Stat3 activation is required for cellular transformation by v-src - PubMed (original) (raw)
Stat3 activation is required for cellular transformation by v-src
J F Bromberg et al. Mol Cell Biol. 1998 May.
Abstract
Stat3 activation has been associated with cytokine-induced proliferation, anti-apoptosis, and transformation. Constitutively activated Stat3 has been found in many human tumors as well as v-abl- and v-src-transformed cell lines. Because of these correlations, we examined directly the relationship of activated Stat3 to cellular transformation and found that wild-type Stat3 enhances the transforming potential of v-src while three dominant negative Stat3 mutants inhibit v-src transformation. Stat3 wild-type or mutant proteins did not affect v-ras transformation. We conclude that Stat3 has a necessary role in v-src transformation.
Figures
FIG. 1
Wild-type Stat3 and dominant negative Stat3 constructs. (A) Wild-type Stat3 was cloned into RcCMV-Neo and tagged at the 3′ end with a FLAG epitope (7, 33). (B) Stat3Y705-F was generated by PCR and cloned into RcCMV-Neo and tagged at the 3′ end with a FLAG epitope. The expressed protein can be detected with FLAG (M2) monoclonal antiserum or Stat3-C antiserum. It cannot be phosphorylated on Y705 and can presumably compete with wild-type Stat3 for v-src or other kinases (5, 6). (C) Stat3DB was tagged at the 3′ end with a FLAG epitope which contains slightly fewer amino acids than those of the above constructs. Within the Stat3 DNA-binding domain, multiple changes (VVV461–463AAA and EE434–435AA), which abolish the ability of this protein to bind DNA while it retains the ability to be phosphorylated, form dimers, and translocate to the nucleus (15, 16), were made. (D) Stat3S contains an alanine instead of a serine at position 727 (34). This protein can be phosphorylated on tyrosine, form dimers, and bind DNA, but it cannot fully activate transcription (34). Stat3S was cloned into RcCMV and does not contain a FLAG epitope (34).
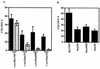
FIG. 2
Stat3 potentiates v-src transformation in soft agar colony-forming assays. (A) NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected with RcCMV plus v-src and Stat3 plus v-src. One microgram of RcCMV-based vectors was used per transfection; 500 ng of pBabe/v-src (24) was used per transfection. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were trypsinized and plated (5 × 105 cells in 3ml) into soft agar with 10% BCS (20) containing 2 μg of puromycin per ml and 800 μg of G418 per ml. Seventeen days later, colonies were counted. Each bar represents the average of 10 independent transfections performed simultaneously; each error bar indicates the standard deviation. (B) v-_src_-transformed NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected (as above) with RcCMV and Stat3. One microgram of the RcCMV-based vectors was used per transfection. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were trypsinized and plated (105 cells in 3 ml) into soft agar containing 2 μg of puromycin per ml and 800 μg of G418 per ml. Fourteen days later, colonies were counted. Each bar corresponds to the average of six independent transfections performed simultaneously; each error bar indicates the standard deviation.
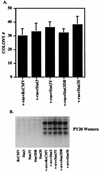
FIG. 3
Stat3 dominant negative mutants abrogate v-src transformation. (A) The black columns indicate NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) transfected with a 1:2 ratio (500 ng to 1 μg) of v-src plus RcCMV, v-src plus Stat3Y, v-src plus Stat3DB, or v-src plus Stat3S and plated in soft agar, as described in the legend to Fig. 2A. Seventeen days later, colonies were counted. Each black bar represents the average of 10 independent transfections performed simultaneously; each error bar indicates the standard deviation. The grey columns indicate NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) transfected with a 1:10 ratio (0.2 to 1.8 μg) of v-src plus RcCMV, v-src plus Stat3Y, v-src plus Stat3DB, or v-src plus Stat3S and plated in soft agar, as described in the legend to Fig. 2A. Twenty-five days later, colonies were counted. Each grey bar represents the average of three independent transfections performed simultaneously; each error bar indicates the standard deviation. (B) v-_src_-transformed NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected with RcCMV, Stat3Y, Stat3DB, or Stat3S and plated in soft agar, as described in the legend to Fig. 2B. Fourteen days later, colonies were counted. Each bar corresponds to the average of six independent transfections performed simultaneously; each error bar indicates the standard deviation.
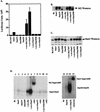
FIG. 4
In vitro assays. (A) Luciferase assay. NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transiently transfected (as described in the legend to Fig. 3A) with RcCMV, Stat3, Stat3Y, Stat3DB, Stat3S, v-src plus RcCMV, v-src plus Stat3, v-src plus Stat3Y, v-src plus Stat3DB, or v-src plus Stat3S. All were transfected with 1 μg of Ly6E (three copies) Luc (34) and a β-galactosidase expression vector. One microgram of RcCMV-based vectors was used per transfection; 500 ng of pBabe/v-src was used in appropriate transfections. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were lysed and luciferase assays were performed. Each bar represents the average of eight individual transfections with standard deviations, each performed in duplicate and normalized to β-galactosidase activity. (B) FLAG Western blotting. Thirty micrograms of protein per lane from whole-cell extracts from transiently transfected NIH 3T3 cells or v-_src_-transformed cells (as in Fig. 5A) were analyzed by Western blot analyses using chemiluminescence. The Stat3DB-FLAG construct is slightly smaller (see Fig. 1C) and therefore migrates faster. Stat3S expression was not determined due to its lack of a FLAG epitope. (C) Stat3 Western blotting. As in panel B, 30 μg of protein per lane was analyzed for the relative levels of Stat3 protein. Lanes 1 and 6 reveal endogenous levels of Stat3 protein, while the other lanes demonstrate that the transiently transfected cells are producing additional Stat3 protein. (D) Gel shift. NIH 3T3 cells were transiently transfected as described in the legend to Fig. 5A. Nuclear extracts from these cells were used in an EMSA using a 32P-labeled Stat3 binding site (M67) (31). Stat3-Stat3 homodimers form when Stat3 is phosphorylated on tyrosine 705. M2 (FLAG) monoclonal antiserum was used at a 1:100 dilution for supershifting of Stat3-FLAG complexes. A 1:100 dilution of Stat3-C antiserum was used for supershifting of Stat3S complexes.
FIG. 5
Stat3 and Stat3 dominant negative mutants do not affect v-ras transformation or overall tyrosine phosphorylation by v-src. (A) NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected (as in the legend to Fig. 2A) with 500 ng of v-ras (pRSV Ha-rasLeu61) (11) in conjunction with 1 μg of RcCMV, Stat3, Stat3Y, Stat3DB, or Stat3S. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were trypsinized and plated (5 × 105 cells) into soft agar. Seventeen days later, colonies were counted. Each bar represents the average of four independent transfections. (B) NIH 3T3 cells were transfected (as in the legend to Fig. 2A) with 500 ng of v-src and 1 μg of RcCMV-based Stat3 constructs. Whole-cell extracts were isolated, and 100 μg per lane was analyzed by Western blotting with an antiserum to phosphotyrosine (PY20).
Similar articles
- Stat3 activation by Src induces specific gene regulation and is required for cell transformation.
Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Turkson J, et al. Mol Cell Biol. 1998 May;18(5):2545-52. doi: 10.1128/MCB.18.5.2545. Mol Cell Biol. 1998. PMID: 9566874 Free PMC article. - Activation of Stat3 in v-Src-transformed fibroblasts requires cooperation of Jak1 kinase activity.
Zhang Y, Turkson J, Carter-Su C, Smithgall T, Levitzki A, Kraker A, Krolewski JJ, Medveczky P, Jove R. Zhang Y, et al. J Biol Chem. 2000 Aug 11;275(32):24935-44. doi: 10.1074/jbc.M002383200. J Biol Chem. 2000. PMID: 10823829 - Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein.
Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, Sekharam M, Frank DA, Holzman LB, Wu J, Sebti S, Jove R. Turkson J, et al. Mol Cell Biol. 1999 Nov;19(11):7519-28. doi: 10.1128/MCB.19.11.7519. Mol Cell Biol. 1999. PMID: 10523640 Free PMC article. - G protein coupled receptor signaling through the Src and Stat3 pathway: role in proliferation and transformation.
Ram PT, Iyengar R. Ram PT, et al. Oncogene. 2001 Mar 26;20(13):1601-6. doi: 10.1038/sj.onc.1204186. Oncogene. 2001. PMID: 11313907 Review. - The role of STATs in transcriptional control and their impact on cellular function.
Bromberg J, Darnell JE Jr. Bromberg J, et al. Oncogene. 2000 May 15;19(21):2468-73. doi: 10.1038/sj.onc.1203476. Oncogene. 2000. PMID: 10851045 Review.
Cited by
- STAT3 Protein-Protein Interaction Analysis Finds P300 as a Regulator of STAT3 and Histone 3 Lysine 27 Acetylation in Pericytes.
Kundu G, Ghasemi M, Yim S, Rohil A, Xin C, Ren L, Srivastava S, Akinfolarin A, Kumar S, Srivastava GP, Sabbisetti VS, Murugaiyan G, Ajay AK. Kundu G, et al. Biomedicines. 2024 Sep 14;12(9):2102. doi: 10.3390/biomedicines12092102. Biomedicines. 2024. PMID: 39335615 Free PMC article. - PRMT5-mediated methylation of STAT3 is required for lung cancer stem cell maintenance and tumour growth.
Abe Y, Sano T, Otsuka N, Ogawa M, Tanaka N. Abe Y, et al. Commun Biol. 2024 May 17;7(1):593. doi: 10.1038/s42003-024-06290-7. Commun Biol. 2024. PMID: 38760429 Free PMC article. - TRPM7 transactivates the FOSL1 gene through STAT3 and enhances glioma stemness.
Guo S, Ramar V, Guo AA, Saafir T, Akpobiyeri H, Hudson B, Li J, Liu M. Guo S, et al. Cell Mol Life Sci. 2023 Aug 29;80(9):270. doi: 10.1007/s00018-023-04921-6. Cell Mol Life Sci. 2023. PMID: 37642779 Free PMC article. - Potential Therapeutic Effects of Thiazolidinedione on Malignant Glioma.
Sheu ML, Pan LY, Hu HY, Su HL, Sheehan J, Tsou HK, Pan HC. Sheu ML, et al. Int J Mol Sci. 2022 Nov 4;23(21):13510. doi: 10.3390/ijms232113510. Int J Mol Sci. 2022. PMID: 36362294 Free PMC article. - Roads to Stat3 Paved with Cadherins.
Adan H, Daniel J, Raptis L. Adan H, et al. Cells. 2022 Aug 16;11(16):2537. doi: 10.3390/cells11162537. Cells. 2022. PMID: 36010614 Free PMC article. Review.
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1994.
- Balkwill F, Taylor-Papadimitriou J. Interferon affects both G1 and S+G2 in cells stimulated from quiescence to growth. Nature. 1978;274:798–801. - PubMed
- Bromberg, J., and J. E. Darnell, Jr. Unpublished observations.
- Campbell G S, Yu C L, Jove R, Carter-Su C. Constitutive activation of JAK1 in Src-transformed cells. J Biol Chem. 1997;272:2591–2594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI032489/AI/NIAID NIH HHS/United States
- AI34420/AI/NIAID NIH HHS/United States
- R37 AI034420/AI/NIAID NIH HHS/United States
- AI32489/AI/NIAID NIH HHS/United States
- K08 CA67950/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous