Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair - PubMed (original) (raw)
Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair
B J Hwang et al. Proc Natl Acad Sci U S A. 1999.
Abstract
In human cells, efficient global genomic repair of DNA damage induced by ultraviolet radiation requires the p53 tumor suppressor, but the mechanism has been unclear. The p48 gene is required for expression of an ultraviolet radiation-damaged DNA binding activity and is disrupted by mutations in the subset of xeroderma pigmentosum group E cells that lack this activity. Here, we show that p48 mRNA levels strongly depend on basal p53 expression and increase further after DNA damage in a p53-dependent manner. Furthermore, like p53(-/-) cells, xeroderma pigmentosum group E cells are deficient in global genomic repair. These results identify p48 as the link between p53 and the nucleotide excision repair apparatus.
Figures
Figure 1
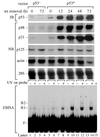
UV induces p48 transcription by a p53-dependent pathway. Primary human fibroblast cell lines, either p53+/+ (WI38) or p53−/− (041 mut), were treated with 10 J/m2 UV radiation and were harvested after different time intervals. The level of p53 protein was measured by immunoblot (IB) of protein extract. The levels of p48, p21, p125, actin mRNA, and 28S rRNA were measured by Northern blot (NB) of total RNA. The levels of 28S rRNA confirmed equal transfer of total RNA to the membrane.
Figure 2
IR induces p48 transcription by a p53-dependent pathway. Cells were exposed to 2 Gy of IR and were analyzed 0, 1, 2, 4, and 8 h later for levels of p53 protein, p48, p21, p125, actin mRNA, and 28S rRNA.
Figure 3
Expression of p53 induces p48 transcription and the activity of UV-damaged DNA binding protein. We used p53−/− (041 mut) cells that had been stably transfected with a control vector not containing p53 (p53−) or with a vector containing wild-type p53 cDNA under the control of a tetracycline regulated promoter (p53+) (2). Cells were analyzed for levels of p53 protein, p48, p21, p125, actin mRNA, and 28S rRNA at different times after inducing p53 expression by removal of tetracycline. UV-DDB was measured by an electrophoretic mobility-shift assay (EMSA) with UV-damaged DNA probe.
Figure 4
GGR and TCR in XPE cells. DDB− (XP2RO) and DDB+ (XP89TO) cells were analyzed at different times after exposure to 10 J/m2 UV. GGR of 6-4 photoproducts (A) and CPDs (B) were measured by an immunoblot assay with monoclonal antibodies specific for 6-4 photoproducts or CPDs. TCR of CPDs on the transcribed strand (C) and nontranscribed strand (D) of the dihydrofolate reductase gene were measured by restored resistance to T4 endonuclease V, which specifically cleaves CPDs. GGR and TCR also are shown for other primary fibroblast cell lines: wild type (WI38), p53−/− (041 mut), and XPC (XP10BE), as determined in previous experiments (2, 3, 34, 35). To facilitate comparison, repair in wild type (dotted lines) and XPE (solid lines) are shown over the entire time course.
Figure 5
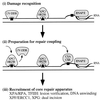
Model for photoproduct recognition in nucleotide excision repair. (i) Damage recognition. CPDs (square) are recognized by UV-DDB when p48 transcription increases in a p53-dependent manner. Although 6-4 photoproducts (triangle) can be recognized by UV-DDB (32), they may distort the DNA sufficiently to be recognized directly by XPC/HR23B. Thus, the absence of UV-DDB in XPE cells causes only a delay in the global repair of 6-4 photoproducts rather than an absolute deficiency. Either photoproduct is recognized in the transcribed strand of expressed genes by the arrest of RNA polymerase II (RNAP II) translocation (31). (ii) Preparation for repair coupling. XPC/HR23B is postulated to recognize UV-DDB bound to DNA, perhaps replacing it at the lesion site and then partially opening the DNA. A complex of the Cockayne Syndrome proteins, CSA/CSB, is postulated to recognize and displace the arrested RNA polymerase II, opening the DNA at that site. (iii) Recruitment of core repair apparatus. XPC/HR23B recruits the core nucleotide excision repair apparatus for GGR whereas CSA/CSB recruits the core apparatus for TCR. The helicases in TFIIH create a bubble of unwound DNA and, along with XPA/RPA, may participate in lesion verification before permitting the structure-specific nucleases XPF/ERCC1 and XPG to make incisions on each side of the lesion.
Similar articles
- Increased expression of p53 enhances transcription-coupled repair and global genomic repair of a UVC-damaged reporter gene in human cells.
Dregoesc D, Rybak AP, Rainbow AJ. Dregoesc D, et al. DNA Repair (Amst). 2007 May 1;6(5):588-601. doi: 10.1016/j.dnarep.2006.11.008. Epub 2006 Dec 28. DNA Repair (Amst). 2007. PMID: 17196445 - p53 responsive nucleotide excision repair gene products p48 and XPC, but not p53, localize to sites of UV-irradiation-induced DNA damage, in vivo.
Fitch ME, Cross IV, Ford JM. Fitch ME, et al. Carcinogenesis. 2003 May;24(5):843-50. doi: 10.1093/carcin/bgg031. Carcinogenesis. 2003. PMID: 12771027 - Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation.
Rapić-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS. Rapić-Otrin V, et al. Nucleic Acids Res. 2002 Jun 1;30(11):2588-98. doi: 10.1093/nar/30.11.2588. Nucleic Acids Res. 2002. PMID: 12034848 Free PMC article. - Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein.
Tang J, Chu G. Tang J, et al. DNA Repair (Amst). 2002 Aug 6;1(8):601-16. doi: 10.1016/s1568-7864(02)00052-6. DNA Repair (Amst). 2002. PMID: 12509284 Free PMC article. Review. - DNA repair-deficient Xpa and Xpa/p53+/- knock-out mice: nature of the models.
van Steeg H, de Vries A, van Oostrom CTh, van Benthem J, Beems RB, van Kreijl CF. van Steeg H, et al. Toxicol Pathol. 2001;29 Suppl:109-16. doi: 10.1080/019262301753178519. Toxicol Pathol. 2001. PMID: 11695546 Review.
Cited by
- The role of altered nucleotide excision repair and UVB-induced DNA damage in melanomagenesis.
Budden T, Bowden NA. Budden T, et al. Int J Mol Sci. 2013 Jan 9;14(1):1132-51. doi: 10.3390/ijms14011132. Int J Mol Sci. 2013. PMID: 23303275 Free PMC article. Review. - Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation.
Christmann M, Kaina B. Christmann M, et al. Nucleic Acids Res. 2013 Oct;41(18):8403-20. doi: 10.1093/nar/gkt635. Epub 2013 Jul 27. Nucleic Acids Res. 2013. PMID: 23892398 Free PMC article. Review. - Retinal degeneration and ionizing radiation hypersensitivity in a mouse model for Cockayne syndrome.
Gorgels TG, van der Pluijm I, Brandt RM, Garinis GA, van Steeg H, van den Aardweg G, Jansen GH, Ruijter JM, Bergen AA, van Norren D, Hoeijmakers JH, van der Horst GT. Gorgels TG, et al. Mol Cell Biol. 2007 Feb;27(4):1433-41. doi: 10.1128/MCB.01037-06. Epub 2006 Dec 4. Mol Cell Biol. 2007. PMID: 17145777 Free PMC article. - A gene signature-based approach identifies mTOR as a regulator of p73.
Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. Rosenbluth JM, et al. Mol Cell Biol. 2008 Oct;28(19):5951-64. doi: 10.1128/MCB.00305-08. Epub 2008 Aug 4. Mol Cell Biol. 2008. PMID: 18678646 Free PMC article. - Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage.
Alekseev S, Luijsterburg MS, Pines A, Geverts B, Mari PO, Giglia-Mari G, Lans H, Houtsmuller AB, Mullenders LH, Hoeijmakers JH, Vermeulen W. Alekseev S, et al. Mol Cell Biol. 2008 Dec;28(24):7402-13. doi: 10.1128/MCB.01108-08. Epub 2008 Oct 20. Mol Cell Biol. 2008. PMID: 18936169 Free PMC article.
References
- Hanawalt P. Science. 1994;266:1957–1958. - PubMed
- Ford J M, Hanawalt P C. J Biol Chem. 1997;272:28073–28080. - PubMed
- Levine A J. Cell. 1997;88:323–331. - PubMed
- Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous