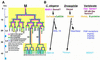Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning - PubMed (original) (raw)
Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning
B D Harfe et al. Genes Dev. 1998.
Abstract
Mesodermal development is a multistep process in which cells become increasingly specialized to form specific tissue types. In Drosophila and mammals, proper segregation and patterning of the mesoderm involves the bHLH factor Twist. We investigated the activity of a Twist-related factor, CeTwist, during Caenorhabditis elegans mesoderm development. Embryonic mesoderm in C. elegans derives from a number of distinct founder cells that are specified during the early lineages; in contrast, a single blast cell (M) is responsible for all nongonadal mesoderm formation during postembryonic development. Using immunofluorescence and reporter fusions, we determined the activity pattern of the gene encoding CeTwist. No activity was observed during specification of mesodermal lineages in the early embryo; instead, the gene was active within the M lineage and in a number of mesodermal cells with nonstriated muscle fates. A role for CeTwist in postembryonic mesodermal cell fate specification was indicated by ectopic expression and genetic interference assays. These experiments showed that CeTwist was responsible for activating two target genes normally expressed in specific subsets of nonstriated muscles derived from the M lineage. In vitro and in vivo assays suggested that CeTwist cooperates with the C. elegans E/Daughterless homolog in directly activating these targets. The two target genes that we have studied, ceh-24 and egl-15, encode an NK-2 class homeodomain and an FGF receptor (FGFR) homolog, respectively. Twist activates FGFR and NK-homeodomain target genes during mesodermal patterning of Drosophila and similar target interactions have been proposed to modulate mesenchymal growth during closure of the vertebrate skull. These results suggest the possibility that a conserved pathway may be used for diverse functions in mesodermal specification.
Figures
Figure 1
Postembryonic mesodermal lineage M in C. elegans. Times indicated are post-hatching at 25°C (lineage redrawn from Sulston and Horvitz 1979).
Figure 2
Sequence requirements for in vivo activation by the NdE-box motif. (A) Wild-type and mutated forms of the ceh-24 NdE-box vulval muscle enhancer were concatamerized and assayed for activation of a truncated pes-10 promoter fragment. Activity was measured using a lacZ or GFP reporter construct. The pes-10 promoter used in this assay is not intrinsically active, but can be activated in a variety of tissues by use of additional enhancer sequences for activation (A. Fire, S. Xu, and G. Seydoux, pers. comm.). NdE boxes are underlined. Mutations are indicated with lowercase letters. All constructs were injected into wild-type hermaphrodite animals and were assayed in at least two independent transgenic lines. A wild-type ceh-24 NdE box enhancer assayed in front of the deleted pes-10 promoter was capable of activating reporter transcription only when more than two NdE boxes were present. ceh-24 NdE box enhancer mutations 1m, 2m, 4m, 5m, 8m, 11m, and all wild-type ceh-24 NdE box enhancers were assayed with both lacZ and GFP reporter genes. All others were assayed with lacZ. A construct containing nine copies of a wild-type ceh-24 NdE box enhancer, in addition to vulval, intestinal, uterine, and anal depressor muscle expression had additional expression in one cell in the head. This cell has been tentatively identified as the head mesodermal cell. Expression in this cell has not been observed with any other construct. (B) Summary of NdE box mutations. The two NdE boxes are underlined. Base pairs that were required for reporter expression are boxed. Base pairs highlighted in gray resulted in loss of expression only when mutated concurrently. Separate mutation of these base pairs had no evident effect on reporter expression (Fig. 2A, 5m, 6m, 7m, and 8m).
Figure 3
_Cis_-acting elements controlling egl-15 promoter activity. Deletion constructs derived from an egl-15::gfp translational fusion are diagrammed. All constructs were tested for activity in at least two independent lines. Larvae, embryos, and adults were examined for expression of GFP in early M lineage descendants, head neurons, and vm1 vulval muscles, respectively. (+) GFP expression levels similar to that of the full-length egl-15 promoter construct (NH#300). (−) no observed reporter expression; (+/−) expression that was only marginally detectable. Each construct contained the egl-15 ATG codon (+1) and was fused in-frame with GFP. NH#300 was a gift of C. Branda and M. Stern (Yale University, New Haven, CT). Construct pBH56.17 contained the pes-10 promoter in place of the first 196 bp of the egl-15 promoter. The five NdE-like E boxes are numbered 1 (−602: taaCATATGcac), 2 (−572: ctcCAGATGtga), 3 (−471: taaCATCTGgac); 4 (−371: caaCATATGcgt), and 5 (−239: ttaCATATGgtg). 1Reporter expression in the vm1 vulval muscles of constructs pBH55.40 and pBH56.01 was greatly reduced relative to the full length egl-15::gfp fusion. A similar decrease in M lineage activity (which is much less evident in the parent construct) would likely result in a complete failure to detect the reporter.
Figure 4
bHLH comparisons among Twist family members and related factors. Amino acids identical to CeTwist are indicated by dashes. Sources for bHLH domain sequence are: Dtwist (Drosophila, Thisse et al. 1988); Mtwist; (Mus, Wolf et al. 1991); Htwist (Human, Howard et al. 1997; Ghouzzi et al. 1997); Xetwist (Xenopus, Hopwood et al. 1989). Dermo-1 (Mus, Li et al. 1995), and dHand (Mus, Hollenberg et al. 1995; Srivastava et al. 1995). No close homolog of CeTwist exists in the C. elegans genome database (80% completion; 5/98). On the basis of BLAST similarity searches, the distantly-related CeMyoD (Krause et al. 1990) may be the nearest homolog of CeTwist in C. elegans.
Figure 5
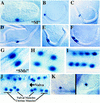
Activity pattern of hlh-8 reporter fusions. (A–K). Pattern of expression from an integrated hlh-8::lacZ reporter construct (pBH47.74; see Fig. 8). An identical expression pattern was observed with GFP as a reporter (data not shown). (A) Threefold embryo with M labeled; (B) L1 animal with M labeled; (C) M daughters; (D) M granddaughters; (E) eight descendants of the M blast cell; (F) eighteen descendants of the M blast cell; (G) SMs; (H–I) Reporter expression in sex myoblast descendants; (J) Ventral view of L4 larva with 16 sex muscles (8 vulval and 8 uterine) arranged around the vulva; (K) An hlh-8::lacZ fusion with additional 5′ sequences (pBH47.72, see Fig. 8) showed reporter activity in coelomocytes. The reporter activity in these cells was nuclear, and was seen with both hlh-8::lacZ and hlh-8::gfp fusion constructs. These two properties confirm that reporter expression was intrinsic to these cells (coelomocytes can also accumulate GFP secreted by other cells, but this process produces vesicular GFP and is not observed with lacZ; Fire et al. 1998b).
Figure 6
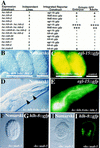
Consequences of ectopic CeTwist and CeE/DA expression. Ectopic heat shock expression constructs were injected into animals already containing an integrated GFP reporter construct (see Materials and Methods). Heritable extrachromosomal arrays of each ectopic expression construct were produced before phenotypic effects were assayed. Genes listed correspond to the following proteins: (hlh-8) CeTwist; (hlh-2) CeE/DA; (hlh-1) CeMyoD; (bhlh-8) CeTwist missing its basic domain and helix I. (A) Ectopic protein was produced from heat shock constructs by placement of stable transgenic lines at 37°C for 2 hr. Heat-shocked animals were allowed to recover at room temperature for >2 hr before being assayed for expression of the integrated GFP reporter. (+) Modest levels of GFP expression. Each additional + indicates a dramatic increase in GFP expression. Animals negative for ectopic reporter expression are indicated by a −. In heat-shocked animals with at least one +, reporter expression was seen in most cells of embryos and adults. (B,C) Example of hsp16-2::hlh-8+hsp16-2::hlh-2 embryos examined 4 hr after the initial heat shock. (B) Nomarski photograph. (C) CeTwist–CeE/DA driven ectopic expression from an egl-15::gfp integrated reporter construct. No expression was seen in embryos without the hs::hlh-8 construct. (D,E) Example of hsp16-2::hlh-8+hsp16-2::hlh-2 adult animals 24 hr after the initial heat shock. (D) Nomarski photograph. (E) CeTwist–CeE/DA driven ectopic expression from an egl-15::gfp integrated reporter construct. Large vacuoles were seen in the pharynx (arrowheads). (F–I) MAB-5-driven ectopic expression from an hlh-8::gfp reporter construct. An hsp16-1::mab-5 fusion construct (Salser et al. 1992) was introduced into a strains with an integrated hlh-8::gfp reporter. (F,G) Embryo in the absence of heat shock; (H,I) embro following heat shock.
Figure 7
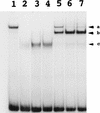
In vitro DNA-binding activities of CeTwist and CeE/DA. CeTwist and CeE/DA preferentially bound E boxes as heterodimers in vitro. CeE/DA-fusion protein was obtained as described previously (Krause et al. 1997). Equimolar starting concentrations of CeE/DA and CeTwist fusion proteins were arbitrarily defined as 1×. (Lane 1) CeE/DA (1×); (lanes 2–4) CeTwist at 1×, 2×, and 3×, respectively; (Lanes 5–7) CeE/DA at 1× and CeTwist at 1×, 2×, and 3×, respectively. Arrowheads: (a) CeE/DA homodimer; (b) CeTwist–CeE/DA heterodimer; (c) CeTwist homodimer. Gel shifts for this figure were performed with a probe that contained the E-box sequence CAGGTG. Equivalent binding results were obtained with a consensus NdE box (CATATG) with the exception that little or no binding was seen with the CeE/DA homodimer, while no binding by either CeTwist or CeTwist–CeE/DA was seen with a mutated E box (gATATG; data not shown).
Figure 8
_Cis_-acting signals controlling hlh-8. All constructs were assayed in at least two independent transgenic lines. Reporter activity was scored in all M lineage cells and in all six coelomocytes (four embryonic and two postembryonic). (+) Reporter-positive cells, (−) reporter-negative cells. In − constructs reporter expression was absent in the entire M lineage or all coelomocytes. All translational fusion constructs contained the first nine amino acids of hlh-8 (black boxes) fused in frame to GFP. (A) Identification of _hlh-8 cis_-acting sequences. Gray boxes represent the entire predicted coding region of an uncharacterized putative gene upstream of hlh-8. Black boxes and white boxes represent hlh-8 and GFP coding regions, respectively. The location of DNA elements required for expression in coelomocytes and the M lineage are indicated (horizontal lined boxes). Two NdE box elements were found, at −128 (acgCATATGttg) and −48 (tttCATATGttt). 1Antibody staining indicated CeTwist protein accumulated in defecation-associated muscles. This activity was evident in tagged constructs with the complete CeTwist coding region (pPD47.08), but not in any of the _hlh-8_–reporter fusions. 2Insertion of GFP into an ORF (gray boxes) of the predicted gene upstream of hlh-8 produced reporter activity in hypodermal cells (pBH45.26; data not shown). (B) Point mutation analysis of a 21-bp segment essential for M-lineage activation of the hlh-8 promoter. All constructs contained 315 bp of DNA 5′ of the start of hlh-8 transcription and the first nine amino acids of hlh-8 fused in frame to GFP. A construct containing 315 bp of wild-type sequence was active in the M lineage (pBH52.01; Fig. 8A). Deletion of 21 bp of 5′ hlh-8 promoter sequence resulted in an inactive construct (pBH49.33; Fig. 8A). The GAGA motif, putative NF-κB site, and putative _HOX_-binding site are underlined. Mutationsare indicated with lowercase letters. All mutations tested abolished reporter activity.
Figure 8
_Cis_-acting signals controlling hlh-8. All constructs were assayed in at least two independent transgenic lines. Reporter activity was scored in all M lineage cells and in all six coelomocytes (four embryonic and two postembryonic). (+) Reporter-positive cells, (−) reporter-negative cells. In − constructs reporter expression was absent in the entire M lineage or all coelomocytes. All translational fusion constructs contained the first nine amino acids of hlh-8 (black boxes) fused in frame to GFP. (A) Identification of _hlh-8 cis_-acting sequences. Gray boxes represent the entire predicted coding region of an uncharacterized putative gene upstream of hlh-8. Black boxes and white boxes represent hlh-8 and GFP coding regions, respectively. The location of DNA elements required for expression in coelomocytes and the M lineage are indicated (horizontal lined boxes). Two NdE box elements were found, at −128 (acgCATATGttg) and −48 (tttCATATGttt). 1Antibody staining indicated CeTwist protein accumulated in defecation-associated muscles. This activity was evident in tagged constructs with the complete CeTwist coding region (pPD47.08), but not in any of the _hlh-8_–reporter fusions. 2Insertion of GFP into an ORF (gray boxes) of the predicted gene upstream of hlh-8 produced reporter activity in hypodermal cells (pBH45.26; data not shown). (B) Point mutation analysis of a 21-bp segment essential for M-lineage activation of the hlh-8 promoter. All constructs contained 315 bp of DNA 5′ of the start of hlh-8 transcription and the first nine amino acids of hlh-8 fused in frame to GFP. A construct containing 315 bp of wild-type sequence was active in the M lineage (pBH52.01; Fig. 8A). Deletion of 21 bp of 5′ hlh-8 promoter sequence resulted in an inactive construct (pBH49.33; Fig. 8A). The GAGA motif, putative NF-κB site, and putative _HOX_-binding site are underlined. Mutationsare indicated with lowercase letters. All mutations tested abolished reporter activity.
Figure 9
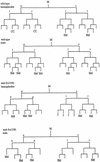
Requirements for MAB-5 in M lineage patterning. Divisions and cell fates in the M lineage are shown for wild-type (Sulston and Horvitz 1979) and sample animals of genotype mab-5(e1239); him-5(e1490) (SM) Sex myoblast; (CC) Coelomocyte; (?) not followed; Cells that apparently adopted a body wall muscle fate are unmarked. To characterize the extent of lineage variability and identify consistent features in the mutant, lineages were followed from the L1 to L3 stage in three hermaphrodites and two males. General features of the mutant lineages are summarized as follows: at hatching, M had migrated toward the posterior (as in wild type) in all mutant animals but was often situated dorsal to wild type. The first division of M was generally normal (although M divided along the anterior–posterior axis in one animal). In the second division, an abnormality was seen in the cleavage plane of Md: this cell frequently divided along the anterior–posterior instead of left–right body axis (n = 5/9 divisions observed in hermaphrodites and 2/3 divisions observed in males). The most striking and consistent phenotypes seen in mab-5(−) hermaphrodites were the loss of the two M-derived coelomocytes (100% of spot-checked animals, with n > 200) and the production of cells that appeared to be supernumary SMs. Like wild-type SMs, these supernumerary myoblasts became enlarged and divided during the L3 stage. The putative SMs arose in multiple, variable positions after M had undergone four divisions (generally without the additional round of cell division that normally occurs in the Mvl/rpa position). The following behavior was seen in the two lineaged hermaphrodites not shown in the figure: The first generated one dorsal SM (Md.ppa) and one ventral SM (Mv.rpa) on the right and two SMs (origins unknown) on the left. All of these cells migrated toward the gonadal anchor cell, enlarged, and underwent multiple cell divisions during the L3. In a second animal in which te Md/vl lineages were followed to mid-L2, the M.vlpa cell became an SM (which migrated toward the gonad) and there were no apparent dorsal SMs. In the additional male lineage, M.drpa and M.vrpa both became apparent SMs that migrated toward the gonad, enlarged, and divided during the L3. To augment this analysis, the early M cell division pattern and the pattern of SMs were observed in larger numbers of animals by spot-checking cell number and position (by Nomarski microscopy) in appropriately staged larvae. Among 27 left or right sides observed in L3 hermaphrodites, 1 side had zero SMs, 7 sides had one SM, 11 sides had two SMs, 6 sides had three SMs, 1 side had four SMs, and 1 side had six SMs. Among 4 left or right sides spot-checked in males, 1 had one SM, 2 had two SMs, and 1 side had four SMs. Generally the resulting cells could be seen in the ventral quadrant, having migrated to flank the center of the gonad; in a minority of cases, the cells apparently migrated to dorsal midbody region (presumably dorsally generated cells that migrated anteriorly but not ventrally) or, less frequently, in more posterior ventral or dorsal positions (presumably SM cells that failed to migrate). The male SMs normally migrate posteriorly, indicating that mab-5 controls the direction of migration of these cells in males. M-derived male mating structures are generally absent in mab-5(0) mutants (see Kenyon 1986).
Figure 10
Aspects of a mesodermal patterning pathway are conserved. (A) A proposed pathway of genes active in the M lineage. Expression patterns for mab-5 and hlh-2 are derived from antibody staining (S. Salser and C. Kenyon, unpubl.; Krause et al. 1997, this work). mab-5 expression may extend to later stages in the SM lineages. Expression patterns for ceh-24 and egl-15 are predicted from reporter constructs (Harfe and Fire 1998; C. Branda and M. Stern, unpubl.). The hlh-8 pattern is inferred from both types of data (see text). The proposed pathway, inferred from genetic and interference experiments as described in the text, includes a requirement for MAB-5 in the early activation of hlh-8 and a role for the resulting CeTwist–CeE/DA heterodimer in the activation of ceh-24 and egl-15. Also as expected from this model (data not shown), MAB-5 was required for early (but not late) M lineage activity of egl-15 reporter constructs. (B) Comparative aspects of a Twist-dependent mesodermal diversification pathway. The vertical location of each protein name corresponds to the proposed time of action. In Drosophila, Dorsal can directly activate Dtwist by binding NF-κB/Dorsal sites upstream of Dtwist (Thisse et al. 1991). Dtwist can activate heartless (FGFR) expression (Shishido et al. 1997), although it is not known if this interaction is direct (dashed arrow). hlh-8 requires a consensus NF-κB/Dorsal site for promoter activity. No dorsal gene has been identified in C. elegans. Dominant mutations in human Twist (Htwist), FGFRs, and the homeodomain MSX2 can result in craniosynostosis (Jabs et al. 1993; Muenke et al. 1994; Meyers et al. 1995; Przylepa et al. 1996; Ghouzzi et al. 1997; Howard et al. 1997).
Similar articles
- Caenorhabditis elegans twist plays an essential role in non-striated muscle development.
Corsi AK, Kostas SA, Fire A, Krause M. Corsi AK, et al. Development. 2000 May;127(10):2041-51. doi: 10.1242/dev.127.10.2041. Development. 2000. PMID: 10769229 - Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm.
Liu J, Fire A. Liu J, et al. Development. 2000 Dec;127(23):5179-90. doi: 10.1242/dev.127.23.5179. Development. 2000. PMID: 11060243 - Characterization of a dominant negative C. elegans Twist mutant protein with implications for human Saethre-Chotzen syndrome.
Corsi AK, Brodigan TM, Jorgensen EM, Krause M. Corsi AK, et al. Development. 2002 Jun;129(11):2761-72. doi: 10.1242/dev.129.11.2761. Development. 2002. PMID: 12015302 - A Twist in fate: evolutionary comparison of Twist structure and function.
Castanon I, Baylies MK. Castanon I, et al. Gene. 2002 Apr 3;287(1-2):11-22. doi: 10.1016/s0378-1119(01)00893-9. Gene. 2002. PMID: 11992718 Review. - Intersecting signalling and transcriptional pathways in Drosophila heart specification.
Frasch M. Frasch M. Semin Cell Dev Biol. 1999 Feb;10(1):61-71. doi: 10.1006/scdb.1998.0279. Semin Cell Dev Biol. 1999. PMID: 10355030 Review.
Cited by
- Free long-chain fatty acids trigger early postembryonic development in starved Caenorhabditis elegans by suppressing mTORC1.
Ruan M, Xu F, Li N, Yu J, Teng F, Tang J, Huang C, Zhu H. Ruan M, et al. PLoS Biol. 2024 Oct 22;22(10):e3002841. doi: 10.1371/journal.pbio.3002841. eCollection 2024 Oct. PLoS Biol. 2024. PMID: 39436954 Free PMC article. - UBR-5 and UBE2D mediate timely exit from stem fate via destabilization of poly(A)-binding protein PABP-2 in cell state transition.
Calva Moreno JF, Jose G, Weaver YM, Weaver BP. Calva Moreno JF, et al. Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2407561121. doi: 10.1073/pnas.2407561121. Epub 2024 Oct 15. Proc Natl Acad Sci U S A. 2024. PMID: 39405353 Free PMC article. - SEM-2/SoxC regulates multiple aspects of C. elegans postembryonic mesoderm development.
Baccas M, Ganesan V, Leung A, Pineiro L, McKillop AN, Liu J. Baccas M, et al. bioRxiv [Preprint]. 2024 Jul 4:2024.07.04.602042. doi: 10.1101/2024.07.04.602042. bioRxiv. 2024. PMID: 39005444 Free PMC article. Preprint. - Accelerated hermaphrodite maturation on male pheromones suggests a general principle of coordination between larval behavior and development.
Faerberg DF, Aprison EZ, Ruvinsky I. Faerberg DF, et al. Development. 2024 Jul 1;151(13):dev202961. doi: 10.1242/dev.202961. Epub 2024 Jul 8. Development. 2024. PMID: 38975828 Free PMC article. - Bacterial vitamin B6 is required for post-embryonic development in C. elegans.
Feng M, Gao B, Ruiz D, Garcia LR, Sun Q. Feng M, et al. Commun Biol. 2024 Mar 26;7(1):367. doi: 10.1038/s42003-024-05992-2. Commun Biol. 2024. PMID: 38532074 Free PMC article.
References
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. - PubMed
- Chen ZF, Behringer RR. Twist is required in head mesenchyme for cranial tube morphogenesis. Genes & Dev. 1995;9:686–699. - PubMed
- Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. - PubMed
- Chitwood BG, Chitwood MB. Introduction to nematology. Baltimore, Maryland: University Park Press; 1974.
- Clark S, Chisholm AD, Horvitz HR. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 HD008331/HD/NICHD NIH HHS/United States
- R01GM37706/GM/NIGMS NIH HHS/United States
- T32GM07231/GM/NIGMS NIH HHS/United States
- R01GM37053/GM/NIGMS NIH HHS/United States
- R01 GM037706/GM/NIGMS NIH HHS/United States
- Z01 DK036117/ImNIH/Intramural NIH HHS/United States
- T32 GM007231/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials