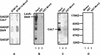Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis - PubMed (original) (raw)
Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis
M Lei et al. Genes Dev. 1997.
Abstract
The initiation of DNA synthesis is an important cell cycle event that defines the beginning of S phase. This critical event involves the participation of proteins whose functions are regulated by cyclin dependent protein kinases (Cdks). The Mcm2-7 proteins are a family of six conserved proteins that are essential for the initiation of DNA synthesis in all eukaryotes. In Saccharomyces cerevisiae, members of the Mcm2-7 family undergo cell cycle-specific phosphorylation. Phosphorylation of Mcm proteins at the beginning of S phase coincides with the removal of these proteins from chromatin and the onset of DNA synthesis. In this study, we identified DBF4, which encodes the regulatory subunit of a Cdk-like protein kinase Cdc7-Dbf4, in a screen for second site suppressors of mcm2-1. The dbf4 suppressor mutation restores competence to initiate DNA synthesis to the mcm2-1 mutant. Cdc7-Dbf4 interacts physically with Mcm2 and phosphorylates Mcm2 and three other members of the Mcm2-7 family in vitro. Blocking the kinase activity of Cdc7-Dbf4 at the G1-to-S phase transition also blocks the phosphorylation of Mcm2 at this defined point of the cell cycle. Taken together, our data suggest that phosphorylation of Mcm2 and probably other members of the Mcm2-7 proteins by Cdc7-Dbf4 at the G1-to-S phase transition is a critical step in the initiation of DNA synthesis at replication origins.
Figures
Figure 1
The DBF4 gene complements both the heat-sensitive suppression and the cold-sensitive growth phenotype of mts2-1. mts2-1::DBF4 contains a wild-type DBF4 gene integrated at the dbf4 locus of mts2-1. Yeast cells were streaked onto YEPD plates and grown at 38.5°C for 3 days (A), or at 14°C for 7 days (B). (C) The result of the tetrad analysis of a diploid strain that is mcm2-1 homozygous and dbf4-3 heterozygous (mcm2-1 dbf4-3/mcm2-1 DBF4; see Materials and Methods for construction of the strain). Tetrads were dissected on YEPD plates and grown at 30°C for 3 days.
Figure 1
The DBF4 gene complements both the heat-sensitive suppression and the cold-sensitive growth phenotype of mts2-1. mts2-1::DBF4 contains a wild-type DBF4 gene integrated at the dbf4 locus of mts2-1. Yeast cells were streaked onto YEPD plates and grown at 38.5°C for 3 days (A), or at 14°C for 7 days (B). (C) The result of the tetrad analysis of a diploid strain that is mcm2-1 homozygous and dbf4-3 heterozygous (mcm2-1 dbf4-3/mcm2-1 DBF4; see Materials and Methods for construction of the strain). Tetrads were dissected on YEPD plates and grown at 30°C for 3 days.
Figure 2
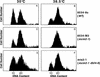
Growth arrest of mcm2-1 cells with nearly 2C DNA at 38.5°C is reversed by the dbf4-6 mutation. 8534-8c (WT), 8534-M2 (mcm2-1) and mts2-1 (mcm2-1 dbf4-6) cells grown at 30°C or 38.5°C were stained with propidium iodide, and the DNA content of the cells were analyzed by flow cytometry. (A,C,E) DNA content profiles of the wild-type, mutant and suppressor strains at 30°C. (B,D,F) DNA content profiles of the three strains at 38.5°C.
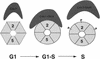
Figure 3
Two-dimensional DNA gel analysis of the initiation of DNA synthesis at replication origins in 8534-8c (WT), 8534-M2 (mcm2-1) and mts2-1 (mcm2-1 dbf4-6) cells. (A–F) Analysis of replication events at ORI121. (G–L) Analysis of replication events at ORI1.
Figure 4
Physical interactions between Cdc7–Dbf4 and Mcm2. (A) Two-hybrid analysis. BTM116 and BTM116.Mcm2 denote plasmids that express LexA and LexA–Mcm2 proteins, respectively. GAD2F, GAD2F.Cdc7, and GAD2F.Dbf4 denote plasmids that express GAL4, GAL4–Cdc7, and GAL4–Dbf4 proteins, respectively. Yeast transformants carrying each pair of the plasmids and a LexAop–lacZ reporter gene were patched on 5-bromo-4-chloro-3-indolyl-β-
d
-galactoside (X-gal) plates. The expression of β-galactosidase is shown by the blue color of colonies. (B,C) GST–Mcm2 fusion affinity column chromatography. (B) Western blot with anti-LexA antibodies. (Lane 1) Total yeast protein extract from BJ2168 cells carrying pKH125; (lane 2,3) proteins eluted from glutathione Sepharose 4B beads coupled with GST (lane 2) and GST.Mcm2 (lane 3). (C) Western blot with anti-Cdc7 antibodies. (Lane 1) Total yeast protein extract from BJ2168 cells; (lanes 2,3) proteins eluted from glutathione Sepharose 4B beads coupled with GST (lane 2) and GST.Mcm2 (lane 3). (D) Coomassie Brilliant Blue staining of a SDS-PAGE showing the GST or GST–Mcm2 released from the beads by 10 m
m
glutathione after elution of the interacting proteins.
Figure 5
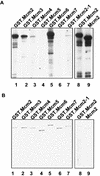
Mcm2, Mcm3, Mcm4, and Mcm6 are substrates of the Cdc7–Dbf4 kinase in vitro. The kinase assay was performed with Cdc7–Dbf4 protein kinase purified from Sf9 cells, and GST–Mcm proteins purified from yeast. The reactions were resolved in SDS–polyacrylamide gels for autoradiography (A), or for Coomassie brilliant blue staining (B). GST.Mcm2, (lane 1); GST.Mcm3, (lane 2); GST.Mcm4, (lane 3); GST.Mcm5, (lane 4); GST.Mcm6, (lane 5); GST.Mcm7, (lane 6); GST.Mcm2-1, (lane 7); GST.Mcm2, (lane 8); Mcm2, (lane 9).
Figure 6
Phosphorylation of Mcm2 at the G1-to-S-phase transition is blocked when Cdc7–Dbf4 kinase is inactivated. The isoform distributions of Mcm2 (left) and Mcm5 (right) were analyzed by immunoblots of two-dimensional protein gels with monospecific antibodies. dbf4-1 cells used in this assay were first arrested at Start by α factor at 30°C. Proteins were isolated from cells released from the α factor block and then arrested at G1-to-S-phase transition by shifting to 37°C (top) and from cells released from the α factor block and then arrested at early S by adding HU to the culture (bottom). In the first dimension gel, proteins are separated by isoelectrofocusing (IEF). In the second dimension gel, proteins are separated by molecular weight (MW). The two-dimensional gels were aligned with chemiluminescence signals from protein aggregates that did not enter the first dimension gel (Young and Tye 1997).
Figure 7
A model for the regulation of Mcm2 by the Cdc7–Dbf4 kinase in the initiation of DNA synthesis at replication origins. Cdc7–Dbf4 interacts with and phosphorylates Mcm2, and likely several other members of the Mcm2–7 proteins. We propose that during the G1-to-S-phase transition, Cdc7–Dbf4 interacts with Mcm2, which is in a complex with other members of the Mcm2–7 family. Phosphorylation of Mcm2 and other subunits in this complex drives the complex into an active conformation that can also be achieved by a single base change mutation in MCM5 (Hardy et al. 1997). This active conformation signals subsequent steps in the replication initiation process.
References
- Adams AEM, Botstein D, Drubin DG. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Nature. 1989;243:231–233. - PubMed
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
- Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous