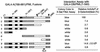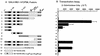Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML - PubMed (original) (raw)
Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML
J H Ahn et al. Mol Cell Biol. 1998 Aug.
Abstract
Both of the major immediate-early (IE) proteins IE1 and IE2 of human cytomegalovirus (HCMV) as well as input viral DNA and sites of viral IE transcription colocalize with or adjacent to punctate PML domains (PML oncogenic domains [PODs] or nuclear domain 10) in the nucleus within the first few hours after infection of permissive human fibroblasts. However, colocalization of IE1 and PML in PODs is only transient, with both proteins subsequently redistributing into a nuclear diffuse form. These processes are believed to promote efficient viral IE transcription and initiation of DNA synthesis especially at low multiplicities of infection. To examine the mechanism of PML displacement by IE1, we carried out indirect immunofluorescence assay experiments with plasmids expressing intact or deleted forms of PML and IE1 in DNA-transfected cells. The results demonstrated that deletion of the C-terminal acidic region of IE1 uncouples the requirements for displacement of both endogenous and coexpressed PML from those needed to target to the PODs. Mutant PML proteins containing either a Cys point mutation within the N-terminal RING finger domain or a small deletion (of positions 281 to 304) within the coiled-coil region did not localize to the PODs but instead gave a nuclear diffuse distribution, similar to that produced by intact PML in the presence of IE1. Endogenous PML also colocalized with IE1 in metaphase chromosomes in HCMV or recombinant adenovirus type 5-IE1-infected HF cells undergoing mitosis, implying that there may be a direct physical interaction between IE1 and PML. Indeed, a specific interaction between IE1 and PML was observed in a yeast two-hybrid assay, and the strength of this interaction was comparable to that of IE2 with the retinoblastoma protein. The RING finger mutant form of PML showed a threefold-lower interaction with IE1 in the yeast system, and deletion of the N-terminal RING finger domain of PML abolished the interaction. Consistent with the IFA results, a mutant IE1 protein that lacks the C-terminal acidic region was sufficient for interaction with PML in the yeast system. The two-hybrid interaction assay also showed that both the N-terminal RING finger domain and the intact coiled-coil region of PML are required cooperatively for efficient self-interactions involving dimerization or oligomerization. Furthermore, truncated or deleted GAL4/PML fusion proteins that retained the RING finger domain but lacked the intact coiled-coil region displayed an unmasked cryptic transactivator function in both yeast and mammalian cells, and the RING finger mutation abolished this transactivation property of PML. Therefore, we suggest that a direct interaction between IE1 and the N-terminal RING finger domain of PML may inhibit oligomerization and protein-protein complex formation by PML, leading to displacement of PML and IE1 from the PODs, and that this interaction may also modulate a putative conditional transactivator function of PML.
Figures
FIG. 1
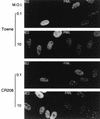
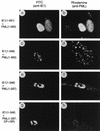
Cells infected with an HCMV mutant that does not express IE1 fail to disperse the PML protein from punctate domains (PODs) into a nuclear diffuse pattern. The photographs show a comparison of PML staining patterns in HF cells infected with either HCMV(Towne) (upper two panels) or the CR208(ΔIE1) virus (lower two panels) at a low MOI of 0.1 and a high MOI of 10. At 6 h after infection, the cells were fixed with paraformaldehyde followed by double-label IFA. IE2 was detected with mouse MAb 12E2 and FITC-labeled anti-mouse IgG (left). PML was detected in the same fields with rabbit anti-PML PML-C PAb and rhodamine-coupled anti-rabbit IgG (right). Note the typical punctate POD patterns of PML in adjacent uninfected cells at low MOI.
FIG. 2
Comparison of the PML protein levels between mock-infected, transfected, and HCMV-infected cells. (A) Detection of the in vitro-translated pCMX-PML cDNA protein product (560 aa). [35S]Met-labeled PML proteins were synthesized in reticulocyte extracts from template RNA transcribed in vitro with T7 polymerase from _Bam_HI-linearized plasmid pCMX-PML and analyzed on SDS–10% polyacrylamide gels. (B) Detection of nuclear forms of the PML protein by mouse MAb 5E10, using extracts prepared from pCMX-PML-transfected Vero cells and HCMV-infected HF cells. Total extracts (35 μg of protein) prepared from untransfected (lane 1) or pCMX-PML-transfected Vero cells (lane 2) and from mock-infected (lane 3) or HCMV-infected HF cells (lane 4) were electrophoretically fractionated on SDS–10% polyacrylamide gels, and Western blot analysis was performed by incubating the membrane with MAb 5E10 directed against PML. (C) Comparison of the levels of all forms of the PML protein detectable in mock-infected and HCMV-infected HF cells. Gel fractionated extracts from mock-infected (30 μg for lane 2 and 8 μg for lane 3) and HCMV-infected HF cells (30 μg for lane 2 and 8 μg for lane 4) were subjected to Western blot analysis using either rabbit PAb PML-C directed against the C terminus of PML (lanes 1 and 2) or mouse MAb 6E1 against IE1 (lanes 3 and 4).
FIG. 3
Summary of the localization patterns of IE1, IE2, and PML in Vero cells transiently transfected with genomic plasmids expressing deleted versions of IE1. At the top is an illustration of the overlapping five exon structure (solid bar) of the MIE gene transcription unit in the inverted (i.e., viral) genomic orientation. The positions of key restriction sites used to generate the deleted or truncated versions of IE1 are indicated above the diagram. Bg, _Bgl_II; EV, _Eco_RV; Sp, _Spe_I. The enhancer/promoter region of the MIE locus (ENH; hatched bar) and the translation start (ATG) and termination (TAA) sites as well as polyadenylation sites (pA) are also indicated. Below is a comparison of the structures of the proteins encoded by the variant MIE expression gene plasmids used. Open bars represent coding regions, with gaps denoting in-frame deletions; diamonds indicate inserted triple-terminator oligonucleotides. The estimated map locations for the epitopes recognized by MAbs 6E1, 12E2, and CH810 are shown at the bottom (hatched bars). To detect IE1, IE2, and PML, FITC-labeled MAb 6E1 (for IE1), 12E2 (for IE2), and CH810 (for both IE1 and IE2) were used in double-label IFA experiments together with rhodamine-coupled rabbit PAb against PML. IFA patterns; ND, nuclear diffuse; P, punctate; ND/P, mixture of nuclear diffuse and punctate; G, nuclear granular structures.
FIG. 4
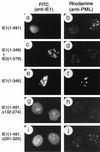
Effects of wild-type or mutant IE1 expression on distribution of the endogenous PML proteins in transient expression assays. Vero cells were transfected with plasmids encoding various mutant IE1 proteins and fixed with paraformaldehyde followed by double-label IFA at 48 h after transfection. (a and b) Paired photographs of cells receiving plasmid pMP17 encoding wild-type IE1(1-491); (c and d) pRL55 encoding both IE1(1-346) and wild-type IE2; (e and f) pRL74 encoding IE1(1-346); (g and h) pRL60 encoding IE1(Δ132-274); (i and j) pRL61 encoding IE1(Δ291-320). (a, c, e, g, and i) Detection of IE1 with mouse MAb 6E1 (a, c, e, and i) or MAb CH810 (g) and FITC-labeled anti-mouse IgG. (b, d, f, h, and j) Detection of PML in the same fields with rabbit anti-PML-C PAb and rhodamine-coupled anti-rabbit IgG.
FIG. 5
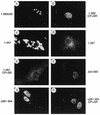
Localization patterns of wild-type and mutant PML in transfected Vero cells. Vero cells were transfected with plasmids expressing the wild-type (wt) PML(1-560) protein (pCMX-PML) (a), PML(1-560, C88P89→S88R89) (pGH623-5) (b), PML(1-447) (pJHA286) (c), PML(1-267) (pJHA287) (d), PML(1-267, C88P89→S88R89) (pJHA288) (e), PML(224-560) (pJHA289) (f), PML(1-560, Δ282-304) (pJHA290) (g), and PML(1-560, C88P89→S88R89 Δ282-304) (pJHA291) (h). The cells were fixed with paraformaldehyde at 48 h after transfection followed by IFA with anti-PML-C PAb (a, b, g, and h) or anti-PML-N PAb (c, d, e, and f) and rhodamine-coupled anti-rabbit IgG. Phase-contrast images confirming the nuclear plus cytoplasmic locations of PML(1-447) are available upon request.
FIG. 6
Effects of mutant PML proteins on self-interaction measured with a yeast two-hybrid assay. (Left) Diagram illustrating the structure of the GAL4-A/PML fusion proteins used. The two major translocation fusion points occurring within the PML protein (at positions 552 and 955) in PML/RARα fusions in APL are indicated by arrows. The proposed NLS (at positions 467 to 490) is indicated (36). The location of the paired RING finger point mutations (C88P89→S88R89) is indicated by a star. The amino acid positions of the restriction enzyme cleavage sites used to generate the GAL4-A/mutant PML fusion are indicated. Dotted bars, N-terminal Pro-rich domain and C-terminal Ser-rich domain; black bars, RING finger domain (left) and two adjacent Cys/His-rich domains (right); hatched bars, putative α-helical region. Av, _Avr_II; Pv, _Pvu_II; Sm, _Sma_I. (Right) Qualitative and quantitative results of the yeast self-interaction assay. No detectable β-galactosidase activity was measured in Y190 cells transformed with the plasmid encoding the GAL4-DB/PML(1-560) fusion protein alone. Plasmids encoding a variety of GAL4-A/PML fusion proteins were then introduced together with GAL4-DB/PML(1-560) into Y190 cells. Transformants were selected on plates lacking Trp and Leu, and β-galactosidase activity of the transformants was measured as described in Materials and Methods. Lines: 1, GAL4-A/PML(1-560) in pJHA266; 2, GAL4-A/PML(1-560, C88P89→S88R89) (pJHA267); 3, GAL4-A/PML(1-447) (pJHA268); 4, GAL4-A/PML(1-267) (pJHA269); 5, GAL4-A/PML(1-96) (pJHA270); 6, GAL4-A/PML(224-560) (pJHA281); 7, GAL4-A/PML(447-560) (pJHA271); 8, GAL4-A/PML(1-560, Δ281-304) (pEB5); 9, GAL4-A/PML(1-560, C88P89→S88R89 Δ281-304) (pEB6). aMean values for β-galactosidase units for interaction between GAL4-DB/PML(1-560) and GAL4-A/PML(1-560) in duplicated assays are indicated as 100%. The relative activity of control SNF1/SNF4 interaction in the same assay was 148%. bSelf-interaction with the GAL4-DB/PML(96-560, Δ281-304) fusion protein encoded by plasmid pJHA294.
FIG. 7
Effects of wild-type or mutant IE1 expression on distribution of the PML proteins in cotransfection assays. Vero cells were cotransfected with the following plasmid pairs: (a and b) pMP18 encoding wild-type IE1(1-491) and pCMX-PML encoding intact PML(1-560); (c and d) pJHA300 encoding IE1(1-346) and pCMX-PML; (e and f) pJHA300 and pJHA261 encoding mutant PML(1-267); (g and h) pJHA300 and pJHA275 encoding mutant PML(1-267, C88P89→S88R89). At 48 h after transfection, cells were fixed with paraformaldehyde followed by double-label IFA. (a, c, e, and g) Detection of IE1 with mouse MAb 6E1 and FITC-labeled anti-mouse IgG. (b, d, f, and h) Detection of PML in the same fields with rabbit anti-PML PML-C (b and d) or PML-N (f and h) PAb and rhodamine-coupled anti-rabbit IgG. Phase-contrast images confirming the nuclear plus cytoplasmic locations of PML(1-560) in the presence of IE1(1-346) are available upon request.
FIG. 8
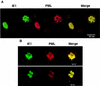
Confocal microscopy images demonstrating colocalization between PML and IE1 in metaphase chromosomes in HCMV- or Ad5-IE1-infected cells undergoing mitosis. HF cells were infected with either HCMV(Towne) (A) or Ad5-IE1 (B) at an MOI of 0.5. At 72 h after infection, the cells were permeabilized in absolute methanol at −20°C followed by double-label IFA. Left-hand panels (green fluorescence), detection of IE1 with mouse MAb 6E1 and FITC-labeled anti-mouse IgG; center panels (red fluorescence), detection of PML in the same fields with rabbit anti-PML-C PAb and rhodamine-coupled anti-rabbit IgG; Right-hand panels (yellow merge fluorescence), confocal images from each fluorochrome were recorded and superimposed to demonstrate colocalization. Two representative cells are shown in the upper and lower sections of panel B.
FIG. 9
Yeast two-hybrid assays demonstrating specific interactions between IE1 and PML. The GAL4-DB fusion proteins used were GAL4-DB/EBNA-1(1-641, Δ102-325) (encoded by plasmid pYW18), GAL4-DB/PML(1-560) (pJHA238), GAL4-DB/PML(1-560, C88P89→S88R89) (pJHA247), GAL4-DB/PML(97-267) (pJHA277), GAL4-DB/PML(224-560) (pJHA280), GAL4-DB/SNF1 (pSE1112), and GAL4-DB/Rb(301-928) (pRb2). The GAL4-A fusion proteins used were GAL4-A/IE1(1-491) (in pJHA239), GAL4-A/IE2(1-579) (pJHA140), GAL4-A/IE1(1-346) (pJHA300), GAL4-A/IE1(1-231) (pJHA255), GAL4-A/IE1(232-491) (pJHA249), GAL4-A/IE1(Δ132-274) (pJHA251), GAL4-A/IE1(132-274) (pJHA257), and GAL4-A/SNF4 (pSE1111). Paired plasmids encoding the GAL4-DB and GAL4-A fusion proteins were introduced together into Y190 cells. Transformants were selected on plates lacking Trp and Leu, and β-galactosidase activity of the transformants were measured. Mean values for β-galactosidase units in duplicated assays are denoted by the black bars with error range indicated.
FIG. 10
Unmasking of a cryptic transactivator property within the N-terminal domain of PML proteins in yeast cells. (A) Diagram illustrating the structure of the GAL4-DB/PML (lines 1 to 12) and other control GAL4-DB/IE2 (line 13) or GAL4-DB/IE110 (lines 14 and 15) fusion proteins used. See Fig. 6 for details of PML protein features. (B) β-Galactosidase activity measured in Y190 cells transformed with plasmids encoding the GAL4-DB/PML or other control fusion proteins. Single-transformants were selected on plates lacking Trp, and β-galactosidase activity of the transformants was measured. Lines: 1, GAL4-DB/PML(1-560) in pJHA238; 2, GAL4-DB/PML(1-560, C88P89→S88R89) (pJHA247); 3, GAL4-DB/PML(1-447) (pJHA252); 4, GAL4-DB/PML(1-267) (pJHA253); 5, GAL4-DB/PML(1-267, C88P89→S88R89) (pJHA273); 6, GAL4-DB/PML(1-96) (pJHA254); 7, GAL4-DB/PML(1-96, C88P89→S88R89) (pJHA274); 8, GAL4-DB/PML96-267) (pJHA277); 9, GAL4-DB/PML(224-560) (pJHA280); 10, GAL4-DB/PML(447-560) (pJHA250); 11, GAL4-DB/PML(1-560, Δ281-304) (pEB1); 12, GAL4-DB/PML(1-560, C88P89→S88R89 Δ281-304) (pEB2); 13, GAL4-DB/IE2(290-579) (pCJC420); 14, GAL4-DB/IE110(104-240) (pLZ59); 15, GAL4-DB/IE110(104-240, C152P153→S152R153) (pLZ60).
FIG. 11
Assays for activator domains in PML using GAL4 fusion proteins expressed in transient assays in mammalian cells. Vero cells were cotransfected with plasmids containing either the parent E1b-CAT or GAL45/E1b-CAT reporter target gene together with plasmids encoding the GAL4-DB (pGH250) or GAL4-DB/PML fusion proteins. A representative autoradiograph of a transient CAT assay is shown. The basal samples show the levels of E1b-CAT or GAL45/E1b-CAT expression in the presence of vector plasmid DNA only. GAL4/IE2(544-579) in plasmid pMP54a containing the C-terminal IE2 transactivator domain described previously (59) and GAL4/CBF1(1-500) in plasmid pJH93 were used as positive and negative controls, respectively (32). GAL4-DB/PML fusion proteins tested were GAL4-DB/PML(1-560) in pJHA258, GAL4-DB/PML(1-560, C88P89→S88R89) in pJHA259, GAL4-DB/PML(1-447) in pJHA260, GAL4-DB/PML(1-267) in pJHA261, GAL4-DB/PML(1-267, C88P89→S88R89) in pJHA275, GAL4-DB/PML(97-267) in pJHA278, and GAL4-DB/PML(447-560) in pJHA263.
Similar articles
- Characterisation of the PML/RAR alpha rearrangement associated with t(15;17) acute promyelocytic leukaemia.
Grimwade D, Solomon E. Grimwade D, et al. Curr Top Microbiol Immunol. 1997;220:81-112. doi: 10.1007/978-3-642-60479-9_6. Curr Top Microbiol Immunol. 1997. PMID: 9103677 Review. - PML and the oncogenic nuclear domains in regulating transcriptional repression.
Li H, Chen JD. Li H, et al. Curr Opin Cell Biol. 2000 Oct;12(5):641-4. doi: 10.1016/s0955-0674(00)00144-7. Curr Opin Cell Biol. 2000. PMID: 10978902 Review.
Cited by
- Efficient proliferation and mitosis of glioblastoma cells infected with human cytomegalovirus is mediated by RhoA GTPase.
Al-Qahtani AA, Alarifi S, Alkahtani S, Stournaras C, Sourvinos G. Al-Qahtani AA, et al. Mol Med Rep. 2020 Oct;22(4):3066-3072. doi: 10.3892/mmr.2020.11434. Epub 2020 Aug 19. Mol Med Rep. 2020. PMID: 32945485 Free PMC article. - Expression of Human Cytomegalovirus IE1 Leads to Accumulation of Mono-SUMOylated PML That Is Protected from Degradation by Herpes Simplex Virus 1 ICP0.
Hou W, Cruz-Cosme R, Wen F, Ahn JH, Reeves I, Luo MH, Tang Q. Hou W, et al. J Virol. 2018 Nov 12;92(23):e01452-18. doi: 10.1128/JVI.01452-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30258013 Free PMC article. - Importance of Promyelocytic Leukema Protein (PML) for Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication.
Hossain MG, Ohsaki E, Honda T, Ueda K. Hossain MG, et al. Front Microbiol. 2018 Oct 8;9:2324. doi: 10.3389/fmicb.2018.02324. eCollection 2018. Front Microbiol. 2018. PMID: 30349510 Free PMC article. - Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx.
Hofmann H, Sindre H, Stamminger T. Hofmann H, et al. J Virol. 2002 Jun;76(11):5769-83. doi: 10.1128/jvi.76.11.5769-5783.2002. J Virol. 2002. PMID: 11992005 Free PMC article. - The carboxy terminal region of the human cytomegalovirus immediate early 1 (IE1) protein disrupts type II inteferon signaling.
Raghavan B, Cook CH, Trgovcich J. Raghavan B, et al. Viruses. 2014 Apr 2;6(4):1502-24. doi: 10.3390/v6041502. Viruses. 2014. PMID: 24699362 Free PMC article.
References
- Ahn J-H, Chiou C-J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. - PubMed
- Ahn, J.-H. Unpublished data.
- Barlow P N, Luisi B, Milner A, Elliott M, Everett R D. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. J Mol Biol. 1994;237:201–211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous