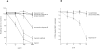Identification of Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) as the rosetting ligand of the malaria parasite P. falciparum - PubMed (original) (raw)
Identification of Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) as the rosetting ligand of the malaria parasite P. falciparum
Q Chen et al. J Exp Med. 1998.
Abstract
Severe Plasmodium falciparum malaria is characterized by excessive sequestration of infected and uninfected erythrocytes in the microvasculature of the affected organ. Rosetting, the adhesion of P. falciparum-infected erythrocytes to uninfected erythrocytes is a virulent parasite phenotype associated with the occurrence of severe malaria. Here we report on the identification by single-cell reverse transcriptase PCR and cDNA cloning of the adhesive ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1). Rosetting PfEMP1 contains clusters of glycosaminoglycan-binding motifs. A recombinant fusion protein (Duffy binding-like 1-glutathione S transferase; Duffy binding-like-1-GST) was found to adhere directly to normal erythrocytes, disrupt naturally formed rosettes, block rosette reformation, and bind to a heparin-Sepharose matrix. The adhesive interactions could be inhibited with heparan sulfate or enzymes that remove heparan sulfate from the cell surface whereas other enzymes or similar glycosaminoglycans of a like negative charge did not affect the binding. PfEMP1 is suggested to be the rosetting ligand and heparan sulfate, or a heparan sulfate-like molecule, the receptor both for PfEMP1 binding and naturally formed erythrocyte rosettes.
Figures
Figure 1
Identification of rosetting PfEMP1. (A) Rosetting, single _P. falciparum_–infected erythrocyte is seen by light microscopy held by a 5-μm micropipette (A, 1). The uninfected erythrocytes are stripped of the infected cell and careful examination confirms that it indeed is infected by a single parasite (A, 2–3). B shows the amplification of a 434-bp band in four (from reactions 3, 4, 5, and 7) out of eight single-infected, rosetting erythrocytes using degenerate primers generated from the primary sequence of the DBL-1 domain of PfEMP1. C shows the amplification pattern with the same primers as in B of bulk cultures of rosetting (R+) FCR3S1.2 cultures and the R− FCR3s/a parasites. Note that the 434-bp product is only seen with the R+ parasites. D shows the hybridization pattern in Northern blotting of the 434-bp sequence to mRNA extracted from the highly rosetting parasite FCR3S1.2 (84% R+) and the weak hybridization to the R− FCR3S/a parasite (9% R+). E shows the autoradiogarph of a Triton X-100 insoluble, SDS-soluble extract of FCR3S1.2-infected erythrocytes after radio-iodination labeling. PfEMP1 (arrow) is labeled on FCR3S1.2-infected erythrocytes and is cleaved by low concentrations of trypsin.
Figure 1
Identification of rosetting PfEMP1. (A) Rosetting, single _P. falciparum_–infected erythrocyte is seen by light microscopy held by a 5-μm micropipette (A, 1). The uninfected erythrocytes are stripped of the infected cell and careful examination confirms that it indeed is infected by a single parasite (A, 2–3). B shows the amplification of a 434-bp band in four (from reactions 3, 4, 5, and 7) out of eight single-infected, rosetting erythrocytes using degenerate primers generated from the primary sequence of the DBL-1 domain of PfEMP1. C shows the amplification pattern with the same primers as in B of bulk cultures of rosetting (R+) FCR3S1.2 cultures and the R− FCR3s/a parasites. Note that the 434-bp product is only seen with the R+ parasites. D shows the hybridization pattern in Northern blotting of the 434-bp sequence to mRNA extracted from the highly rosetting parasite FCR3S1.2 (84% R+) and the weak hybridization to the R− FCR3S/a parasite (9% R+). E shows the autoradiogarph of a Triton X-100 insoluble, SDS-soluble extract of FCR3S1.2-infected erythrocytes after radio-iodination labeling. PfEMP1 (arrow) is labeled on FCR3S1.2-infected erythrocytes and is cleaved by low concentrations of trypsin.
Figure 1
Identification of rosetting PfEMP1. (A) Rosetting, single _P. falciparum_–infected erythrocyte is seen by light microscopy held by a 5-μm micropipette (A, 1). The uninfected erythrocytes are stripped of the infected cell and careful examination confirms that it indeed is infected by a single parasite (A, 2–3). B shows the amplification of a 434-bp band in four (from reactions 3, 4, 5, and 7) out of eight single-infected, rosetting erythrocytes using degenerate primers generated from the primary sequence of the DBL-1 domain of PfEMP1. C shows the amplification pattern with the same primers as in B of bulk cultures of rosetting (R+) FCR3S1.2 cultures and the R− FCR3s/a parasites. Note that the 434-bp product is only seen with the R+ parasites. D shows the hybridization pattern in Northern blotting of the 434-bp sequence to mRNA extracted from the highly rosetting parasite FCR3S1.2 (84% R+) and the weak hybridization to the R− FCR3S/a parasite (9% R+). E shows the autoradiogarph of a Triton X-100 insoluble, SDS-soluble extract of FCR3S1.2-infected erythrocytes after radio-iodination labeling. PfEMP1 (arrow) is labeled on FCR3S1.2-infected erythrocytes and is cleaved by low concentrations of trypsin.
Figure 1
Identification of rosetting PfEMP1. (A) Rosetting, single _P. falciparum_–infected erythrocyte is seen by light microscopy held by a 5-μm micropipette (A, 1). The uninfected erythrocytes are stripped of the infected cell and careful examination confirms that it indeed is infected by a single parasite (A, 2–3). B shows the amplification of a 434-bp band in four (from reactions 3, 4, 5, and 7) out of eight single-infected, rosetting erythrocytes using degenerate primers generated from the primary sequence of the DBL-1 domain of PfEMP1. C shows the amplification pattern with the same primers as in B of bulk cultures of rosetting (R+) FCR3S1.2 cultures and the R− FCR3s/a parasites. Note that the 434-bp product is only seen with the R+ parasites. D shows the hybridization pattern in Northern blotting of the 434-bp sequence to mRNA extracted from the highly rosetting parasite FCR3S1.2 (84% R+) and the weak hybridization to the R− FCR3S/a parasite (9% R+). E shows the autoradiogarph of a Triton X-100 insoluble, SDS-soluble extract of FCR3S1.2-infected erythrocytes after radio-iodination labeling. PfEMP1 (arrow) is labeled on FCR3S1.2-infected erythrocytes and is cleaved by low concentrations of trypsin.
Figure 1
Identification of rosetting PfEMP1. (A) Rosetting, single _P. falciparum_–infected erythrocyte is seen by light microscopy held by a 5-μm micropipette (A, 1). The uninfected erythrocytes are stripped of the infected cell and careful examination confirms that it indeed is infected by a single parasite (A, 2–3). B shows the amplification of a 434-bp band in four (from reactions 3, 4, 5, and 7) out of eight single-infected, rosetting erythrocytes using degenerate primers generated from the primary sequence of the DBL-1 domain of PfEMP1. C shows the amplification pattern with the same primers as in B of bulk cultures of rosetting (R+) FCR3S1.2 cultures and the R− FCR3s/a parasites. Note that the 434-bp product is only seen with the R+ parasites. D shows the hybridization pattern in Northern blotting of the 434-bp sequence to mRNA extracted from the highly rosetting parasite FCR3S1.2 (84% R+) and the weak hybridization to the R− FCR3S/a parasite (9% R+). E shows the autoradiogarph of a Triton X-100 insoluble, SDS-soluble extract of FCR3S1.2-infected erythrocytes after radio-iodination labeling. PfEMP1 (arrow) is labeled on FCR3S1.2-infected erythrocytes and is cleaved by low concentrations of trypsin.
Figure 2
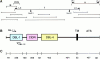
Map of cDNA structure, sequencing clones, deduced amino acid sequence, and the location of GAG-binding motifs in the rosetting PfEMP1 of FCR3S1.2. A shows the location of the 434-bp fragment and the three fragments (I, II, and III) that were initially cloned for sequencing. Restriction enzyme digestion sites are indicated by arrows. Additional overlapping clones used for sequencing are shown below. B shows the primary structure of the rosetting FCR3S1.2-PfEMP1. It has two DBL domains (DBL-1 and -4), one CIDR, one TM region, and one ATS. C shows the distribution of aa in different regions of FCR3S1.2-PfEMP1. D shows the complete aa sequence of FCR3S1.2-PfEMP1. The location of potential GAG-binding motifs are shown in pink. Motifs No. 4, 5 and 9, 10 (aa 221–232 and 533–549, respectively) are seen as a single stretch since they are located next to each other. (See Materials and Methods for description of identification of GAG-binding motifs.) These sequence data are available from EMBL/GenBank/ DDBJ under accession number AF003473.
Figure 2
Map of cDNA structure, sequencing clones, deduced amino acid sequence, and the location of GAG-binding motifs in the rosetting PfEMP1 of FCR3S1.2. A shows the location of the 434-bp fragment and the three fragments (I, II, and III) that were initially cloned for sequencing. Restriction enzyme digestion sites are indicated by arrows. Additional overlapping clones used for sequencing are shown below. B shows the primary structure of the rosetting FCR3S1.2-PfEMP1. It has two DBL domains (DBL-1 and -4), one CIDR, one TM region, and one ATS. C shows the distribution of aa in different regions of FCR3S1.2-PfEMP1. D shows the complete aa sequence of FCR3S1.2-PfEMP1. The location of potential GAG-binding motifs are shown in pink. Motifs No. 4, 5 and 9, 10 (aa 221–232 and 533–549, respectively) are seen as a single stretch since they are located next to each other. (See Materials and Methods for description of identification of GAG-binding motifs.) These sequence data are available from EMBL/GenBank/ DDBJ under accession number AF003473.
Figure 3
Rosetting FCR3S1.2-PfEMP1 binds to heparan sulfate. All the gels are 10% SDS-PAGE stained with Coomassie. A shows the expressed GST, DBL-1–GST, or ATS–GST after purification on glutathione–Sepharose and SDS-PAGE. B shows the binding capacity of different fusion proteins to heparin–Sepharose after SDS-PAGE. C shows the inhibition produced by different GAGs (20 μl, 5 mg/ml) on the binding of DBL-1–GST to heparin–Sepharose followed by SDS-PAGE. D and E show the binding of DBL-1–GST (D) and ATS–GST (E) to monolayers of normal RBCs as visualized by an mAb to GST labeled with biotin and FITC-avidin.
Figure 3
Rosetting FCR3S1.2-PfEMP1 binds to heparan sulfate. All the gels are 10% SDS-PAGE stained with Coomassie. A shows the expressed GST, DBL-1–GST, or ATS–GST after purification on glutathione–Sepharose and SDS-PAGE. B shows the binding capacity of different fusion proteins to heparin–Sepharose after SDS-PAGE. C shows the inhibition produced by different GAGs (20 μl, 5 mg/ml) on the binding of DBL-1–GST to heparin–Sepharose followed by SDS-PAGE. D and E show the binding of DBL-1–GST (D) and ATS–GST (E) to monolayers of normal RBCs as visualized by an mAb to GST labeled with biotin and FITC-avidin.
Figure 3
Rosetting FCR3S1.2-PfEMP1 binds to heparan sulfate. All the gels are 10% SDS-PAGE stained with Coomassie. A shows the expressed GST, DBL-1–GST, or ATS–GST after purification on glutathione–Sepharose and SDS-PAGE. B shows the binding capacity of different fusion proteins to heparin–Sepharose after SDS-PAGE. C shows the inhibition produced by different GAGs (20 μl, 5 mg/ml) on the binding of DBL-1–GST to heparin–Sepharose followed by SDS-PAGE. D and E show the binding of DBL-1–GST (D) and ATS–GST (E) to monolayers of normal RBCs as visualized by an mAb to GST labeled with biotin and FITC-avidin.
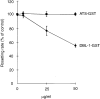
Figure 4
Disruption of preformed, natural rosettes with either DBL-1–GST or ATS–GST. Results are means and standard errors of three experiments.
Figure 5
Effect of GAGs on P. falciparum rosetting. A shows disruption of rosettes exerted by different GAGs. FCR3S1.2 cultures were incubated with GAGs for 1 h at 37°C and compared to control culture. Results are the means and standard error of three separate experiments. B shows the effect of enzyme treatment of uninfected, C-FDA–labeled erythrocytes in a competitive assay of rosette reformation in the presence of normal erythrocytes and FCR3S1.2-infected pRBCs. Results are the means and standard error of three separate experiments, or two experiments for neuraminidase. Neuraminidase and chondroitinase ABC concentrations are in IU, whereas the heparinase III concentration is in Sigma units. (One Sigma unit corresponds to ∼1.7 × 10−3 IU.)
Similar articles
- P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1.
Rowe JA, Moulds JM, Newbold CI, Miller LH. Rowe JA, et al. Nature. 1997 Jul 17;388(6639):292-5. doi: 10.1038/40888. Nature. 1997. PMID: 9230440 - Heparan sulfate on endothelial cells mediates the binding of Plasmodium falciparum-infected erythrocytes via the DBL1alpha domain of PfEMP1.
Vogt AM, Barragan A, Chen Q, Kironde F, Spillmann D, Wahlgren M. Vogt AM, et al. Blood. 2003 Mar 15;101(6):2405-11. doi: 10.1182/blood-2002-07-2016. Epub 2002 Nov 14. Blood. 2003. PMID: 12433689 - Molecular aspects of severe malaria.
Chen Q, Schlichtherle M, Wahlgren M. Chen Q, et al. Clin Microbiol Rev. 2000 Jul;13(3):439-50. doi: 10.1128/CMR.13.3.439. Clin Microbiol Rev. 2000. PMID: 10885986 Free PMC article. Review. - Molecular mechanisms and biological importance of Plasmodium falciparum erythrocyte rosetting.
Wahlgren M, Carlson J, Helmby H, Hedlund I, Treutiger CJ. Wahlgren M, et al. Mem Inst Oswaldo Cruz. 1992;87 Suppl 3:323-9. doi: 10.1590/s0074-02761992000700054. Mem Inst Oswaldo Cruz. 1992. PMID: 1285315 Review.
Cited by
- Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum.
Kyes SA, Rowe JA, Kriek N, Newbold CI. Kyes SA, et al. Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):9333-8. doi: 10.1073/pnas.96.16.9333. Proc Natl Acad Sci U S A. 1999. PMID: 10430943 Free PMC article. - Whole-body imaging of sequestration of Plasmodium falciparum in the rat.
Pettersson F, Vogt AM, Jonsson C, Mok BW, Shamaei-Tousi A, Bergström S, Chen Q, Wahlgren M. Pettersson F, et al. Infect Immun. 2005 Nov;73(11):7736-46. doi: 10.1128/IAI.73.11.7736-7746.2005. Infect Immun. 2005. PMID: 16239578 Free PMC article. - Plasmodium falciparum rosette formation is uncommon in isolates from pregnant women.
Rogerson SJ, Beeson JG, Mhango CG, Dzinjalamala FK, Molyneux ME. Rogerson SJ, et al. Infect Immun. 2000 Jan;68(1):391-3. doi: 10.1128/IAI.68.1.391-393.2000. Infect Immun. 2000. PMID: 10603414 Free PMC article. - Age-dependent increase in antibodies that inhibit Plasmodium falciparum adhesion to a subset of endothelial receptors.
Attaher O, Mahamar A, Swihart B, Barry A, Diarra BS, Kanoute MB, Dembele AB, Keita S, Gaoussou S, Issiaka D, Dicko A, Duffy PE, Fried M. Attaher O, et al. Malar J. 2019 Apr 11;18(1):128. doi: 10.1186/s12936-019-2764-4. Malar J. 2019. PMID: 30971252 Free PMC article. - var gene transcription and PfEMP1 expression in the rosetting and cytoadhesive Plasmodium falciparum clone FCR3S1.2.
Albrecht L, Moll K, Blomqvist K, Normark J, Chen Q, Wahlgren M. Albrecht L, et al. Malar J. 2011 Jan 25;10:17. doi: 10.1186/1475-2875-10-17. Malar J. 2011. PMID: 21266056 Free PMC article.
References
- Miller LH, Good F, Milon G. Malaria pathogenesis. Science. 1994;264:1878–1883. - PubMed
- Pasloske BL, Howard RJ. Malaria, the red cell, and the endothelium. Annu Rev Med. 1994;45:283–295. - PubMed
- Marsh K, English M, Crawley J, Peshu N. The pathogenesis of severe malaria in African children. Ann Trop Med Parasitol. 1996;90:395–402. - PubMed
- Atkinson, C.T., and M. Aikawa. 1990. Ultrastructure of malaria-infected erythrocytes. Blood Cells (NY). 16:351–368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials