: 10.1038/ng0797-277. Nat Genet. 1997. PMID: 9207794 - Selection and characterization of large collections of peptide aptamers through optimized yeast two-hybrid procedures.
Bickle MB, Dusserre E, Moncorgé O, Bottin H, Colas P. Bickle MB, et al. Nat Protoc. 2006;1(3):1066-91. doi: 10.1038/nprot.2006.32. Nat Protoc. 2006. PMID: 17406388 - [Decade of genomics--methods for genome investigation in yeast Saccharomyces cerevisiae].
Skoneczna A. Skoneczna A. Postepy Biochem. 2006;52(4):435-47. Postepy Biochem. 2006. PMID: 17536513 Review. Polish.
Cited by
- LCK-phosphorylated human killer cell-inhibitory receptors recruit and activate phosphatidylinositol 3-kinase.
Marti F, Xu CW, Selvakumar A, Brent R, Dupont B, King PD. Marti F, et al. Proc Natl Acad Sci U S A. 1998 Sep 29;95(20):11810-5. doi: 10.1073/pnas.95.20.11810. Proc Natl Acad Sci U S A. 1998. PMID: 9751747 Free PMC article. - "Mutagenesis" by peptide aptamers identifies genetic network members and pathway connections.
Geyer CR, Colman-Lerner A, Brent R. Geyer CR, et al. Proc Natl Acad Sci U S A. 1999 Jul 20;96(15):8567-72. doi: 10.1073/pnas.96.15.8567. Proc Natl Acad Sci U S A. 1999. PMID: 10411916 Free PMC article. - Targeted modification and transportation of cellular proteins.
Colas P, Cohen B, Ko Ferrigno P, Silver PA, Brent R. Colas P, et al. Proc Natl Acad Sci U S A. 2000 Dec 5;97(25):13720-5. doi: 10.1073/pnas.97.25.13720. Proc Natl Acad Sci U S A. 2000. PMID: 11106396 Free PMC article. - Next-Generation Sequencing for Binary Protein-Protein Interactions.
Suter B, Zhang X, Pesce CG, Mendelsohn AR, Dinesh-Kumar SP, Mao JH. Suter B, et al. Front Genet. 2015 Dec 17;6:346. doi: 10.3389/fgene.2015.00346. eCollection 2015. Front Genet. 2015. PMID: 26734059 Free PMC article. - Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal.
Wu G, Qiu XL, Zhou L, Zhu J, Chamberlin R, Lau J, Chen PL, Lee WH. Wu G, et al. Cancer Res. 2008 Oct 15;68(20):8393-9. doi: 10.1158/0008-5472.CAN-08-1915. Cancer Res. 2008. PMID: 18922912 Free PMC article.
References
- Hartman P E, Roth J R. Adv Genet. 1973;17:1–105. - PubMed
- Botstein D, Maurer R. Annu Rev Genet. 1982;16:61–83. - PubMed
- Herskowitz I. Nature (London) 1987;329:219–222. - PubMed
- Hereford L M, Hartwell L H. J Mol Biol. 1974;84:445–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases