Drosophila myb is required for the G2/M transition and maintenance of diploidy - PubMed (original) (raw)
Drosophila myb is required for the G2/M transition and maintenance of diploidy
A L Katzen et al. Genes Dev. 1998.
Abstract
The myb proto-oncogenes are thought to have a role in the cell division cycle. We have examined this possibility by genetic analysis in Drosophila melanogaster, which possesses a single myb gene. We have described previously two temperature-sensitive, recessive lethal mutants in Drosophila myb (Dm myb). The phenotypes of these mutants revealed a requirement for myb in diverse cellular lineages throughout the course of Drosophila development. We now report a cellular explanation for these findings by showing that Dm myb is required for both mitosis and prevention of endoreduplication in wing cells. Myb apparently acts at or near the time of the G2/M transition. The two mutant alleles of Dm myb produce the same cellular phenotype, although the responsible mutations are located in different functional domains of the gene product. The mutant phenotype can be partially suppressed by ectopic expression of either cdc2 or string, two genes that are known to promote the transition from G2 to M. We conclude that Dm myb is required for completion of cell division and may serve two independent functions: promotion of mitosis, on the one hand, and prevention of endoreduplication when cells are arrested in G2, on the other.
Figures
Figure 1
Mutant DMyb proteins contain amino acid substitutions at evolutionarily conserved positions. (Top) A schematic representation of the mouse c-Myb protein. The four regions of conservation shared between vertebrate and Drosophila Myb proteins are indicated by Roman numerals. (R1, R2, and R3) Three imperfect tandem repeats that comprise the DNA-binding domain (region I); (TA) transcriptional activator domain; (LZ) leucine zipper; (NR) negative regulatory domain. Also depicted is an additional region encoded by an alternatively spliced exon that contains the majority of conserved region II (Lyon et al. 1994). (Middle) A schematic representation of the DMyb protein. Positions affected by the myb1 and myb2 mutations are indicated. Mouse and Drosophila amino acid sequences for the region in the DNA-binding domain that contains the myb2 mutation are shown by labeled arrows (Gonda et al. 1985; Katzen et al. 1985; Peters et al. 1987). In this region, chicken and human sequences are identical to the mouse sequence (Gerondakis and Bishop 1986; Majello et al. 1986). Mouse, chicken, and Drosophila amino acid sequences for region IV, which includes the myb1 mutation, are shown at bottom. Identical amino acids are boxed, and conservative amino acid differences are underlined. Affected amino acids and the substitutions are shown in bold. Mutations are myb1: GGC → AGC, Gly → Ser, amino acid 613; myb2: AGA → AAA, Arg → Lys, amino acid 177. We also found three nucleotides that differed from the published sequence (Peters et al. 1987) in all three of our strains, only one of which affected the amino acid sequence. At base 281, located in the 5′-untranslated region, cytosine was replaced by guanine. At base 1016, which corresponds to the third base of codon 137 located in the DNA-binding domain, thymine is replaced by cytosine; the amino acid (glycine) is unchanged. At base 1714, which corresponds to the second base of codon 370 located in an unconserved region of the protein (amino-terminal to region II), thymine is replaced by cytosine; this results in a codon for alanine (GCC) instead of the published valine (GTC).
Figure 2
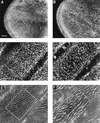
Mutant myb wings were cruder than wild-type wings and had fewer, larger hairs that were not uniformly oriented. Wings were dissected from female flies raised at 18°C unless otherwise specified. Bars in A and D, 0.1 mm. Complete wings for white (A), myb1 (B), and myb1; P(w+,myb+)/+ (C) are shown at the same magnification. D–I are shown at the same magnification, and the number of hairs located within the boxed area of each panel is indicated below in brackets. When compared with the parental w strain (84 hairs) (D), the number of hairs in myb1 mutants (39 hairs) (E) was decreased by approximately half. This phenotype could be fully rescued by a wild-type Dm myb transgene, shown in F [myb1; P(w+,myb+)/+ (86 hairs)]. The reduction in hair number was significantly less in myb2 flies raised at 18°C (68 hairs) (G) but was stronger when myb2 was carried over a deficiency chromosome at 18°C myb2/Df(1)sd (43 hairs) (H), or when myb2 flies were raised at 25°C (46 hairs) (I).
Figure 3
myb1 mutant wings had half the number of nuclei as wild-type wings, a defect that occurs during the last cell division. Shown are regions of developing wings dissected from animals raised at 25°C. Wings were treated with either DAPI alone or DAPI and rhodamine-labeled phalloidin (a stain that highlights wing prehairs because they contain high levels of F-actin). The number of nuclei or hairs located within the box in each panel is indicated below in parentheses. Bars, 0.025 mm. DAPI staining of prepupal wings at 6 hr APF showed that at this stage, the nuclear density in A [w (parental strain) (152 nuclei)] and B [myb1 (155 nuclei)] was the same. DAPI staining of pupal wings at 36–37 hr APF showed that at this stage, the nuclear density in C [w (153 nuclei)] was approximately twice the density in myb1 (83 nuclei) (D). Note that the myb1 nuclei were larger than the w nuclei. Phalloidin staining of the same wings as shown in C and D, E (155 hairs) and F (82 hairs), respectively, showed a one-to-one correspondence between numbers of nuclei and hairs for both wild-type and mutant animals.
Figure 4
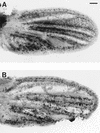
myb1 mutant wing cells entered S phase during the final cell cycle. Shown are developing wings from pupae that were raised at 25°C, injected with BrdU at 16 hr APF, allowed to continue developing at 25°C until 36 hr APF, and then dissected and processed to visualize BrdU incorporation: (A) w (parental strain); (B) myb1. Bar, 0.05 mm.
Figure 5
Overexpression of Dm cdc2 or string during pupal development can suppress the mutant Dm myb wing defect. Wings were dissected from flies that were raised at 25°C, heat shock treated for 20 min at 37°C when they were between 18 and 22 hr APF, and then returned to 25°C. Bars in A and D, 0.05 mm. The number of hairs within the box in each panel is indicated below in parentheses. Shown in A–C at the same magnification is a region near the distal tip between longitudinal veins III and IV. When compared with myb2, overexpression of either Dm cdc2AF or stg in myb2 mutants resulted in an increase in the number of hairs in myb2 mutants: (A) myb 2 (103 hairs); (B) myb 2; HS–Dm cdc2AF/+ (135 hairs); (C) myb2; HS–stg/+ (134 hairs). The wild-type version of cdc2 under the heat shock promoter also suppressed the myb phenotype. To demonstrate that suppression occurred in multiple regions of the wing, a more proximal section is shown at a lower magnification: (D) myb2 (105 hairs); (E) myb2; HS–Dm cdc2/+ (137 hairs).
Figure 6
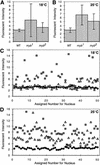
Quantitative microscopic analysis of nuclei from mutant and wild-type wings. Developing wings were dissected from wild-type (WT), myb1, and myb2 pupae at 72 hr AFP for animals raised at 18°C, or 36 hr APF for animals raised at 25°C. Wings were then fixed and stained with DAPI. Four samples from each genotype and temperature were optically sectioned using high-resolution, three-dimensional wide-field fluorescence microscopy (see Materials and Methods). Relative fluorescent intensities and volumes of at least 30 nuclei for each sample (⩾120/genotype) were analyzed. Shown are the calculated averages (means) of nuclear fluorescent intensities for each genotype at 18°C (A) and 25°C (B) with standard deviations indicated by vertical bars. To illustrate the heterogeneity within the population of mutant nuclei, the fluorescent intensities of 50 nuclei from each genotype at 18°C (C) and 25°C (D) are plotted individually. (Solid diamond) Wild-type; (shaded square) myb1; (open triangle) myb2.
Figure 7
The mutant myb2 wing phenotype is enhanced by shifting from permissive to restrictive temperature for 1 day during pupal wing development. Bar in A, 0.05 mm. The number of hairs within the box in each panel is indicated below. Shown is a region near the distal tip between longitudinal veins IV and V. Wings were dissected from white flies that were raised at 18°C until 26 hr APF, transferred to 28°C for 24 hr, and then returned to 18°C, 222 hairs (A), myb2 flies that were raised continuously at 18°C, 178 hairs (B), or myb2 flies that were raised at 18°C until 26 hr APF, transferred to 28°C for 24 hr and then returned to 18°C, 115 hairs (C).
Figure 8
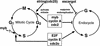
Schematic of gene products known to play key roles in regulating the mitotic and endoreplicative cell cycles. The data presented in this paper demonstrate that Dm myb is a positive regulator of progression from G2 into M and a negative regulator of endoreduplication, thereby maintaining diploidy. These dual functions are shared with Dm cdc2 and Dm cyclin A (Sauer et al. 1995; Hayashi 1996). In contrast, string and escargot, function to either activate the G2/M transition or suppress endoreduplication, respectively, but not both (Edgar and O’Farrell 1989; Smith and Orr-Weaver 1991; Hayashi 1996). The transcription factor E2F, Dm cyclin E, and the Dm cdc2c kinase are required for S phase in both mitotic and endoreplicative cell cycle (Duronio et al. 1995; Sauer et al. 1995; Lilly and Spradling 1996). By analogy to the role that vertebrate myb genes are thought to play in cell cycle regulation, Dm myb may also participate in regulation of the G1/S transition, but our studies indicate that it is not required for DNA synthesis in endocycling cells.
Similar articles
- Mutations in Drosophila myb lead to centrosome amplification and genomic instability.
Fung SM, Ramsay G, Katzen AL. Fung SM, et al. Development. 2002 Jan;129(2):347-59. doi: 10.1242/dev.129.2.347. Development. 2002. PMID: 11807028 - Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes.
Ohi R, Feoktistova A, McCann S, Valentine V, Look AT, Lipsick JS, Gould KL. Ohi R, et al. Mol Cell Biol. 1998 Jul;18(7):4097-108. doi: 10.1128/MCB.18.7.4097. Mol Cell Biol. 1998. PMID: 9632794 Free PMC article. - MYB and CBP: physiological relevance of a biochemical interaction.
Fung SM, Ramsay G, Katzen AL. Fung SM, et al. Mech Dev. 2003 Jun;120(6):711-20. doi: 10.1016/s0925-4773(03)00044-3. Mech Dev. 2003. PMID: 12834870 - Myb and oncogenesis.
Ganter B, Lipsick JS. Ganter B, et al. Adv Cancer Res. 1999;76:21-60. doi: 10.1016/s0065-230x(08)60773-3. Adv Cancer Res. 1999. PMID: 10218098 Review. No abstract available. - Structure and function of the proteins encoded by the myb gene family.
Kanei-Ishii C, Nomura T, Ogata K, Sarai A, Yasukawa T, Tashiro S, Takahashi T, Tanaka Y, Ishii S. Kanei-Ishii C, et al. Curr Top Microbiol Immunol. 1996;211:89-98. doi: 10.1007/978-3-642-85232-9_9. Curr Top Microbiol Immunol. 1996. PMID: 8585968 Review. No abstract available.
Cited by
- Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK.
Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, Matsumoto K, Ishii S. Kanei-Ishii C, et al. Genes Dev. 2004 Apr 1;18(7):816-29. doi: 10.1101/gad.1170604. Genes Dev. 2004. PMID: 15082531 Free PMC article. - Conservation and diversification of three-repeat Myb transcription factors in plants.
Ito M. Ito M. J Plant Res. 2005 Feb;118(1):61-9. doi: 10.1007/s10265-005-0192-8. Epub 2005 Feb 10. J Plant Res. 2005. PMID: 15703854 Review. - Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex.
Sim CK, Perry S, Tharadra SK, Lipsick JS, Ray A. Sim CK, et al. Genes Dev. 2012 Nov 15;26(22):2483-98. doi: 10.1101/gad.201665.112. Epub 2012 Oct 26. Genes Dev. 2012. PMID: 23105004 Free PMC article. - Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability.
Manak JR, Mitiku N, Lipsick JS. Manak JR, et al. Proc Natl Acad Sci U S A. 2002 May 28;99(11):7438-43. doi: 10.1073/pnas.122231599. Proc Natl Acad Sci U S A. 2002. PMID: 12032301 Free PMC article. - A long lost key opens an ancient lock: Drosophila Myb causes a synthetic multivulval phenotype in nematodes.
Vorster PJ, Goetsch P, Wijeratne TU, Guiley KZ, Andrejka L, Tripathi S, Larson BJ, Rubin SM, Strome S, Lipsick JS. Vorster PJ, et al. Biol Open. 2020 May 7;9(5):bio051508. doi: 10.1242/bio.051508. Biol Open. 2020. PMID: 32295830 Free PMC article.
References
- Adler PN. The genetic control of tissue polarity in Drosophila. BioEssays. 1992;14:735–741. - PubMed
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. - PubMed
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes & Dev. 1994;8:770–782. - PubMed
- Bennett JD, Farlie PG, Watson RJ. E2F binding is required but not sufficient for repression of B-myb transcription in quiescent fibroblasts. Oncogene. 1996;13:1073–1082. - PubMed
- Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer KH. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous