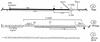In vivo addition of poly(A) tail and AU-rich sequences to the 3' terminus of the Sindbis virus RNA genome: a novel 3'-end repair pathway - PubMed (original) (raw)
In vivo addition of poly(A) tail and AU-rich sequences to the 3' terminus of the Sindbis virus RNA genome: a novel 3'-end repair pathway
R Raju et al. J Virol. 1999 Mar.
Abstract
Alphaviruses are mosquito-transmitted RNA viruses that cause important diseases in both humans and livestock. Sindbis virus (SIN), the type species of the alphavirus genus, carries a 11.7-kb positive-sense RNA genome which is capped at its 5' end and polyadenylated at its 3' end. The 3' nontranslated region (3'NTR) of the SIN genome carries many AU-rich motifs, including a 19-nucleotide (nt) conserved element (3'CSE) and a poly(A) tail. This 3'CSE and the adjoining poly(A) tail are believed to regulate the synthesis of negative-sense RNA and genome replication in vivo. We have recently demonstrated that the SIN genome lacking the poly(A) tail was infectious and that de novo polyadenylation could occur in vivo (K. R. Hill, M. Hajjou, J. Hu, and R. Raju, J. Virol. 71:2693-2704, 1997). Here, we demonstrate that the 3'-terminal 29-nt region of the SIN genome carries a signal for possible cytoplasmic polyadenylation. To further investigate the polyadenylation signals within the 3'NTR, we generated a battery of mutant genomes with mutations in the 3'NTR and tested their ability to generate infectious virus and undergo 3' polyadenylation in vivo. Engineered SIN genomes with terminal deletions within the 19-nt 3'CSE were infectious and regained their poly(A) tail. Also, a SIN genome carrying the poly(A) tail but lacking a part or the entire 19-nt 3'CSE was also infectious. Sequence analysis of viruses generated from these engineered SIN genomes demonstrated the addition of a variety of AU-rich sequence motifs just adjacent to the poly(A) tail. The addition of AU-rich motifs to the mutant SIN genomes appears to require the presence of a significant portion of the 3'NTR. These results indicate the ability of alphavirus RNAs to undergo 3' repair and the existence of a pathway for the addition of AU-rich sequences and a poly(A) tail to their 3' end in the infected host cell. Most importantly, these results indicate the ability of alphavirus replication machinery to use a multitude of AU-rich RNA sequences abutted by a poly(A) motif as promoters for negative-sense RNA synthesis and genome replication in vivo. The possible roles of cytoplasmic polyadenylation machinery, terminal transferase-like enzymes, and the viral polymerase in the terminal repair processes are discussed.
Figures
FIG. 1
(A) Gene organization of Tapa, a SIN cDNA. (B) 3′ sequence of the TT21qa; NS, nonstructural genes; S, structural genes; (A)n, poly(A) tail. It is important to note that linearization of TT21qa with _Xho_I cleaves the DNA between A and G. Therefore, the RNA transcribed from the TT21qa/Xho construct may contain 1 to 5 bases of the _Xho_I motif. (C) 3′ sequence of the SIN genome recovered from BHK cells transfected with TT21qa/Xho RNA. The 11 PCR products corresponding to the viral RNA (Fig. 3A) were sequenced with JC1000-1H. The size of the poly(A) tail ranged from 17 to 39 nt. All 11 virus isolates were identical with respect to the 3′ terminal 73 nt.
FIG. 2
Electrophoretic analysis of the in vitro-synthesized RNA transcripts. Two percent of the total RNA synthesized in a 30-μl reaction mixture was denatured with glyoxal and dimethyl sulfoxide and separated on a 1.25% gel. The gel was soaked in methanol–2,5-diphenyloxazole, dried, and fluorographed. The mock transcription reaction mixture contained no DNA template.
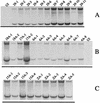
FIG. 3
Expression of genomic and subgenomic SIN RNAs from representative virus isolates. Cultures of BHK cells were infected (MOI of 0.1 to 2.4) with the indicated virus isolates and labelled with [3H]uridine in the presence of dactinomycin. Approximately 4 to 6 μg of [3H]uridine-labelled cytoplasmic RNA was denatured with glyoxal, separated on a 1.25% agarose gel, and fluorographed. The upper band in each lane corresponds to the genomic RNA (49S), and the lower band corresponds to the subgenomic RNA (26S). The identity of the virus isolate is indicated at the top of each lane. UI, uninfected cell RNA. (A) Isolates 29-1 to 29-11, viral plaques derived from TT21qa/Xho; (B) plaques derived from T3′18(A)n, T3′17(A)n, and T3′6(A)n; (C) plaques derived from T3′15(A)n and T3′0(A)n. It is important to note that the amount of RNA made by each isolate is due mostly to the MOI, although base changes in recovered viruses could also be partly responsible.
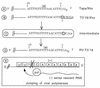
FIG. 4
Polyadenylation of the SIN genome carrying a G residue at the −1 position of the 3′CSE. (A) Sequence of the 3′CSE in Tapa. (B) Sequence of the 3′ terminus in T3′18/Xho. Insertion of a G residue in the −1 position of the 3′CSE and of a _Xho_I site at the 3′ terminus results in the introduction of a C residue in the +1 position. (C) Proposed intermediate during the formation of RV-T3′18 type revertant viruses. (D) Sequence of the 3′ terminus of all six viral isolates indicating the loss of the G residue and polyadenylation at the C residue. (E) Proposed polymerase jumping event during negative-strand synthesis and the loss of the G residue.
FIG. 5
Terminal repair of a 3′ truncated SIN genome. (A) Wild-type sequence of the 3′CSE in Tapa; (B) sequence of the 3′ terminus of T3′15/Xho indicating the deletion of the −1 to −4 positions and the insertion of a _Xho_I site at the terminus; (C) names and sequences of viral isolates generated from T3′15/Xho. The single underline indicates the AU-rich sequences and the poly(A) tail added at the 3′ terminus. The remnants of the _Xho_I motif are highlighted with a double underline. The number in parentheses at the end of each sequence corresponds to the number of viral isolates containing the same sequence. The sequences for isolates 15.1, 15.2, 15.3, and 15.7 (two separate isolates) were from experiment 1, those for isolates 15.4, 15.5, 15.7, 15.8, and 15.9 were from experiment 2, and those for isolates 15.6 and 15.7 were from experiment 3.
FIG. 6
Addition of AU-rich motifs to 3′ mutants of polyadenylated SIN genome. The 3′ sequence of the template used for transfection of BHK cells and the 3′ sequence of the viral isolates recovered from the cells are given for each group. The positions of the base deletions introduced in the parental template are indicated as filled squares. The circled nucleotides highlight the base changes encountered in the revertant virus. The AU-rich sequences and poly(A) tail added to the 3′ end are marked with an underline. (A) T3′18(A)n and virus isolates derived from it. Isolates 18A-1 to 18A-3 correspond to experiment 1 and isolates 18A-4 to 18A-7 correspond to experiment 2. (B) T3′17(A)n and virus isolates derived from it. Isolates 17A-1 to 17A-4, 17A-7, and 17A-10 were derived from experiment 1. Isolates 17A-5, 17A-6, 17A-8, and 17A-9 were derived from transfection experiment 2. (C) T3′6(A)n and virus isolates derived it. The first three isolates were derived from experiment 1, and the rest were derived from experiment 2.
FIG. 7
Addition of long AU-rich sequences of 3′ mutants of the SIN genome. (A) Sequences of T3′15(A)n and virus isolates generated from it. Isolates 15A-1, 15A-2, and 15A-3 were derived from experiment 1. Isolate 15A-4 was derived from experiment 2. (B) Sequences of T3′0(A)n and the virus isolates recovered from it. Isolates ZA-1, -2, and -3 were derived from experiment 1, and isolate ZA-4 was derived from experiment 2. The identification of residues and motifs is as described in the legend to Fig. 6.
Similar articles
- Efficient replication, and evolution of Sindbis virus genomes with non-canonical 3'A/U-rich elements (NC3ARE) in neonatal mice.
James FD, Hietala KA, Eldar D, Guess TE, Cone C, Mundell NA, Barnett JV, Raju R. James FD, et al. Virus Genes. 2007 Dec;35(3):651-62. doi: 10.1007/s11262-007-0130-z. Epub 2007 Jul 7. Virus Genes. 2007. PMID: 17616797 - Non-canonical poly(A) polymerase in mammalian gametogenesis.
Kashiwabara S, Nakanishi T, Kimura M, Baba T. Kashiwabara S, et al. Biochim Biophys Acta. 2008 Apr;1779(4):230-8. doi: 10.1016/j.bbagrm.2008.01.004. Epub 2008 Feb 12. Biochim Biophys Acta. 2008. PMID: 18294465 Review. - Implications of polyadenylation in health and disease.
Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. Curinha A, et al. Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
- Requirements at the 3' end of the sindbis virus genome for efficient synthesis of minus-strand RNA.
Hardy RW, Rice CM. Hardy RW, et al. J Virol. 2005 Apr;79(8):4630-9. doi: 10.1128/JVI.79.8.4630-4639.2005. J Virol. 2005. PMID: 15795249 Free PMC article. - Widespread 3'-end uridylation in eukaryotic RNA viruses.
Huo Y, Shen J, Wu H, Zhang C, Guo L, Yang J, Li W. Huo Y, et al. Sci Rep. 2016 May 6;6:25454. doi: 10.1038/srep25454. Sci Rep. 2016. PMID: 27151171 Free PMC article. - Development of infectious cDNA clones of Salmonid alphavirus subtype 3.
Karlsen M, Villoing S, Ottem KF, Rimstad E, Nylund A. Karlsen M, et al. BMC Res Notes. 2010 Sep 21;3:241. doi: 10.1186/1756-0500-3-241. BMC Res Notes. 2010. PMID: 20858233 Free PMC article. - Replication of alphaviruses requires a pseudoknot that involves the poly(A) tail.
Olsthoorn RCL. Olsthoorn RCL. RNA. 2022 Oct;28(10):1348-1358. doi: 10.1261/rna.079243.122. Epub 2022 Jul 29. RNA. 2022. PMID: 35906005 Free PMC article. - Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity.
Tomar S, Hardy RW, Smith JL, Kuhn RJ. Tomar S, et al. J Virol. 2006 Oct;80(20):9962-9. doi: 10.1128/JVI.01067-06. J Virol. 2006. PMID: 17005674 Free PMC article.
References
- Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. - PubMed
- Boyer J C, Haenni A L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials