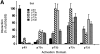Two new p73 splice variants, gamma and delta, with different transcriptional activity - PubMed (original) (raw)
Two new p73 splice variants, gamma and delta, with different transcriptional activity
V De Laurenzi et al. J Exp Med. 1998.
Abstract
p73 has been recently identified as a new structural and functional homologue of the transcription factor p53. It is expressed in either a full-length form, alpha, or a shorter beta mRNA variant, with exon 13 spliced out. Here we report the identification and functional characterization of two new p73 splicing variants, gamma (splicing out exon 11) and delta (splicing out exons 11, 12, and 13). Both gamma and delta p73 variants are expressed in human peripheral blood lymphocytes, primary keratinocytes, and different tumor cell lines, including neuroblastoma, glioblastoma, melanoma, hepatoma, and leukemia. The expression pattern of the four p73 splicing variants differs in both primary cells of different lineage and established cell lines even within the same type of tumor. A two-hybrid assay was used to characterize the homodimeric and heterodimeric interactions between the p73 variants, and showed that neither p73gamma nor p73delta interact with p53, whereas p73gamma showed strong interactions with all p73 isoforms, and p73delta binds efficiently p73alpha and p73gamma but only weakly p73beta. At the functional level, p73gamma is significantly less efficient in activating transcription of the p21(Waf1/Cip1) promoter than p53 or p73beta, whereas the effect of p73delta is intermediate and comparable to that of p73alpha. The ability of the different p73 variants to affect cell growth in p53 null osteosarcoma SAOS-2 cells correlates with their transcriptional activity on the p21(Waf1/Cip1) promoter: p73beta is the most efficient in inhibiting colony formation, whereas p73gamma is almost ineffective. Our results suggest that p73 isoforms may be differentially regulated, with four different isoforms capable of interacting among themselves and with p53. The relative expression level of each splice variant may modulate p73 transcriptional and growth suppression activities by affecting heterodimer formation.
Figures
Figure 1
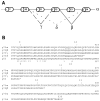
(A) Schematic representation of p73 splicing isoforms. Spliced-out exons are indicated with respect to the full-length form, p73α. (B) Amino acid alignment of the COOH-terminal region of the four p73 splice variants and comparison with p53. The corresponding exons are indicated.
Figure 2
(A) Differential expression of p73 splicing variants and p53 in normal human (PBLs and skin keratinocytes) and cancer cells. Autoradiography of a radioactive PCR performed on several neuroblastoma [BE(2)-M17, SH-Sy5y, BE(2)-2C, SH-EPI, SK-N-BE(2), SMS-KCNR, CHP100, SH-N-SH, and LAN5], glioblastoma (Lipari), lymphoma (Hut 78, Jurkat), hepatoma (Hep-G2), and breast carcinoma (MCF-7) cell lines. The four p73 splice variants (p73α: 535 bp; p73β: 440 bp; p73γ: 386 bp, and p73δ: 154 bp) and the expression of a control housekeeping gene (GAPDH, bottom lane) are shown. (B) Radioactive PCR for p73, p53, and GAPDH performed on mRNA extracted from the BE(2)-M17 cell line for 25, 30, and 35 cycles of amplification, showing the linearity of the reaction. (C) p73 and p53 expression pattern of CD4+ and CD8+ enriched lymphocyte populations sorted by flow cytometry (see Materials and Methods for details on oligonucleotides and PCR cycles). Radioactive PCR gels shown in the three panels were exposed for 12 h; GAPDH was exposed for only 2 h.
Figure 3
(A) Yeast two-hybrid interaction assays among p53, p73α, p73β, p73γ, and p73δ presented as ordinate values relative to β-galactosidase activity of p53–p53 interaction. Control interactions using pGAD or pGBT vectors were negative. (B) Luciferase assay of SAOS-2 cells transfected with p21Waf1/Cip1 promoter–driven luciferase reporter plasmid (4 μg), together with p53 (20 ng) or p73 (20 ng) expression plasmids. When transfected in combination with each other, 10 ng of each indicated expression plasmid was used. (C) Expression of p73 splicing variants. Anti-HA Western blot of whole cell extracts after transfection of SAOS-2 cells with plasmids encoding the indicated proteins. Molecular masses are shown in kD (right).
Figure 3
(A) Yeast two-hybrid interaction assays among p53, p73α, p73β, p73γ, and p73δ presented as ordinate values relative to β-galactosidase activity of p53–p53 interaction. Control interactions using pGAD or pGBT vectors were negative. (B) Luciferase assay of SAOS-2 cells transfected with p21Waf1/Cip1 promoter–driven luciferase reporter plasmid (4 μg), together with p53 (20 ng) or p73 (20 ng) expression plasmids. When transfected in combination with each other, 10 ng of each indicated expression plasmid was used. (C) Expression of p73 splicing variants. Anti-HA Western blot of whole cell extracts after transfection of SAOS-2 cells with plasmids encoding the indicated proteins. Molecular masses are shown in kD (right).
Figure 3
(A) Yeast two-hybrid interaction assays among p53, p73α, p73β, p73γ, and p73δ presented as ordinate values relative to β-galactosidase activity of p53–p53 interaction. Control interactions using pGAD or pGBT vectors were negative. (B) Luciferase assay of SAOS-2 cells transfected with p21Waf1/Cip1 promoter–driven luciferase reporter plasmid (4 μg), together with p53 (20 ng) or p73 (20 ng) expression plasmids. When transfected in combination with each other, 10 ng of each indicated expression plasmid was used. (C) Expression of p73 splicing variants. Anti-HA Western blot of whole cell extracts after transfection of SAOS-2 cells with plasmids encoding the indicated proteins. Molecular masses are shown in kD (right).
Figure 4
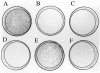
Colony formation assay in SAOS-2 cells grown for 2 wk under G418 selection after transfection with 20 μg of pcDNA3 vector expressing neor alone (A) or with p53 (B), p73α (C), p73β (D), p73γ (E), and p73δ (F). One of three different experiments is shown; the mean colony count (± SEM) of the three experiments was as follows: control, 1,563 ± 120 (A); p53, 74 ± 20 (B); p73α, 95 ± 32 (C); p73β, 26 ± 12 (D); p73γ, 754 ± 149 (E); p73δ, 88 ± 25 (F).
Similar articles
- p73gamma transactivates the p21 promoter through preferential interaction with the p300/CBP-associated factor in human prostate cancer cells.
Momii Y, Izumi H, Shiota M, Onitsuka T, Abe T, Kobayashi H, Miyamoto N, Uchiumi T, Kohno K. Momii Y, et al. Oncol Rep. 2007 Aug;18(2):411-6. Oncol Rep. 2007. PMID: 17611664 - New p73 variants with altered C-terminal structures have varied transcriptional activities.
Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. Ueda Y, et al. Oncogene. 1999 Sep 2;18(35):4993-8. doi: 10.1038/sj.onc.1202817. Oncogene. 1999. PMID: 10490834 - Overexpression of the wild type p73 gene in breast cancer tissues and cell lines.
Zaika AI, Kovalev S, Marchenko ND, Moll UM. Zaika AI, et al. Cancer Res. 1999 Jul 1;59(13):3257-63. Cancer Res. 1999. PMID: 10397274 - Structure, function and regulation of p63 and p73.
Levrero M, De Laurenzi V, Costanzo A, Gong J, Melino G, Wang JY. Levrero M, et al. Cell Death Differ. 1999 Dec;6(12):1146-53. doi: 10.1038/sj.cdd.4400624. Cell Death Differ. 1999. PMID: 10637429 Review. - p73.
Davis PK, Dowdy SF. Davis PK, et al. Int J Biochem Cell Biol. 2001 Oct;33(10):935-9. doi: 10.1016/s1357-2725(01)00073-5. Int J Biochem Cell Biol. 2001. PMID: 11470228 Review.
Cited by
- Human DMTF1β antagonizes DMTF1α regulation of the p14(ARF) tumor suppressor and promotes cellular proliferation.
Tschan MP, Federzoni EA, Haimovici A, Britschgi C, Moser BA, Jin J, Reddy VA, Sheeter DA, Fischer KM, Sun P, Torbett BE. Tschan MP, et al. Biochim Biophys Acta. 2015 Sep;1849(9):1198-208. doi: 10.1016/j.bbagrm.2015.07.009. Epub 2015 Jul 15. Biochim Biophys Acta. 2015. PMID: 26187004 Free PMC article. - PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73.
Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A, Neale MH, Martin SJ, Latchman DS, Knight RA, Melino G, De Laurenzi V. Munarriz E, et al. Mol Cell Biol. 2004 Dec;24(24):10593-610. doi: 10.1128/MCB.24.24.10593-10610.2004. Mol Cell Biol. 2004. PMID: 15572666 Free PMC article. - A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain.
Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. Gaiddon C, et al. Mol Cell Biol. 2001 Mar;21(5):1874-87. doi: 10.1128/MCB.21.5.1874-1887.2001. Mol Cell Biol. 2001. PMID: 11238924 Free PMC article. - Ubiquitin-dependent degradation of p73 is inhibited by PML.
Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, Pandolfi PP. Bernassola F, et al. J Exp Med. 2004 Jun 7;199(11):1545-57. doi: 10.1084/jem.20031943. J Exp Med. 2004. PMID: 15184504 Free PMC article. - Physical interaction of tumour suppressor p53/p73 with CCAAT-binding transcription factor 2 (CTF2) and differential regulation of human high-mobility group 1 (HMG1) gene expression.
Uramoto H, Izumi H, Nagatani G, Ohmori H, Nagasue N, Ise T, Yoshida T, Yasumoto K, Kohno K. Uramoto H, et al. Biochem J. 2003 Apr 15;371(Pt 2):301-10. doi: 10.1042/BJ20021646. Biochem J. 2003. PMID: 12534345 Free PMC article.
References
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. - PubMed
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. - PubMed
- Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708. - PubMed
- Hollstein M, Sidransky D, Vogelstein CC. p53 mutations in human cancers. Science. 1991;253:49–53. - PubMed
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J, Valent A, Minty A, Chalon P, Lelias J, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous