Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses - PubMed (original) (raw)
Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses
J R Xu et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The rice blast fungus, Magnaporthe grisea, generates enormous turgor pressure within a specialized cell called the appressorium to breach the surface of host plant cells. Here, we show that a mitogen-activated protein kinase, Mps1, is essential for appressorium penetration. Mps1 is 85% similar to yeast Slt2 mitogen-activated protein kinase and can rescue the thermosensitive growth of slt2 null mutants. The mps1-1Delta mutants of M. grisea have some phenotypes in common with slt2 mutants of yeast, including sensitivity to cell-wall-digesting enzymes, but display additional phenotypes, including reduced sporulation and fertility. Interestingly, mps1-1Delta mutants are completely nonpathogenic because of the inability of appressoria to penetrate plant cell surfaces, suggesting that penetration requires remodeling of the appressorium wall through an Mps1-dependent signaling pathway. Although mps1-1Delta mutants are unable to cause disease, they are able to trigger early plant-cell defense responses, including the accumulation of autofluorescent compounds and the rearrangement of the actin cytoskeleton. We conclude that MPS1 is essential for pathogen penetration; however, penetration is not required for induction of some plant defense responses.
Figures
Figure 1
Structure, function, and relatedness of MPS1. (A) Sequence alignment of Mps1 with S. cerevisiae Slt2 and S. pombe Spm1. Identical residues are shown in black boxes and similar residues are shaded. The 11 protein-kinase subdomains are labeled with roman numerals. (B) Relatedness of Mpk1 to yeast MAPKs. GenBank accession nos. used for yeast MAPKs are StyI (X89262), Hog1 (L06279), Spm1 (U65405), Slt2 (X59262), Spk1 (X57334), Kss1 (M26398), Fus3 (M31132), and Smk1 (L35047). Accession nos. for M. grisea MAPKs are Mps1 (AF020316) and Pmk1 (U70134). A third MAPK from M. grisea, Pmk2, is most closely related to yeast Hog1 and StyI kinases (J.-R.X. and N. Talbot, unpublished work). The phylogram was prepared with the
growtree
program from the Genetics Computer Group’s software package (Madison, WI). (C) Complementation of a yeast slt2 mutant. S. cerevisiae strains numbered 2–6 are DL456 (slt2_Δ, ura3_-52); strain 1 is CG219 (ura3–52 SLT2); strains numbered 2, 3, and 4 contain the plasmid pNX9 (MPS1 controlled by the GAL1 promoter in pYES2); strains 5 and 6 contain the vector pYES2. The upper plate (1% yeast extract/1% peptone/1% dextrose) was incubated at 30°C and replicated to 1% yeast extract/1% peptone/1% galactose/1% raffinose (lower right) or 1% yeast extract/1% peptone/1% dextrose (lower left) plates that were incubated at 37°C before being photographed.
Figure 2
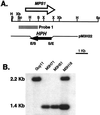
MPS1 gene-replacement vector and transformants. (A) Physical map of the MPS1 genomic region and the gene-replacement vector pM3H22. The restriction enzymes are E, _Eco_RI; H, _Hin_dIII; S, _Sal_I; Sc, _Sac_I; X, _Xho_I; Xb, _Xba_I. (B) Southern blot of wild-type strain Guy11, ectopic-integration transformant M3H18, and MPS1 gene-replacement transformants M3H51 and M3H71. All DNA samples were digested with _Sal_I. The blot was probed with the 1.6-kb _Sac_I–_Sal_I fragment. Guy11 and transformant M3H18 contained the MPS1 2.2-kb _Sal_I fragment. M3H18 also contained a 1.4-kb _Sal_I fragment, indicative of an ectopic insertion event. Transformants M3H51 and M3H71 do not contain the 2.2-kb _Sal_I fragment but have the 1.4-kb _Sal_I fragment, indicative of a gene replacement.
Figure 3
_mps1-1_Δ mutants have defects in cell-wall integrity. (A) Growth of _mps1–1_Δ strains on complete media without 1 M sorbitol (plate 1); growth of MPS1 strains on complete media without 1 M sorbitol (plate 2); growth of _mps1–1_Δ strains on complete media with 1 M sorbitol (plate 3); and growth of MPS1 strains on complete media with 1 M sorbitol (plate 4). _mps1–1_Δ mutants sporulate poorly and undergo progressive autolysis in the absence of osmotic stabilization. Radial growth rates are identical to those of wild-type strains. (B) Sensitivity of _mps1–1_Δ strains to cell-wall-digesting enzymes. Wild-type (Upper) or _mps1–1_Δ (Lower) strains were photographed 15 min after treatment with Novozyme 234. The _mps1–1_Δ mutants are reduced to spheroplasts with very short enzyme treatment. (×230.)
Figure 4
Infection assays. (Upper) Typical infected leaf areas from 14-day-old rice seedlings spray-inoculated with either wild-type (WT) or _mps1–1_Δ (mps1) strains. (Lower) Typical infected areas from rice leaves inoculated by leaf sheath injection of either wild-type (WT) or _mps1–1_Δ (mps1) strains. A wounded control (CON) inoculated with water is also shown. Small areas of fungal growth occur near the injection site in _mps1–1_Δ strains.
Figure 5
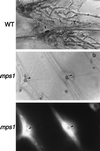
Fungal penetration assays on plant epidermal cells. (Top) Wild-type strains (WT) form appressoria (arrow) that penetrate and produce abundant infectious hyphae. (Middle and Bottom) Strains containing the _mps1–1_Δ allele (mps1) produce numerous appressoria (arrows) but do not penetrate or form infectious hyphae. Lower shows fluorescence (465 nm) illumination of the same area depicted in Middle. Plant-cell-derived autofluorescence surrounds nonpenetrating appressoria. (×465.)
Figure 6
Actin rearrangements in response to _mps1–1_Δ mutants. Onion epidermal cells were left untreated (A), inoculated with wild-type conidia (B and C), or inoculated with _mps1–1_Δ mutants (D and E). Plant cells were visualized with phase-contrast microscopy (B and D) and with epifluorescence (C and E) after staining with rhodamine-phalloidin. Arrows indicate appressoria (B and D) and underlying actin rearrangements (C and E). (×280.)
Similar articles
- The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium.
Kojima K, Kikuchi T, Takano Y, Oshiro E, Okuno T. Kojima K, et al. Mol Plant Microbe Interact. 2002 Dec;15(12):1268-76. doi: 10.1094/MPMI.2002.15.12.1268. Mol Plant Microbe Interact. 2002. PMID: 12481999 - Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea.
Dixon KP, Xu JR, Smirnoff N, Talbot NJ. Dixon KP, et al. Plant Cell. 1999 Oct;11(10):2045-58. doi: 10.1105/tpc.11.10.2045. Plant Cell. 1999. PMID: 10521531 Free PMC article. - Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea.
Li L, Xue C, Bruno K, Nishimura M, Xu JR. Li L, et al. Mol Plant Microbe Interact. 2004 May;17(5):547-56. doi: 10.1094/MPMI.2004.17.5.547. Mol Plant Microbe Interact. 2004. PMID: 15141959 - The role of glycerol in the pathogenic lifestyle of the rice blast fungus Magnaporthe oryzae.
Foster AJ, Ryder LS, Kershaw MJ, Talbot NJ. Foster AJ, et al. Environ Microbiol. 2017 Mar;19(3):1008-1016. doi: 10.1111/1462-2920.13688. Epub 2017 Mar 1. Environ Microbiol. 2017. PMID: 28165657 Review. - Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea.
Howard RJ, Valent B. Howard RJ, et al. Annu Rev Microbiol. 1996;50:491-512. doi: 10.1146/annurev.micro.50.1.491. Annu Rev Microbiol. 1996. PMID: 8905089 Review.
Cited by
- Cucumber hypocotyls respond to cutin monomers via both an inducible and a constitutive H(2)O(2)-generating system.
Kauss H, Fauth M, Merten A, Jeblick W. Kauss H, et al. Plant Physiol. 1999 Aug;120(4):1175-82. doi: 10.1104/pp.120.4.1175. Plant Physiol. 1999. PMID: 10444101 Free PMC article. - PDE1 encodes a P-type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea.
Balhadère PV, Talbot NJ. Balhadère PV, et al. Plant Cell. 2001 Sep;13(9):1987-2004. doi: 10.1105/tpc.010056. Plant Cell. 2001. PMID: 11549759 Free PMC article. - Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi.
Kong Z, Zhang X, Zhou F, Tang L, Chen Y, Li S, Zhang X, Kuai L, Su W, Cui W, Cai J, Wang Y, Yang J, Peng YL, Wang D, Liu J. Kong Z, et al. mBio. 2023 Apr 25;14(2):e0288322. doi: 10.1128/mbio.02883-22. Epub 2023 Feb 13. mBio. 2023. PMID: 36779710 Free PMC article. - MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae.
He M, Xu Y, Chen J, Luo Y, Lv Y, Su J, Kershaw MJ, Li W, Wang J, Yin J, Zhu X, Liu X, Chern M, Ma B, Wang J, Qin P, Chen W, Wang Y, Wang W, Ren Z, Wu X, Li P, Li S, Peng Y, Lin F, Talbot NJ, Chen X. He M, et al. Autophagy. 2018;14(9):1543-1561. doi: 10.1080/15548627.2018.1458171. Epub 2018 Aug 31. Autophagy. 2018. PMID: 29929416 Free PMC article. - Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa.
Park G, Pan S, Borkovich KA. Park G, et al. Eukaryot Cell. 2008 Dec;7(12):2113-22. doi: 10.1128/EC.00466-07. Epub 2008 Oct 10. Eukaryot Cell. 2008. PMID: 18849472 Free PMC article.
References
- Emmett R W, Parbery D G. Annu Rev Phytopathol. 1975;13:147–167.
- Ou S H. Rice Diseases. Surrey, U.K.: Commonwealth Mycological Institute; 1985.
- Howard R J, Bourett T M, Ferrari M A. In: Infection by Magnaporthe grisea: An in Vitro Analysis. Mendgen K, Lesemann D E, editors. Berlin: Springer; 1991. pp. 251–264.
- Howard R J. In: Cell Biology of Pathogenesis. Zeigler R S, Leong S A, Teng P S, editors. Wallingford, U.K.: CAB International; 1994. pp. 3–22.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials