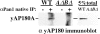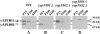Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis - PubMed (original) (raw)
Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis
B Wendland et al. J Cell Biol. 1998.
Abstract
A genetic screen for factors required for endocytosis in the budding yeast Saccharomyces cerevisiae previously identified PAN1. Pan1p is a homologue of the mammalian protein eps15, which has been implicated in endocytosis by virtue of its association with the plasma membrane clathrin adaptor complex AP-2. Pan1p contains two eps15 homology (EH) domains, a protein-protein interaction motif also present in other proteins that function in membrane trafficking. To address the role of Pan1p and EH domains in endocytosis, a yeast two-hybrid screen was performed using the EH domain-containing region of Pan1p. This screen identified yAP180A, one of two yeast homologues of a class of clathrin assembly proteins (AP180) that exhibit in vitro clathrin cage assembly activity. In vitro binding studies using GST fusion proteins and yeast extracts defined distinct binding sites on yAP180A for Pan1p and clathrin. yAP180 proteins and Pan1p, like actin, localize to peripheral patches along the plasma membrane. Mammalian synaptojanin, a phosphatidylinositol polyphosphate-5-phosphatase, also has been implicated in endocytosis recently, and three synaptojanin-like genes have been identified in yeast. We observed genetic interactions between the yeast SJL1 gene and PAN1, which suggest a role for phosphoinositide metabolites in Pan1p function. Together with other studies, these findings suggest that Pan1p coordinates regulatory interactions between proteins required for both endocytosis and actin-cytoskeleton organization; these proteins include the yAP180 proteins, clathrin, the ubiquitin-protein ligase Rsp5p, End3p, and synaptojanin. We suggest that Pan1p (and by extension eps15) serves as a multivalent adaptor around which dynamic interactions between structural and regulatory components of the endocytic pathway converge.
Figures
Figure 1
Two-hybrid screen with Pan1p EH domains identifies yeast members of the AP180 protein family. (A) Schematic of the yeast two-hybrid Pan1p bait plasmids used for the screen, which identified the prey plasmids encoding fusion proteins containing the yAP180A protein. (B) An alignment of the conserved amino-terminal 300 residues of the members of the AP180 protein family, shown in single letter code. The corresponding accession numbers are as follows: (a) mAP180, A44825 (PIR); (b) hCALM, U45976 (Genbank/EMBL/DDBJ); (c) ceAP180, U88308 (Genbank/EMBL/DDBJ); (d) yAP180A, P38856 (Swiss-Prot); (e) yAP180B, P53309 (Swiss-Prot). Amino acids identical in at least four of the proteins are indicated in black, similarities are shown in grey. A predicted leucine zipper in the yAP180 proteins is underlined.
Figure 2
GST–yAP180A fusion protein binding to Pan1p and clathrin in vitro. (A) Schematic of the yAP180A protein with fragments used for GST fusion proteins shown below. •, indicates the location of the tripeptide “NPF,” hatched region delineates the Pan1p-binding domain, cross-hatched region shows the clathrin-binding domain, and Q17 indicates 17 consecutive glutamine residues. (B) Extracts from wild-type yeast cells were incubated with immobilized GST fusion proteins, washed, and bound proteins resolved by SDS-PAGE followed by immunoblotting with a Pan1p-specific antiserum. (C) Bound proteins visualized by silver staining a 7.5% acrylamide gel. (D) A similar experiment as in B, but using a monoclonal antibody specific for yeast clathrin heavy chain and a polyclonal antibody specific for yeast clathrin light chain.
Figure 3
Native coimmunoprecipitation of yA180A with Pan1p. 1% Triton X-100 extracts of wild-type or yap1801Δ yap1802Δ cells were generated and no antibodies (−) or Pan1p antiserum (+) were added. Native immune complexes were isolated, separated on SDS-PAGE, and analyzed by immunoblotting for yAP180 proteins. For comparison, 5% of total extract was loaded (right).
Figure 4
Fractionation of yAP180A and yAP180B. (A) Wild type, (B) yap1801Δ, (C) yap1802Δ, and (D) yap1801Δ yap1802Δ cells were spheroplasted, homogenized, and subjected to differential centrifugation fractionation. The fractions were resolved by SDS-PAGE, and yAP180 proteins were detected by immunoblotting with affinity-purified antibodies that recognize both yAP180A and yAP180B proteins.
Figure 5
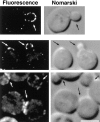
Localization of GFP–yAP180A fusion protein. yap1801Δ yap1802Δ cells expressing GFP–yAP180A from either single copy (CEN; top) or multi-copy (2 μ; bottom) vectors were examined using fluorescence and differential interference (Nomarski) light microscopy. The images were processed using deconvoluting software (Delta Vision) on a Silcon Graphics workstation. Buds and cytokinesis necks containing concentrated spots of signal are indicated by arrows. Bar, 5 μm.
Figure 6
A model describing the interactions between Pan1p and proteins that regulate clathrin (yAP180A/B), the actin cytoskeleton (End3p), the ubiquitination pathway (Rsp5p), and inositol phospholipids (the synaptojanin-like inositol-5-phosphatases). Solid arrows indicate physical interactions between the proteins, and the dashed arrows denote genetic interactions.
Similar articles
- Pan1p: an actin director of endocytosis in yeast.
Huang B, Cai M. Huang B, et al. Int J Biochem Cell Biol. 2007;39(10):1760-4. doi: 10.1016/j.biocel.2006.12.001. Epub 2006 Dec 21. Int J Biochem Cell Biol. 2007. PMID: 17303466 Review. - A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis.
Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. Gagny B, et al. J Cell Sci. 2000 Sep;113 ( Pt 18):3309-19. doi: 10.1242/jcs.113.18.3309. J Cell Sci. 2000. PMID: 10954428 - Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex.
Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Duncan MC, et al. Nat Cell Biol. 2001 Jul;3(7):687-90. doi: 10.1038/35083087. Nat Cell Biol. 2001. PMID: 11433303 - Sac phosphatase domain proteins.
Hughes WE, Cooke FT, Parker PJ. Hughes WE, et al. Biochem J. 2000 Sep 1;350 Pt 2(Pt 2):337-52. Biochem J. 2000. PMID: 10947947 Free PMC article. Review.
Cited by
- Pan1 regulates transitions between stages of clathrin-mediated endocytosis.
Bradford MK, Whitworth K, Wendland B. Bradford MK, et al. Mol Biol Cell. 2015 Apr 1;26(7):1371-85. doi: 10.1091/mbc.E14-11-1510. Epub 2015 Jan 28. Mol Biol Cell. 2015. PMID: 25631817 Free PMC article. - The Early Endocytosis Gene PAL1 Contributes to Stress Tolerance and Hyphal Formation in Candida albicans.
Yu M, Ma D, Eszterhas S, Rollenhagen C, Lee SA. Yu M, et al. J Fungi (Basel). 2023 Nov 10;9(11):1097. doi: 10.3390/jof9111097. J Fungi (Basel). 2023. PMID: 37998902 Free PMC article. - The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme.
Kee Y, Lyon N, Huibregtse JM. Kee Y, et al. EMBO J. 2005 Jul 6;24(13):2414-24. doi: 10.1038/sj.emboj.7600710. Epub 2005 Jun 2. EMBO J. 2005. PMID: 15933713 Free PMC article. - Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p.
Zeng G, Yu X, Cai M. Zeng G, et al. Mol Biol Cell. 2001 Dec;12(12):3759-72. doi: 10.1091/mbc.12.12.3759. Mol Biol Cell. 2001. PMID: 11739778 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous