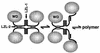A role for Groucho tetramerization in transcriptional repression - PubMed (original) (raw)
A role for Groucho tetramerization in transcriptional repression
G Chen et al. Mol Cell Biol. 1998 Dec.
Abstract
The Drosophila Groucho (Gro) protein is a corepressor required by a number of DNA-binding transcriptional repressors. Comparison of Gro with its homologues in other eukaryotic organisms reveals that Gro contains, in addition to a conserved C-terminal WD repeat domain, a conserved N-terminal domain, which has previously been implicated in transcriptional repression. We determined, via a variety of hydrodynamic measurements as well as protein cross-linking, that native Gro is a tetramer in solution and that tetramerization is mediated by two putative amphipathic alpha-helices (termed leucine zipper-like motifs) found in the N-terminal region. Point mutations in the leucine zipper-like motifs that block tetramerization also block repression by Gro, as assayed in cultured Drosophila cells with Gal4-Gro fusion proteins. Furthermore, the heterologous tetramerization domain from p53 fully substitutes for the Gro tetramerization domain in transcriptional repression. These findings suggest that oligomerization is essential for Gro-mediated repression and that the primary function of the conserved N-terminal domain is to mediate this oligomerization.
Figures
FIG. 1
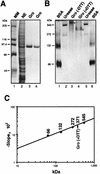
Determination of the molecular mass of native Gro. (A) Expression and purification of FLAG-tagged Gro. A Coomassie blue-stained SDS–8% polyacrylamide gel is shown. Lane 1, protein mass markers (MM) with sizes indicated on the left; lane 2, 10 μl of nuclear extract (NE) prepared from Sf9 insect cells infected with the baculovirus expressing FLAG-tagged Gro; lane 3, 1.0 μg of purified Gro; lane 4, 0.5 μg of purified Gro. (B) ND-PAGE analysis of Gro. A Coomassie blue-stained ND–8% polyacrylamide gel is shown. Lanes 1 and 6, bovine serum albumin (66-kDa monomer and 132-kDa dimer); lanes 2 and 5, urease (272-kDa trimer and 545-kDa hexamer); lanes 3 and 4, 1.0 μg of purified Gro (marked by asterisks) without (−DTT) or with (+DTT) treatment with 40 mM DTT prior to electrophoresis. (C) Molecular mass determination. The calibration curve prepared from Ferguson plots is shown (see text for details). The known masses of standards and the calculated mass of native Gro are given in kilodaltons above the curve.
FIG. 2
Analysis of Gro quaternary structure by gel filtration, cross-linking, and velocity sedimentation. (A) Superdex 200 gel filtration of DTT-treated Gro. A silver-stained SDS-PAGE gel is shown with gel filtration fraction numbers labeled at the top of each lane. Fractions with peak levels of native protein standards that were run in a parallel gel filtration experiment are indicated above the gel: thyroglobulin, 669 kDa (Stokes radius, 88 Å); ferritin, 440 kDa (62 Å); catalase, 232 kDa (52.2 Å); aldolase, 158 kDa (48 Å); and bovine serum albumin, 67 kDa (35 Å). The position of the Gro peak is indicated by an arrow. (B) Protein cross-linking analysis of Gro. Equal amounts of Gro plus DTT were incubated with various concentrations of glutaraldehyde (indicated above each lane). Cross-linked Gro products were resolved by SDS-PAGE and visualized by Coomassie blue staining. The Gro multimers resulting from cross-linking with sizes corresponding to Gro dimer, trimer, and tetramer are marked with asterisks. (C and D) Sucrose gradient (5 to 20%) centrifugation of non-cross-linked (C) or cross-linked (D) Gro. Silver-stained SDS-PAGE gels to analyze gradient fractions for the presence of Gro are shown, with fraction numbers indicated at the top of each lane. The peak positions of native protein standards centrifuged through parallel gradients are indicated above the gel: ferritin (440 kDa), catalase (232 kDa), and bovine serum albumin (67 kDa). The positions of Gro or Gro incubated with 0.05% glutaraldehyde (GroCL) prior to centrifugation are indicated.
FIG. 3
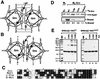
Mapping the tetramerization domain of Gro. (A) Schematic diagram of various full-length or truncated [35S]methionine-labeled Gro proteins that were produced by in vitro translation. The conserved N-terminal glutamine-rich domain (Q) and C-terminal WD repeat domain (WD) are indicated. GP and SP, glycine-proline and serine-proline, respectively, which are predominant in these regions; CcN, CcN motif containing putative cdc2 and casein kinase II phosphorylation sites as well as a nuclear localization signal (44); aa, amino acids. (B) In vitro coimmunoprecipitation assays. Purified FLAG-tagged Gro (1 μg) (M2-Gro) immobilized on anti-FLAG affinity resin was incubated with 10 μl of each of the 35S-labeled Gro variants. Lanes 1 to 10 show an amount of each input protein equal to 20% of the amount used in the binding reactions shown in lanes 11 to 20. After being extensively washed, the bound 35S-Gro was eluted with SDS-PAGE sample buffer, resolved by SDS-PAGE, and visualized by autoradiography (lanes 12 to 20). As a negative control, the anti-FLAG affinity bead (M2) alone was examined for interaction with full-length 35S-Gro (11). (C) Cross-linking analysis of truncated 35S-Gro proteins. Cross-linking reactions conducted as described in the legend to Fig. 2B were analyzed by SDS-PAGE and autoradiography. The left and right panels show the cross-linking profiles of 35S-labeled Gro(1–194) and Gro(134–719), respectively. The percent glutaraldehyde (G%) used in the cross-linking reactions is shown above the lanes. The 35S-Gro monomers are indicated by lines, and cross-linked dimer, trimer, and tetramer species are indicated by arrowheads. The sizes of prestained protein markers are indicated on the left. (D) Yeast two-hybrid analysis of Gro oligomerization. The top panels show three independent yeast colonies cotransformed with the LexA DNA binding domain-Gro fusion proteins (LexAGro, indicated on the left of each panel) and GAD-Gro fusion proteins (GADGro, indicated on the top of each panel). The bottom panels show the results of growing these colonies in the presence of the chromogenic β-galactosidase substrate X-Gal. Cells expressing β-galactosidase turn blue.
FIG. 4
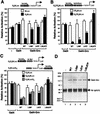
Sequence analysis and mutagenesis of the tetramerization domain. (A to C) Sequence analyses of the N-terminal tetramerization region of Gro. (A and B) Helical-wheel projections of the two putative LZL segments (residues 24 to 52 and 73 to 100) found in this region. Heptad positions are labeled by letters a through g in the wheels, where hydrophobic residues at positions a and d constitute the cores of proposed Gro homodimeric coiled coils. Potential interhelical salt bridges between charged residues at positions e and g are indicated by dashed lines. The two leucine residues (L38 and L87) mutated to prolines are boxed. (C) Sequence alignment of Gro LZL motif (residues 20 to 54) with leucine zippers found in the proto-oncogene products Maf-1 (9), c-Fos (47), and N-Myc (14). Identical residues are shaded in black and conserved residues are boxed. The hydrophobic residue at the d position of each heptad repeat is labeled with an asterisk. (D) The double point mutation (L38,87P) abolishes Gro oligomerization in vitro. With the coimmunoprecipitation assays described in the legend to Fig. 3B, wild-type (WT) and mutant forms of full-length 35S-Gro (indicated at the top of the gel) were examined for interaction with purified FLAG-tagged Gro (M2-Gro). The upper panel shows 10% of the input 35S-Gro proteins used for assays and the lower panel indicates 35S-Gro retained on the anti-FLAG M2 beads alone or beads with purified M2-Gro. The percentage of input protein bound is indicated on the bottom of each lane. (E) Cross-linking analyses of wild-type (WT) and mutant Gro tetramerization domains. Wild-type and mutant forms of the six-histidine-tagged N-terminal region (residues 2 to 194) of Gro (6HGro) were purified and subjected to protein cross-linking assays as described in the legend to Fig. 2B. Cross-linked products were analyzed by SDS-PAGE and Coomassie blue staining. The cross-linking patterns of wild-type and L38,87P mutant forms of 6HGro are shown. The asterisks indicate the dimer, trimer, and tetramer species.
FIG. 5
LZL motifs are essential for Gro-mediated repression in Drosophila SL2 cells. (A) Gal4-GroL38,87P fails to repress basal transcription. Expression constructs encoding the Gal4-DNA binding domain (residues 1 to 147) fused to wild-type or mutant forms of Gro were cotransfected into SL2 cells with one of the firefly luciferase reporters (either tkLuc or G5tkLuc) and an internal control reporter (pRL-TK) encoding Renilla luciferase. All firefly luciferase activities (measured by Promega’s dual-luciferase assay system and further normalized to Renilla luciferase activities) driven by Gal4-Gro fusion proteins were normalized to that mediated by Gal4(1–147) alone, which was set at 100%. Each bar represents the average + standard deviation of three independent duplicate experiments. (B) Gal4-GroL38,87P fails to repress transcription activated by the combination of Dorsal and Twist. SL2 cells were transfected with one of the luciferase reporters (DE5tkLuc or G5DE5tkLuc) and DNA constructs expressing Dorsal, Twist, and one of the indicated Gal4-Gro fusion proteins. All activities were normalized to the activity observed in the presence of Dorsal, Twist, and Gal4(1–147), which was set at 100%. (C) Gal4-GroL38,87P fails to repress transcription activated by Sp1. SL2 cells were transfected with one of the luciferase reporters (S4tkLuc, G5S4tkLuc, or S4tkLucG5) and with expression constructs encoding Sp1 and one of the indicated Gal4-Gro fusion proteins. All activities were normalized to the activity observed in the presence of Sp1 and Gal4(1–147), which was set at 100%. (D) Immunoblot analysis of the expression level of Gal4-Gro fusion proteins in the transient-transfection experiments described above. Gal4-Gro fusion proteins were first precipitated from total cell lysates with the anti-Gal4 DNA binding domain antibody, and the immunoprecipitates were then resolved by SDS-PAGE and immunoblotted with antibody against the Gal4 DNA binding domain. The positions of the Gal4-Gro fusion proteins and of the immunoglobulin G heavy chain [Ab IgG(H)] are indicated.
FIG. 6
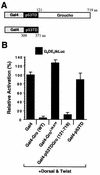
Functional analysis of the Gro tetramerization domain. (A) Schematic diagram of the p53 tetramerization domain (p53TD)-containing transcription factor chimeras used in this experiment. p53TD is the region of p53 from residue 309 to residue 371. aa, amino acids. (B) The Gro and p53 tetramerization domains are functionally interchangeable for repression. Transient-transfection assays were conducted essentially as described in legend to Fig. 5B with the reporter G5DE5tkLuc and expression vectors expressing Dorsal, Twist, and each indicated Gal4 fusion protein. WT, wild type.
FIG. 7
Speculative model for Gro tetramerization and Gro polymerization. Since leucine zippers are dimerization motifs, we postulate that the LZL motifs are dimerization motifs. The polymerization of Gro could be facilitated by the same interactions that promote tetramerization. In accord with this idea, we have modeled the Gro tetramer as a dimer of dimers (left). Breaking one of the coiled-coil interactions holding the tetramer together (middle) would expose hydrophobic surfaces that could be used in the further oligomerization (right) of Gro via contacts with the similar hydrophobic surfaces on another similarly disrupted tetramer. The postulated process is related to the phenomenon of domain swapping (3).
Similar articles
- Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression.
Flores-Saaib RD, Courey AJ. Flores-Saaib RD, et al. Nucleic Acids Res. 2000 Nov 1;28(21):4189-96. doi: 10.1093/nar/28.21.4189. Nucleic Acids Res. 2000. PMID: 11058116 Free PMC article. - Groucho/TLE family proteins and transcriptional repression.
Chen G, Courey AJ. Chen G, et al. Gene. 2000 May 16;249(1-2):1-16. doi: 10.1016/s0378-1119(00)00161-x. Gene. 2000. PMID: 10831834 Review. - Groucho oligomerization is required for repression in vivo.
Song H, Hasson P, Paroush Z, Courey AJ. Song H, et al. Mol Cell Biol. 2004 May;24(10):4341-50. doi: 10.1128/MCB.24.10.4341-4350.2004. Mol Cell Biol. 2004. PMID: 15121853 Free PMC article. - Mad proteins contain a dominant transcription repression domain.
Ayer DE, Laherty CD, Lawrence QA, Armstrong AP, Eisenman RN. Ayer DE, et al. Mol Cell Biol. 1996 Oct;16(10):5772-81. doi: 10.1128/MCB.16.10.5772. Mol Cell Biol. 1996. PMID: 8816491 Free PMC article.
Cited by
- Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression.
Flores-Saaib RD, Courey AJ. Flores-Saaib RD, et al. Nucleic Acids Res. 2000 Nov 1;28(21):4189-96. doi: 10.1093/nar/28.21.4189. Nucleic Acids Res. 2000. PMID: 11058116 Free PMC article. - A new Groucho TLE4 protein may regulate the repressive activity of Pax5 in human B lymphocytes.
Milili M, Gauthier L, Veran J, Mattei MG, Schiff C. Milili M, et al. Immunology. 2002 Aug;106(4):447-55. doi: 10.1046/j.1365-2567.2002.01456.x. Immunology. 2002. PMID: 12153506 Free PMC article. - The Wnt Transcriptional Switch: TLE Removal or Inactivation?
Ramakrishnan AB, Sinha A, Fan VB, Cadigan KM. Ramakrishnan AB, et al. Bioessays. 2018 Feb;40(2):10.1002/bies.201700162. doi: 10.1002/bies.201700162. Epub 2017 Dec 18. Bioessays. 2018. PMID: 29250807 Free PMC article. Review. - Functional dissection of the global repressor Tup1 in yeast: dominant role of the C-terminal repression domain.
Zhang Z, Varanasi U, Trumbly RJ. Zhang Z, et al. Genetics. 2002 Jul;161(3):957-69. doi: 10.1093/genetics/161.3.957. Genetics. 2002. PMID: 12136003 Free PMC article. - Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis.
Dasen JS, Martinez Barbera JP, Herman TS, Connell SO, Olson L, Ju B, Tollkuhn J, Baek SH, Rose DW, Rosenfeld MG. Dasen JS, et al. Genes Dev. 2001 Dec 1;15(23):3193-207. doi: 10.1101/gad.932601. Genes Dev. 2001. PMID: 11731482 Free PMC article.
References
- Akimaru H, Hou D X, Ishii S. Drosophila CBP is required for dorsal-dependent twist gene expression. Nat Genet. 1997;17:211–214. - PubMed
- Bryan J K. Molecular weight of protein multimers from polyacrylamide gel electrophoresis. Anal Biochem. 1977;78:513–519. - PubMed
- Campos-Ortega J A. Mechanisms of early neurogenesis in Drosophila melanogaster. J Neurobiol. 1993;24:1305–1327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous