Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I - PubMed (original) (raw)
Comparative Study
. 1997 Dec;8(12):2553-62.
doi: 10.1091/mbc.8.12.2553.
Affiliations
- PMID: 9398675
- PMCID: PMC25727
- DOI: 10.1091/mbc.8.12.2553
Free PMC article
Comparative Study
Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I
D E Warnock et al. Mol Biol Cell. 1997 Dec.
Free PMC article
Abstract
To begin to understand mechanistic differences in endocytosis in neurons and nonneuronal cells, we have compared the biochemical properties of the ubiquitously expressed dynamin-II isoform with those of neuron-specific dynamin-I. Like dynamin-I, dynamin-II is specifically localized to and highly concentrated in coated pits on the plasma membrane and can assemble in vitro into rings and helical arrays. As expected, the two closely related isoforms share a similar mechanism for GTP hydrolysis: both are stimulated in vitro by self-assembly and by interaction with microtubules or the SH3 domain-containing protein, grb2. Deletion of the C-terminal proline/arginine-rich domain from either isoform abrogates self-assembly and assembly-dependent increases in GTP hydrolysis. However, dynamin-II exhibits a approximately threefold higher rate of intrinsic GTP hydrolysis and higher affinity for GTP than dynamin-I. Strikingly, the stimulated GTPase activity of dynamin-II can be >40-fold higher than dynamin-I, due principally to its greater propensity for self-assembly and the increased resistance of assembled dynamin-II to GTP-triggered disassembly. These results are consistent with the hypothesis that self-assembly is a major regulator of dynamin GTPase activity and that the intrinsic rate of GTP hydrolysis reflects a dynamic, GTP-dependent equilibrium of assembly and disassembly.
Figures
Figure 1
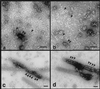
Guanine nucleotides influence the distribution of dynII on coated pits in A431 cells. Upper panels show coated pits with various degrees of curvature detected at steady state on ripped-off plasma membrane preparations from perforated A431 cells. Endogenous dynII was detected using hudy-1 mAb followed by 10 nm gold-conjugated secondary antibodies. Dynamin is uniformly distributed on flat lattices (a, b) and on curved coated pits (b–d) but is localized peripherally at the base around the more deeply invaginated coated pits (e, f) distinguished by the pool of stain encircling their base. Lower panels show representative fields from perforated A431 cells incubated in the presence of GTP (g), GTPγS (h), or GDPβS (i). ∼39,000×.
Figure 2
Self-assembly of dynI and dynII is dependent on an intact PRD. Self-assembly of dynI and dynII and their ΔPRD counterparts was followed by a sedimentation assay that separates dynamin structures that pellet (P) from unassembled dynamin that remains in the supernatant (S) after centrifugation at 100,000 × g for 15 min. Dynamins (in HCB150) were diluted 10-fold to a final protein concentration of 0.2 mg/ml, into either HCB150 (high-salt) or PH (low-salt) buffer. Equal aliquots of soluble and pelleted dynamin were analyzed by SDS-PAGE on 7.5% acrylamide gels and detected by Coomassie brilliant blue staining. The full gels are shown to demonstrate the purity of the protein preparation. Molecular weight protein standards are as indicated.
Figure 3
DynI and dynII self-assemble into similar oligomeric structures. DynI (a) and dynII (b) were self-assembled by dialysis into HCB30 containing 1% ethylene glycol and examined by electron microscopy in negative stain. Arrowheads indicate rings and small stacks of rings. In c and d, dynI and dynII, respectively, were incubated with taxol-stabilized microtubules at 0.1 mg/ml in PH buffer. Final dynamin concentration in each condition was 0.1 mg/ml. Arrowheads show dynamin rings encircling the microtubule template. Bars, 100 nm.
Figure 4
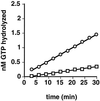
DynII has a higher endogenous rate of GTP hydrolysis than dynI. Intrinsic GTPase activities of dynI (□) and dynII (○) were measured in 20 μl of PH buffer as described in EXPERIMENTAL PROCEDURES and containing 0.2 μg of dynamin and 0.25 mM GTP. Backgrounds obtained without dynamin have been subtracted. A representative experiment is shown; average rates determined from four independent experiments are given in the text.
Figure 5
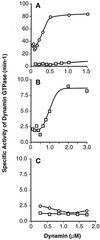
The positive cooperative behavior of dynII GTPase activity requires the C-terminal PRD. GTPase activities of increasing dynamin concentrations were measured as described in EXPERIMENTAL PROCEDURES. (A) Concentration dependence of the specific activity of dynII as compared with dynI. Data for dynI are replotted on an expanded scale in B to illustrate the cooperative behavior of dynI specific activity. C compares the specific activities of truncated dynI and dynII, lacking their C-terminal PRD. Each point represents the linear rate of GTP hydrolysis derived from five time points. Data are representative of three separate experiments.
Figure 6
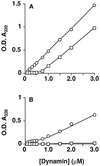
DynII shows a greater propensity for self-assembly than dynI. Light scattering measured at _A_320 nm was used to follow dynamin self-assembly as described in EXPERIMENTAL PROCEDURES. DynI (□) or dynII (○) at 2 mg/ml in HCB150 was diluted to the indicated protein concentrations in PH buffer in the absence (A) or presence (B) of 250 μM GTP. Assays were in a final volume of 200 μl of PH buffer, fixed at 50 mM ionic strength. Steady-state assembly was reached after 20 min even at low protein concentrations, at which point the absorbance values shown were recorded. Care was taken at higher protein concentrations that readings of light scattering were taken well before complete hydrolysis of the GTP. Each point represents the mean derived from three independent determinations with all standard deviations smaller than the symbols. Data are representative of three separate experiments.
Figure 7
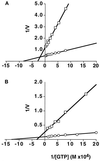
Michaelis-Menton kinetics of GTP hydrolysis by dynI and dynII. The assembly-independent rates of GTP hydrolysis (A) were determined using ΔPRD mutants of dynI (□) and dynII (○) (at 1.3 μM and 0.88 μM, respectively) in the presence of increasing concentrations of GTP and a plot of 1/V (nmol/l/min) versus 1/[GTP] (M) is shown. Assembly-dependent rates of GTP hydrolysis (B) were determined for dynI (□) and dynII (○) (both at 0.1 μM) in the presence of 0.1 mg/ml taxol-stabilized microtubules.
Similar articles
- Dynamin self-assembly stimulates its GTPase activity.
Warnock DE, Hinshaw JE, Schmid SL. Warnock DE, et al. J Biol Chem. 1996 Sep 13;271(37):22310-4. doi: 10.1074/jbc.271.37.22310. J Biol Chem. 1996. PMID: 8798389 - Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis.
Song BD, Leonard M, Schmid SL. Song BD, et al. J Biol Chem. 2004 Sep 24;279(39):40431-6. doi: 10.1074/jbc.M407007200. Epub 2004 Jul 19. J Biol Chem. 2004. PMID: 15262989 - Domain structure and intramolecular regulation of dynamin GTPase.
Muhlberg AB, Warnock DE, Schmid SL. Muhlberg AB, et al. EMBO J. 1997 Nov 17;16(22):6676-83. doi: 10.1093/emboj/16.22.6676. EMBO J. 1997. PMID: 9362482 Free PMC article. - Dynamin GTPase, a force-generating molecular switch.
Warnock DE, Schmid SL. Warnock DE, et al. Bioessays. 1996 Nov;18(11):885-93. doi: 10.1002/bies.950181107. Bioessays. 1996. PMID: 8939066 Review. - Building a fission machine--structural insights into dynamin assembly and activation.
Chappie JS, Dyda F. Chappie JS, et al. J Cell Sci. 2013 Jul 1;126(Pt 13):2773-84. doi: 10.1242/jcs.108845. Epub 2013 Jun 18. J Cell Sci. 2013. PMID: 23781021 Free PMC article. Review.
Cited by
- Evidence that dynamin-2 functions as a signal-transducing GTPase.
Fish KN, Schmid SL, Damke H. Fish KN, et al. J Cell Biol. 2000 Jul 10;150(1):145-54. doi: 10.1083/jcb.150.1.145. J Cell Biol. 2000. PMID: 10893263 Free PMC article. - Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis.
Kirchhausen T, Macia E, Pelish HE. Kirchhausen T, et al. Methods Enzymol. 2008;438:77-93. doi: 10.1016/S0076-6879(07)38006-3. Methods Enzymol. 2008. PMID: 18413242 Free PMC article. - The Lipid-Binding Defective Dynamin 2 Mutant in Charcot-Marie-Tooth Disease Impairs Proper Actin Bundling and Actin Organization in Glomerular Podocytes.
Hamasaki E, Wakita N, Yasuoka H, Nagaoka H, Morita M, Takashima E, Uchihashi T, Takeda T, Abe T, Lee JW, Iimura T, Saleem MA, Ogo N, Asai A, Narita A, Takei K, Yamada H. Hamasaki E, et al. Front Cell Dev Biol. 2022 May 10;10:884509. doi: 10.3389/fcell.2022.884509. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35620056 Free PMC article. - A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells.
Srinivasan S, Burckhardt CJ, Bhave M, Chen Z, Chen PH, Wang X, Danuser G, Schmid SL. Srinivasan S, et al. PLoS Biol. 2018 Apr 18;16(4):e2005377. doi: 10.1371/journal.pbio.2005377. eCollection 2018 Apr. PLoS Biol. 2018. PMID: 29668686 Free PMC article. - Single Intramuscular Injection of AAV-shRNA Reduces DNM2 and Prevents Myotubular Myopathy in Mice.
Tasfaout H, Lionello VM, Kretz C, Koebel P, Messaddeq N, Bitz D, Laporte J, Cowling BS. Tasfaout H, et al. Mol Ther. 2018 Apr 4;26(4):1082-1092. doi: 10.1016/j.ymthe.2018.02.008. Epub 2018 Feb 14. Mol Ther. 2018. PMID: 29506908 Free PMC article.
References
- Chen MS, Ober RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, Vallee RB. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. - PubMed
- Cook T, Mesa K, Urrutia R. Three dynamin encoding genes are differentially expressed in developing rat brain. J Neurochem. 1996;67:927–931. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous