Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites - PubMed (original) (raw)
Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites
R E Johnson et al. Genes Dev. 1998.
Abstract
Abasic (AP) sites arise in DNA through spontaneous base loss and enzymatic removal of damaged bases. APN1 encodes the major AP-endonuclease of Saccharomyces cerevisiae. Human HAP1 (REF1) encodes the major AP endonuclease which, in addition to its role in DNA repair, functions as a redox regulatory protein. We identify APN2, the yeast homolog of HAP1 and provide evidence that Apn1 and Apn2 represent alternate pathways for repairing AP sites. The apn1Delta apn2Delta strain displays a highly elevated level of MMS-induced mutagenesis, which is dependent on the REV3, REV7, and REV1 genes. Our findings indicate that AP sites are highly cytotoxic and mutagenic in eukaryotes, and that the REV3, REV7-encoded DNA polymerase zeta mediates the mutagenic bypass of AP sites.
Figures
Figure 1
Apn2 is a member of the Exo III/HAP1 family of proteins. (A) Alignment of human HAP1, S. cerevisiae Apn2, and E. coli Exo III (Xth) proteins. Identical and highly conserved residues are highlighted. Amino acid positions are indicated by numbers in parentheses. The conserved residues thought to be involved in catalytic activity (Glu-96, Asp-283, and His-309 in HAP1 protein) are indicated by arrows. The Cys-65 residue in HAP1 essential for the redox activity is circled and indicated by an asterisk. (B) Schematic alignment of Exo III-type nucleases from various organisms. Boxes represent primary amino acid sequence. Narrow regions indicate unique sequences, whereas larger, shaded, or stippled regions indicate regions of homology among the proteins. The different shades within the boxes indicate independent domains; spaces indicate gaps that were introduced to optimize alignment of the proteins. Protein lengths are indicated by numbers at right. Positions of the highly conserved amino acid sequences QE(T/L/I)K, R(L/I)D, and SDH(C/A)P are indicated. Positions of the redox cysteine residue in human HAP1, and of cysteine residues present at the corresponding position in Drosophila RRP and Arabidopsis thaliana ARP proteins are indicated by the letter C within the alignments.
Figure 1
Apn2 is a member of the Exo III/HAP1 family of proteins. (A) Alignment of human HAP1, S. cerevisiae Apn2, and E. coli Exo III (Xth) proteins. Identical and highly conserved residues are highlighted. Amino acid positions are indicated by numbers in parentheses. The conserved residues thought to be involved in catalytic activity (Glu-96, Asp-283, and His-309 in HAP1 protein) are indicated by arrows. The Cys-65 residue in HAP1 essential for the redox activity is circled and indicated by an asterisk. (B) Schematic alignment of Exo III-type nucleases from various organisms. Boxes represent primary amino acid sequence. Narrow regions indicate unique sequences, whereas larger, shaded, or stippled regions indicate regions of homology among the proteins. The different shades within the boxes indicate independent domains; spaces indicate gaps that were introduced to optimize alignment of the proteins. Protein lengths are indicated by numbers at right. Positions of the highly conserved amino acid sequences QE(T/L/I)K, R(L/I)D, and SDH(C/A)P are indicated. Positions of the redox cysteine residue in human HAP1, and of cysteine residues present at the corresponding position in Drosophila RRP and Arabidopsis thaliana ARP proteins are indicated by the letter C within the alignments.
Figure 2
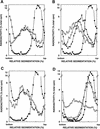
Enhanced MMS sensitivity and MMS mutagenesis in the apn1Δ apn2Δ strain. (A) MMS sensitivity of various yeast strains. Cells grown overnight in YPD medium were treated with MMS at the concentrations indicated for a 20-min period. Appropriate dilutions were spread onto YPD plates. Each curve represents the average of two to three experiments for each strain. (•) EMY74.7, wild type (APN1 APN2); (□) YRP190, apn1Δ; (▵) YRP263, apn2Δ; (○) YRP269, apn1Δ apn2Δ. (B) MMS-induced mutations at the CAN1 locus. Cells grown overnight in YPD medium were treated with MMS at the concentrations indicated for a 20-min period. Appropriate dilutions were spread onto YPD plates for viability determinations and onto synthetic complete medium lacking arginine and containing canavanine for the determination of _CAN1_S to _can1_r mutagenesis. Each curve represents the average of two to three experiments for each strain. (•) EMY74.7, wild type (APN1 APN2); (□) YRP190, apn1Δ; (▵) YRP263, apn2Δ; (○) YRP269, apn1Δ apn2Δ.
Figure 3
Alkaline sucrose gradient analysis of DNA from cells treated with 0.1% MMS for 20 min. (A) YRP276, wild type (APN1 APN2). (B) YRP210, apn1Δ. (C) YRP291, apn2Δ. (D) YRP292, apn1Δ apn2Δ. (○) Untreated cells; (•) cells treated with MMS for 20 min; (▴) cells treated with MMS for 20 min, and then given a 1-hr repair period; (▵) cells treated with MMS for 20 min, and then given a 2-hr repair period.
Figure 4
Requirement of REV genes for MMS-induced mutagenesis in the apn1Δ apn2Δ strains. Methods are as described in the legend to Fig. 2. (A) MMS sensitivity of yeast strains. (•) EMY74.7, wild type (APN1 APN2); (○) YRP269, apn1Δ apn2Δ; (▵) YREV1.15, rev1Δ; (█) YREV3.15, rev3Δ; (▾) YREV7.1, rev7Δ; (▴) YREV1.13, apn1Δ apn2Δ rev1Δ; (□) YREV3.40, apn1Δ apn2Δ rev3Δ; (▿) YREV7.4, apn1Δ apn2Δ rev7Δ. (B) MMS-induced mutations of _CAN1_S to _can1_r. (○) YRP269, apn1Δ apn2Δ; (▴) YREV1.13, apn1Δ apn2Δ rev1Δ; (□) YREV3.40, apn1Δ apn2Δ rev3Δ; (▿) YREV7.4, apn1Δ apn2Δ rev7Δ.
Similar articles
- Apurinic endonuclease activity of yeast Apn2 protein.
Unk I, Haracska L, Johnson RE, Prakash S, Prakash L. Unk I, et al. J Biol Chem. 2000 Jul 21;275(29):22427-34. doi: 10.1074/jbc.M002845200. J Biol Chem. 2000. PMID: 10806210 - Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast.
Torres-Ramos CA, Johnson RE, Prakash L, Prakash S. Torres-Ramos CA, et al. Mol Cell Biol. 2000 May;20(10):3522-8. doi: 10.1128/MCB.20.10.3522-3528.2000. Mol Cell Biol. 2000. PMID: 10779341 Free PMC article. - Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae.
Boiteux S, Guillet M. Boiteux S, et al. DNA Repair (Amst). 2004 Jan 5;3(1):1-12. doi: 10.1016/j.dnarep.2003.10.002. DNA Repair (Amst). 2004. PMID: 14697754 Review. - Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.
Mol CD, Hosfield DJ, Tainer JA. Mol CD, et al. Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
Cited by
- Kinetic Features of 3'-5'-Exonuclease Activity of Apurinic/Apyrimidinic Endonuclease Apn2 from Saccharomyces cerevisiae.
Kuznetsova AA, Gavrilova AA, Ishchenko AA, Saparbaev M, Fedorova OS, Kuznetsov NA. Kuznetsova AA, et al. Int J Mol Sci. 2022 Nov 19;23(22):14404. doi: 10.3390/ijms232214404. Int J Mol Sci. 2022. PMID: 36430884 Free PMC article. - iDamage: a method to integrate modified DNA into the yeast genome.
Masłowska KH, Laureti L, Pagès V. Masłowska KH, et al. Nucleic Acids Res. 2019 Nov 18;47(20):e124. doi: 10.1093/nar/gkz723. Nucleic Acids Res. 2019. PMID: 31418026 Free PMC article. - Mutagenicity of N3-methyladenine: a multi-translesion polymerase affair.
Monti P, Traverso I, Casolari L, Menichini P, Inga A, Ottaggio L, Russo D, Iyer P, Gold B, Fronza G. Monti P, et al. Mutat Res. 2010 Jan 5;683(1-2):50-6. doi: 10.1016/j.mrfmmm.2009.10.007. Mutat Res. 2010. PMID: 19874831 Free PMC article. - Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair.
Kim N, Jinks-Robertson S. Kim N, et al. Mol Cell Biol. 2010 Jul;30(13):3206-15. doi: 10.1128/MCB.00308-10. Epub 2010 Apr 26. Mol Cell Biol. 2010. PMID: 20421413 Free PMC article. - Single-Strand Break End Resection in Genome Integrity: Mechanism and Regulation by APE2.
Hossain MA, Lin Y, Yan S. Hossain MA, et al. Int J Mol Sci. 2018 Aug 14;19(8):2389. doi: 10.3390/ijms19082389. Int J Mol Sci. 2018. PMID: 30110897 Free PMC article. Review.
References
- Abate C, Patel L, Rauscher FJ, III, Curran T. Redox regulation of Fos and Jun DNA-binding activity in vitro. Science. 1990;249:1157–1161. - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Barzilay G, Mol CD, Robson CN, Walker LJ, Cunningham RP, Tainer JA, Hickson ID. Identification of critical active-site residues in the multifuncitonal human DNA repair enzyme HAP1. Nat Struct Biol. 1995;2:561–568. - PubMed
- Bjoras M, Klungland A, Johansen RF, Seeberg E. Purification and properties of the alkylation repair DNA glycosylase encoded MAG gene from Saccharomyces cerevisiae. Biochemistry. 1995;34:4577–4582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM19261/GM/NIGMS NIH HHS/United States
- CA53791/CA/NCI NIH HHS/United States
- CA41261/CA/NCI NIH HHS/United States
- R37 CA041261/CA/NCI NIH HHS/United States
- R01 CA053791/CA/NCI NIH HHS/United States
- R01 CA041261/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous