Particle size determinants in the human immunodeficiency virus type 1 Gag protein - PubMed (original) (raw)
Particle size determinants in the human immunodeficiency virus type 1 Gag protein
L Garnier et al. J Virol. 1998 Jun.
Abstract
The retroviral Gag protein plays the central role in the assembly process and can form membrane-enclosed, virus-like particles in the absence of any other viral products. These particles are similar to authentic virions in density and size. Three small domains of the human immunodeficiency virus type 1 (HIV-1) Gag protein have been previously identified as being important for budding. Regions that lie outside these domains can be deleted without any effect on particle release or density. However, the regions of Gag that control the size of HIV-1 particles are less well understood. In the case of Rous sarcoma virus (RSV), the size determinant maps to the CA (capsid) and adjacent spacer sequences within Gag, but systematic mapping of the HIV Gag protein has not been reported. To locate the size determinants of HIV-1, we analyzed a large collection of Gag mutants. To our surprise, all mutants with defects in the MA (matrix), CA, and the N-terminal part of NC (nucleocapsid) sequences produced dense particles of normal size, suggesting that oncoviruses (RSV) and lentiviruses (HIV-1) have different size-controlling elements. The most important region found to be critical for determining HIV-1 particle size is the p6 sequence. Particles lacking all or small parts of p6 were uniform in size distribution but very large as measured by rate zonal gradients. Further evidence for this novel function of p6 was obtained by placing this sequence at the C terminus of RSV CA mutants that produce heterogeneously sized particles. We found that the RSV-p6 chimeras produced normally sized particles. Thus, we present evidence that the entire p6 sequence plays a role in determining the size of a retroviral particle.
Figures
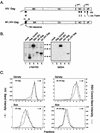
FIG. 1
Control experiments. (A) The wild-type HIV-1 Gag protein is designated HIV Gag. In M1.HIV Gag the black box at its N terminus represents the first 10 residues of pp60v-scr, which were substituted for the first 32 residues of HIV-1 Gag. The squiggle depicts the fatty acid myristate. The names of the Gag cleavage products (MA, CA, NC, and p6) are indicated. The two arrows above the Gag protein show the spacer peptides (SP1 and SP2). The black rectangles beneath M1.HIV Gag protein indicate domains essential for membrane binding (M), a late event in particle budding (L), and interactions between Gag molecules (I) as well as the leucine triplet sequence important for Vpr incorporation. Numbers refer to amino acid residues. (B) COS-1 cells were transfected with the indicated DNAs and were labeled as described in Materials and Methods. For comparison, an assembly-incompetent deletion mutant (RHB.T10C [37], which lacks late domains needed for budding) is shown in lanes 2. Molecular weight standards (in kilodaltons) are shown between the two panels. (C) The graphs represent the measurement of particle density in isopycnic sucrose gradients and the distribution of particle size in rate zonal gradients. COS-1 cells were transfected with pSV.HIV Gag or pSV.M1.HIV Gag. After 48 h, the cells and RSV-infected turkey embryo fibroblasts were labeled with [35S]methionine for 8 h (for isopycnic gradients) or 5 h (for velocity gradients). After the labeling period, particles and densities were analyzed as described in Materials and Methods. Arrows indicate the direction of sedimentation.
FIG. 2
Schematic diagrams of deletion mutants of M1.HIV Gag. The names of the Gag cleavage products (MA, CA, NC, and p6) are indicated. The shaded region within CA marks the MHR. The black boxes at the N termini of the constructs represent the first 10 amino acids from pp60v-scr. The squiggle depicts the fatty acid myristate. The black rectangles above the Gag proteins M1.HIV Gag, M1.HCNCΔ, and M1.HNCΔ mark the cysteine-histidine boxes. The two arrows beneath M1.HIV Gag protein indicate the spacer peptides (SP1 and SP2). Numbers refer to amino acid residues. The column at the right summarizes the size distribution of the mutants: N, homogeneous particles of normal size; N*, homogeneous particles that are slightly smaller; L. H., large particles that are heterogeneous in size; H, particles that are heterogeneous in size; V. L., homogeneous particles of very large size.
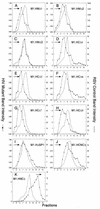
FIG. 3
Distribution of particle size in rate zonal gradients. COS-1 cells were transfected with the indicated deletion mutant DNAs and labeled with [35S]methionine for 5 h. After the labeling period, particle sizes were analyzed as described in Materials and Methods. Arrows indicate the direction of sedimentation.
FIG. 4
Deletion of the HIV-1 (SP2/p6) sequence. (A) The wild-type HIV Gag protein is shown at the top. HIV.Δp6 (_Bgl_II) is a designation of the wild-type HIV Gag protein deleted of SP2 and the p6 sequence by using the _Bgl_II site. In M1.HIV.Δp6 (_Bgl_II), the black box at its N terminus represents the first 10 residues of pp60v-scr. The squiggle depicts the fatty acid myristate. The names of the Gag cleavage products (MA, CA, NC, and p6) are indicated. The two arrows above the wild-type HIV Gag protein show the spacer peptides (SP1 and SP2). Numbers refer to amino acid residues. (B) The graphs represent the measurement of particle density in isopycnic sucrose gradients and the distribution of particle size in rate velocity gradients. COS-1 cells were transfected with pSV.HIV.Δp6 (_Bgl_II) or pSV.M1.HIV.Δp6 (_Bgl_II), and particle sizes and densities were analyzed as described in Materials and Methods. Arrows indicate the direction of sedimentation.
FIG. 5
Schematic diagrams of smaller p6 deletion mutants. M1.HIV Gag protein is shown at the top. The names of the Gag cleavage products (MA, CA, NC, and p6) are indicated. The black box at its N-terminus represents the first 10 amino acids from pp60v-scr. The squiggle represents the fatty acid myristate. The two arrows above the wild-type Gag protein indicate the spacer peptides (SP1 and SP2). The black rectangles beneath the p6 sequence indicate the L domain and the leucine triplet region. The various deletion mutants of M1.HIV Gag are illustrated below, and the residues retained in the p6 sequence are shown. The PEPTAP motif and the mutations within this motif are shown beneath the Gag proteins M1.HIVΔLeuT, M1.Gag.P5A, M1.Gag.P7A, and M1.Gag.P10L. The squares above the Gag proteins M1.Δp6.32-52, M1.Δp6.32-47, M1.Δp6.35-47, M1.Δp6.38-47, and M1.Δp6.43-52 mark the leucine residues. Numbers refer to amino acid residues. The column at the right summarizes the size distribution of the mutants: N, homogeneous particles of normal size; V. L., homogeneous particles of very large size; H*, particles that are heterogeneous in size (normal peak); L, particles of larger size.
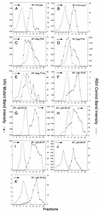
FIG. 6
Distribution of particle size in rate zonal gradients. COS-1 cells were transfected with the indicated smaller p6 deletion mutant DNAs and labeled with [35S]methionine for 5 h. Particle sizes were analyzed as described in Materials and Methods. Arrows indicate the direction of sedimentation.
FIG. 7
Schematic diagrams of chimeric RSV-HIV Gag proteins. The wild-type RSV Gag protein (open box) is shown at the top. The names of the Gag cleavage products (MA, p2, p10, CA, NC, and PR) are indicated. Shaded boxes represent HIV-1 sequences. The black boxes at the N termini of the constructs depict the first 10 amino acids from pp60v-scr. The squiggle indicates the fatty acid myristate. Numbers refer to amino acid residues. The column at the right summarizes the size distribution of the mutants: N, homogeneous particles of normal size; H, particles that are heterogeneous in size; H*, particles more normal but still heterogeneous.
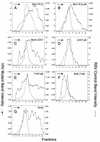
FIG. 8
Distribution of particle size in rate zonal gradients. COS-1 cells were transfected with the indicated chimeric RSV-HIV mutant DNAs and labeled with [35S]methionine for 5 h. Particle sizes were analyzed as described in Materials and Methods. For pSV.Myr0.LOC7, the internal control used was Myr1.D37S (34). Arrows indicate the direction of sedimentation.
Similar articles
- Identification of retroviral late domains as determinants of particle size.
Garnier L, Parent LJ, Rovinski B, Cao SX, Wills JW. Garnier L, et al. J Virol. 1999 Mar;73(3):2309-20. doi: 10.1128/JVI.73.3.2309-2320.1999. J Virol. 1999. PMID: 9971814 Free PMC article. - Genetic determinants of Rous sarcoma virus particle size.
Krishna NK, Campbell S, Vogt VM, Wills JW. Krishna NK, et al. J Virol. 1998 Jan;72(1):564-77. doi: 10.1128/JVI.72.1.564-577.1998. J Virol. 1998. PMID: 9420260 Free PMC article. - N-Terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles.
Gross I, Hohenberg H, Huckhagel C, Kräusslich HG. Gross I, et al. J Virol. 1998 Jun;72(6):4798-810. doi: 10.1128/JVI.72.6.4798-4810.1998. J Virol. 1998. PMID: 9573245 Free PMC article. - Defined amino acids in the gag proteins of human immunodeficiency virus type 1 are functionally active during virus assembly.
Kattenbeck B, Rohrhofer A, Niedrig M, Wolf H, Modrow S. Kattenbeck B, et al. Intervirology. 1996;39(1-2):32-9. doi: 10.1159/000150472. Intervirology. 1996. PMID: 8957667 Review. - Role of the nucleocapsid region in HIV-1 Gag assembly as investigated by quantitative fluorescence-based microscopy.
de Rocquigny H, El Meshri SE, Richert L, Didier P, Darlix JL, Mély Y. de Rocquigny H, et al. Virus Res. 2014 Nov 26;193:78-88. doi: 10.1016/j.virusres.2014.06.009. Epub 2014 Jul 9. Virus Res. 2014. PMID: 25016037 Review.
Cited by
- RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes.
Wang SW, Aldovini A. Wang SW, et al. J Virol. 2002 Dec;76(23):11853-65. doi: 10.1128/jvi.76.23.11853-11865.2002. J Virol. 2002. PMID: 12414928 Free PMC article. - The NH2-terminal domain of the human T-cell leukemia virus type 1 capsid protein is involved in particle formation.
Rayne F, Bouamr F, Lalanne J, Mamoun RZ. Rayne F, et al. J Virol. 2001 Jun;75(11):5277-87. doi: 10.1128/JVI.75.11.5277-5287.2001. J Virol. 2001. PMID: 11333909 Free PMC article. - Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains.
Lindwasser OW, Resh MD. Lindwasser OW, et al. J Virol. 2001 Sep;75(17):7913-24. doi: 10.1128/jvi.75.17.7913-7924.2001. J Virol. 2001. PMID: 11483736 Free PMC article. - HIV-1 release requires Nef-induced caspase activation.
Segura J, Ireland J, Zou Z, Roth G, Buchwald J, Shen TJ, Fischer E, Moir S, Chun TW, Sun PD. Segura J, et al. PLoS One. 2023 Feb 13;18(2):e0281087. doi: 10.1371/journal.pone.0281087. eCollection 2023. PLoS One. 2023. PMID: 36780482 Free PMC article. - Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain.
Accola MA, Strack B, Göttlinger HG. Accola MA, et al. J Virol. 2000 Jun;74(12):5395-402. doi: 10.1128/jvi.74.12.5395-5402.2000. J Virol. 2000. PMID: 10823843 Free PMC article.
References
- Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI36071/AI/NIAID NIH HHS/United States
- CA47482/CA/NCI NIH HHS/United States
- R01 CA047482/CA/NCI NIH HHS/United States
- R37 CA047482/CA/NCI NIH HHS/United States
- AI34736/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials