Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2 - PubMed (original) (raw)
Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2
T Kamijo et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The INK4a-ARF locus encodes two proteins, p16(INK4a) and p19(ARF), that restrain cell growth by affecting the functions of the retinoblastoma protein and p53, respectively. Disruption of this locus by deletions or point mutations is a common event in human cancer, perhaps second only to the loss of p53. Using insect cells infected with baculovirus vectors and NIH 3T3 fibroblasts infected with ARF retrovirus, we determined that mouse p19(ARF) can interact directly with p53, as well as with the p53 regulator mdm2. ARF can bind p53-DNA complexes, and it depends upon functional p53 to transcriptionally induce mdm2 and the cyclin-dependent kinase inhibitor p21(Cip1), and to arrest cell proliferation. Binding of p19(ARF) to p53 requires the ARF N-terminal domain (amino acids 1-62) that is necessary and sufficient to induce cell cycle arrest. Overexpression of p19(ARF) in wild type or ARF-null mouse embryo fibroblasts increases the half-life of p53 from 15 to approximately 75 min, correlating with an increased p53-dependent transcriptional response and growth arrest. Surprisingly, when overexpressed at supra-physiologic levels after introduction into ARF-null NIH 3T3 cells or mouse embryo fibroblasts, the p53 protein is handicapped in inducing this checkpoint response. In this setting, reintroduction of p19(ARF) restores p53's ability to induce p21(Cip1) and mdm2, implying that, in addition to stabilizing p53, ARF modulates p53-dependent function through an additional mechanism.
Figures
Figure 1
ARF stabilizes p53 and induces p53-dependent gene expression. (A) MEFs were lysed 48 hr after infection with retroviruses encoding either p19ARF, C-terminally truncated p19ARF mutants (N84, N62), or an N-terminally truncated ARF mutant (Δ1–62). Proteins were detected by direct immunoblotting using antibodies to p53, p21Cip1, and mdm2 as indicated in the left margin. (B) Northern blot analysis of RNA extracted from MEFs infected for the indicated times with p19ARF or control retrovirus vectors. Uninfected proliferating cells express levels of p53, p21Cip1, and glyceraldehyde 3-phosphate dehydrogenase (GDH) RNAs equal to those detected in cells infected with the control vector. (C) After a 36 hr infection with either a control (Top) or p19ARF retrovirus (Bottom), _ARF_-null MEFs were pulsed for 1 hr with [35S]methionine and chased in medium containing excess unlabeled precursor. Precipitated p53 from cells lysed at the indicated times after labeling was resolved on denaturing gels.
Figure 2
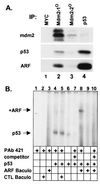
Functional p19ARF binds to both mdm2 and p53 and can form ternary complexes. (A) Sf9 cells coinfected for 48 hr with baculoviruses encoding wild-type p53 and either p19ARF or the indicated p19ARF mutants were lysed and precipitated with control antibody to myc (9E10), p53 (PAb 421), affinity-purified antibody to the ARF C terminus, or anti-HA (to detect HA-tagged N62). Proteins in immune complexes separated on denaturing gels were transferred to filters and detected by immunoblotting with anti-ARF or anti-HA (for N62). (B) Similar experiments to those shown in A were performed using the indicated p53 mutants (top two panels). Sf9 cells were also coinfected with the indicated p53 mutants and mdm2 (Bottom) to document the inability of the 22/23/281 mutant to bind mdm2. Proteins precipitated with PAb421 or antibody 2A10 to mdm2 were electrophoretically resolved, transferred and blotted with 2A10. (C) Sf9 cells infected with the viruses indicated below each panel were lysed and incubated with nonimmune serum (NRS), antibodies to the ARF C terminus, PAb421 (p53), or antibody 2A10 (mdm2) as indicated at the top of each lane. Resolved proteins were blotted with antibodies to mdm2 (Top), p53 (Middle) or ARF (Bottom) as above.
Figure 3
Direct interactions of p19ARF and p53. (A) Sequential precipitations [(IP), lanes 1–4] of lysates from NIH 3T3 cells infected with ARF virus were performed with the indicated antibodies. Precipitated proteins were separated and blotted with antibodies to mdm2, p53, and ARF. (B) EMSA was performed with an end-labeled oligonucleotide containing two consensus p53 binding sites (40). Additions to the binding reactions are indicated below the lanes and included activating antibody PAb-421, 10-fold excess cold unlabeled oligonucleotide (competitor), purified recombinant p53, and Sf9 extracts from cells infected with baculovirus vectors encoding ARF or no recombinant protein ( control, CTL). Arrows indicate positions of the p53-oligonucleotide complex and of that supershifted by ARF.
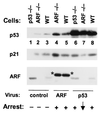
Figure 4
Induction of p21Cip1 by ARF or p53 retroviral vectors. Cells infected with vector alone (lanes 1–3), a retrovirus encoding HA-tagged p19ARF (lanes 4 and 5), or a vector encoding wild-type p53 (lanes 6–8) were lysed 48 hr after infection. Proteins separated on gels were immunoblotted for p53 (Top), p21Cip1 (Middle), and p19ARF (Bottom) as indicated at the left. Infected cells included _p53_-null early passage MEFs (lanes 1 and 6), wild-type MEFs (lanes 3, 5. and 8), or early passage _ARF_-null MEFs (lanes 2, 4, and 7). Endogenous p19ARF, elevated in p53-null cells (lane 1), is repressed after infection with p53 virus (lane 6). HA-tagged ARF (indicated by asterisks) migrates slower than the endogenous protein. Growth arrest was assayed at 48 hr by incorporation of [3H]-thymidine in replica plates.
Figure 5
Transactivation by ARF and p53. NIH 3T3 or 10(1) fibroblasts were transiently transfected with wild-type PG13-CAT (WT) or mutant MG15-CAT (Mut) and increasing amounts p19ARF or p53. In A and B, the ARF plasmid inputs in lanes 2–4 and 6–8 were 1, 2, and 5 μg DNA, whereas only 10 and 100 ng of p53 plasmid were used in B (lanes 5–8). In C, cells received no ARF DNA (lanes 1–4) or 1 μg ARF plasmid (lanes 5–8) plus 1, 2, or 5 μg p53 plasmid (lanes 2–4 and 6–8, respectively). Cell lysates prepared 48 hr after transfection were analyzed for CAT activity. The mono- and diacetylated species are at the middle and top of the plate, respectively. Signal intensities for diacetylated forms computed by densitometry and indicated below the lanes were normalized to 1.0 (A, lane 1).
References
- Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. - PubMed
- Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. - PubMed
- Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. - PubMed
- Duro D, Bernard O, Della Valle V, Berger R, Larsen C-J. Oncogene. 1995;11:21–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA071907/CA/NCI NIH HHS/United States
- CA-21765/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- CA-71907/CA/NCI NIH HHS/United States
- CA-63230/CA/NCI NIH HHS/United States
- R01 CA063230/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous