Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system (original) (raw)
Abstract
The bacterium Vibrio cholerae, like other human pathogens that reside in environmental reservoirs, survives predation by unicellular eukaryotes. Strains of the O1 and O139 serogroups cause cholera, whereas non-O1/non-O139 strains cause human infections through poorly defined mechanisms. Using Dictyostelium discoideum as a model host, we have identified a virulence mechanism in a non-O1/non-O139 V. cholerae strain that involves extracellular translocation of proteins that lack N-terminal hydrophobic leader sequences. Accordingly, we have named these genes “VAS” genes for virulence-associated secretion, and we propose that these genes encode a prototypic “type VI” secretion system. We show that vas genes are required for cytotoxicity of V. cholerae cells toward Dictyostelium amoebae and mammalian J774 macrophages by a contact-dependent mechanism. A large number of Gram-negative bacterial pathogens carry genes homologous to vas genes and potential effector proteins secreted by this pathway (i.e., hemolysin-coregulated protein and VgrG). Mutations in vas homologs in other bacterial species have been reported to attenuate virulence in animals and cultured macrophages. Thus, the genes encoding the VAS-related, type VI secretion system likely play an important conserved function in microbial pathogenesis and represent an additional class of targets for vaccine and antimicrobial drug-based therapies.
Keywords: Dictyostelium discoideum, type VI secretion, virulence-associated secretion
Cholera is a severe, life-threatening diarrheal disease caused by Vibrio cholerae strains of the O1 and O139 serogroups. In contrast, non-O1, non-O139 strains of V. cholerae are primarily associated with isolated cases of extra-intestinal infection or gastroenteritis. An exception to this pattern was a large outbreak of a cholera-like illness that occurred in 1968 in Sudan, where an O37 strain of V. cholerae caused 460 cases and 125 deaths (1). The virulence mechanisms of O1 and O139 strains involve the elaboration of extracellular factors such as cholera enterotoxin and toxin coregulated pili. In contrast, the virulence mechanisms used by non-O1, non-O139 strains remain poorly defined (2). Using the social amoeba Dictyostelium discoideum as a model host, we have developed an experimental system designed to identify novel virulence mechanisms from pathogenic non-O1, non-O139 strains.
D. discoideum is a eukaryotic organism that seeks out and preys on bacteria through its phagocytic feeding behavior. As such, it has been used as a model eukaryotic cell that mimics a mammalian macrophage in aspects of its cell biology and interaction with microbes. Several environmental pathogenic bacteria, including Legionella pneumophila, Mycobacterium marinum, and Pseudomonas aeruginosa (3), resist Dictyostelium predation by producing factors that either kill amoebae or allow successful intracellular survival and multiplication. In these cases, the same virulence mechanisms operative against mammalian cells have also been implicated in resistance to Dictyostelium predation. For example, we have shown that P. aeruginosa can kill Dictyostelium amoebae by using its type III secretion system (T3SS) to deliver the cytotoxic ExoU effector protein into target cells (4) and that this T3SS is also essential for mammalian virulence (5). Thus, we reasoned that screening for resistance to Dictyostelium predation might reveal virulence factors produced by pathogenic non-O1 and non-O139 V. cholerae strains that target eukaryotic host cells.
Here, we report that Dictyostelium amoebae are killed by an isolate of the V. cholerae serogroup O37 from the 1968 Sudan epidemic. The virulence displayed by this V. cholerae O37 strain requires bacterial-amoeboid, cell-cell contact and a set of genes that encode a unique protein secretion machinery. We have named the genes encoding this phenotype “VAS” (for virulence-associated secretion) and further show that this pathway mediates V. cholerae cytotoxicity toward a mammalian macrophage cell line. Previously, bioinformatic analysis (6) identified this cluster of genes as highly conserved in several Gram-negative pathogens. Because the genes in the cluster showed homology to the L. pneumophila icmF, the cluster was designated the IcmF-associated homologous protein (IAHP) gene cluster. However, our analysis indicates that the IAHP gene cluster of V. cholerae encodes a protein secretion system distinct from type III and type IV pathways. Accordingly, we conclude that this system constitutes the prototype of a secretion system that we propose to call “type VI secretion” to clarify its relationship to previously described secretion systems involved in microbial pathogenesis.
Results
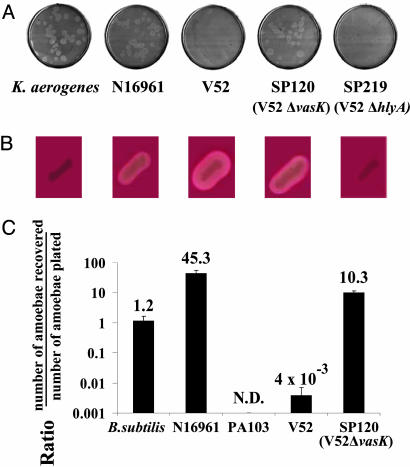
The Non-O1, Non-O139 V. cholerae Strain V52 Resists D. discoideum Predation. Bacterial predation by Dictyostelium is easily scored by plating individual amoebae on nutrient agar plates seeded with bacterial cells. Successful predation by the amoebae is visualized by the appearance of clear plaques corresponding to zones where actively feeding and replicating amoebae have phagocytosed and killed bacteria. An absence of plaques indicates that the bacterial species being tested displays a “virulent” phenotype on Dictyostelium, by either evading amoeboid killing or actively killing Dictyostelium. As shown in Fig. 1_A_, Dictyostelium amoebae readily plaque on Klebsiella aerogenes and an O1 serogroup strain of V. cholerae (N16961). In contrast, amoebae plated on the O37 serogroup V. cholerae strain V52 are killed and fail to form plaques, indicating that this non-O1, non-O139 strain expresses virulence factors active on Dictyostelium (see below). The virulence of this isolate for humans is evidenced by the fact that it was isolated from a victim of a large outbreak of diarrheal disease occurring in 1968 that caused 125 deaths in Sudan (1).
Fig. 1.
V. cholerae cytotoxicity toward the simple eukaryote D. discoideum. (A) Plaque assay. D. discoideum cells were plated on SM/5 with K. aerogenes and V. cholerae strains N16961, V52, SP120 (V52Δ_vasK_), and SP219 (V52Δ_hlyA_) at a density of ≈100 amoebae per plate. Bacterial virulence potential was determined by the number of plaques formed by D. discoideum in bacterial lawns. (B) Hemolytic phenotype of K. aerogenes and V. cholerae strains N16961, V52, SP120, and SP219 on trypticase soy agar containing 5% sheep blood. (C) Killing assay. Virulence of indicated bacteria was determined by enumerating the number of live amoebae recovered from bacterial lawns after a 24-h incubation. Numbers above the columns indicate fold change of number of amoebae in bacterial lawns over a 24-h period. Results shown are the means (±SD) of triplicate determinations.
A Genetic Screen Leads to the Identification of VAS Genes. Transposon mutagenesis was used to define the genes encoding Dictyostelium virulence in V52. In brief, a library of V52 cells carrying random insertions of TnAraOut (7) was screened for colonies with a predation-sensitive phenotype on plates containing a large excess of amoebae. Such bacterial mutants formed notched colonies that reflect the active destruction of bacterial cells by feeding amoebae. Virulent wild-type V52 forms smooth, uniformly round colonies because of their resistance to Dictyostelium predation. _Dictyostelium_-attenuated mutants were purified from notched colonies and characterized for their cytotoxicity toward amoebae in a quantitative assay. Amoebae were mixed with wild-type or mutant bacteria and plated on nutrient agar plates. Bacterial lawns were harvested after 24 h, and surviving amoebae were enumerated by plaque assays in lawns of Escherichia coli strain B/r. As shown in Fig. 1_C_, wild-type strain V52 causes a 250-fold reduction in the recovery of viable amoebae. A nonpathogenic Bacillus subtilis strain, which does not support Dictyostelium replication, caused no detectable decrease in amoebae viability, whereas P. aeruginosa actively killed amoebae as reported (4). The low number of amoebae recovered from lawns of V52 is therefore caused by active killing by V52 and not starvation. In contrast, the isogenic deletion mutant SP120, which is impaired in the VasK gene, one of the genes that emerged from our mutagenesis screen (see below), lost its virulence, and Dictyostelium efficiently used this mutant as a bacterial substrate at efficiencies comparable to K. aerogenes (Fig. 1 A).
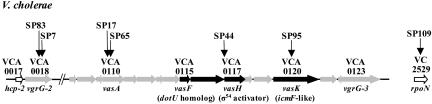
The most interesting group of genes we identified as being involved in Dictyostelium virulence includes vasA (VCA0110), vasH (VCA0117), and vasK (VCA0120), which are all closely linked on the V. cholerae small chromosome (Fig. 2 and Table 1). Two _Dictyostelium-_attenuated mutants, SP17 and SP65, carried independent TnAraOut insertions in gene vasA, which is a homolog of impG of Rhizobium leguminosarum, a gene involved in plant root infection by this bacterial species (8). Another mutant, SP95, carries an insertion in the VasK gene, which encodes a homolog of icmF, a gene involved in intracellular replication of L. pneumophila (see below). vasA and vasK flank vasH, which encodes a predicted activator of Sigma-54, an alternative subunit of RNA polymerase. vasH is disrupted in attenuated mutant SP44 (Fig. 2), and we confirmed that a deletion of vasH in V52 mutant strain SP117 also produced a _Dictyostelium-_attenuated phenotype (data not shown). Interestingly, the _Dictyostelium_-attenuated mutant SP109 carries a TnAraOut insertion on the large chromosome in the gene that encodes Sigma-54 (VC2529). These results suggest that the vasH product and Sigma-54 collaborate to control transcriptional expression of one or more of the Dictyostelium virulence genes expressed by V52.
Fig. 2.
Genetic organization of the VAS pathway of V. cholerae. Horizontal gray arrows designate hypothetical genes, black arrows designate genes with homologues of known function, and empty arrows indicate genes of known function in V. cholerae (drawn to scale). Vertical arrows indicate transposon insertion sites in _Dictyostelium_-attenuated V. cholerae mutants.
Table 1. TnAraOut V. cholerae mutants with _Dictyostelium_-attenuated phenotype.
| Strain | Gene | VC number | Distance from ATG (gene length) | Homology | blastp* (bits/E value) | Function ascribed to homologue |
|---|---|---|---|---|---|---|
| SP17 | vasA | VCA0110 | +510 (1,770) | RL impG | 242/3e-62 | Impaired Rhizobium plant infection |
| SP65 | vasA | VCA0110 | +704 (1,770) | RL impG | 242/3e-62 | See above |
| SP44 | vasH | VCA0117 | +96 (1,593) | EC rtcR | 133/1e-31 | Sigma-54 dependent activator in E. coli |
| SP95 | vasK | VCA0120 | +1477 (3,546) | LP icmF | 154/6e-38 | Type IV protein secretion in L. pneumophila |
| SP7 | _vgrG_-2 | VCA0018 | +985 (2,085) | EC vgrG | 417/8e-117 | Homologous to VgrG-1 that catalyzes action-crosslinking in eukaryotic cells |
| SP83 | _vgrG_-2 | VCA0018 | +704 (2,085) | EC vgrG | 417/8e-117 | See above |
| SP109 | rpoN | VC2529 | +338 (1,464) | VC rpoN | 862/0.0 | Alternative σ54 subunit of RNA polymerase |
Whole-genome microarray-based transcriptome analysis showed that transcriptional expression of two genes, hcp-1 (VC1415) and hcp-2 (VCA0017), was significantly reduced in the vasH deletion mutant SP117 compared with wild-type V52 (see Supporting Text and Table 2, which are published as supporting information on the PNAS web site). This observation is consistent with the fact that these two genes are required for Dictyostelium cytotoxicity (see below). hcp-1 and hcp-2 both encode an identical protein corresponding to hemolysin-coregulated protein (Hcp), a secreted V. cholerae protein that is coexpressed with HlyA hemolysin (9). However, _Dictyostelium_-attenuated mutants were found to still express and secrete HlyA (Fig. 1_B_) and the hlyA mutant SP219 was fully virulent on Dictyostelium (Fig. 1 A).
Two genes in the cluster, vasK (VCA0120) and vasF (VCA0115), show a high degree of similarity to icmF and icmH (dotU), two genes found in L. pneumophila (10). In L. pneumophila, these genes are nonessential components of the type IV secretion system (T4SS) required for cytotoxicity of L. pneumophila toward mammalian and D. discoideum cells (11, 12). IcmF and IcmH (DotU) are thought to be accessory proteins that work in concert to improve the efficiency of T4SS translocation of bacterial effector proteins into the cytosol of eukaryotic target cells (13, 14). VasK and VasF may also cooperate in V. cholerae, because vasF and vasK deletion mutants also show a _Dictyostelium_-attenuated phenotype (data not shown). No other V52 mutant carried insertions in a gene with homology to the group of genes including dotA, dotB, dotC, dotD, icmB, icmC, icmD, icmE, icmG, icmJ, icmK, icmL, icmM, icmN, icmO, icmP, icmQ, icmR, icmS, icmT, icmV, icmW, and icmX, most of which have been shown to be absolutely required for function of the Legionella T4SS (for review, see refs. 15 and 16). To determine whether V52 has a T4SS gene cluster, we sequenced the genome of this strain to times 6.5 coverage. Careful annotation of the sequence found no evidence for the existence of genes encoding homologs of dotA, dotB, dotC, dotD, icmB, icmC, icmD, icmE, icmG, icmJ, icmK, icmL, icmM, icmN, icmO, icmP, icmQ, icmR, icmS, icmT, icmV, icmW, and icmX and thus we conclude that V52 does not carry a recognizable T4SS gene cluster. Also, unlike other non-O1, non-O139 V. cholerae strains (17), the V52 genome does not encode a T3SS other than the typical one required for flagella biosynthesis. Thus, it is also notable that no _Dictyostelium_-attenuated mutants were found in any gene known to be involved in flagellar biosynthesis.
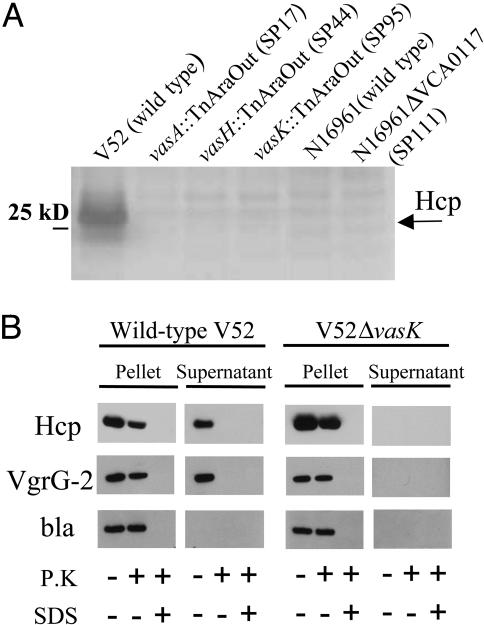
The VAS Pathway Is Responsible for the Secretion of Proteins Lacking N-Terminal Leader Sequences. Because some T3SS and T4SS pathways transport their effector proteins into the bacterial culture supernatant fluids in the absence of eukaryotic target cells, we analyzed culture supernatant fluids of V52 for evidence of such protein export. SDS/PAGE was used to visualize the proteins secreted by V52 and three different _Dictyostelium_-attenuated mutants. We also analyzed supernatant fluids from _Dictyostelium_-sensitive strain N16961 and the isogenic _vasH_-deletion mutant SP111. As shown in Fig. 3_A_, a 28-kDa band that was identified by MS as Hcp appeared as an abundant protein in the supernatant fluid of V52. Interestingly, Hcp was absent in supernatants of V52 mutants with transposon insertions in vasA, vasH, and vasK (strains SP17, SP44, and SP95, respectively), as well as wild-type N16961 and its _vasH_-deletion mutant SP111. Accordingly, we constructed hcp-1 and hcp-2 single- and double-deletion mutants and found that only a double-deletion mutant was avirulent toward Dictyostelium (data not shown). Virulence was restored when a plasmid allowing isopropyl β-d-thiogalactoside-inducible expression of Hcp was introduced. Thus, both hcp alleles are functional and Hcp is apparently essential for VAS-mediated amoeba cytotoxicity. It is also of interest that Hcp has been reported to lack a hydrophobic leader peptide and was previously detected with an unprocessed amino terminus in the supernatant fluids of V. cholerae (9).
Fig. 3.
VAS-dependent secretion. (A) Secretion profiles of V. cholerae VAS mutants. SDS/PAGE of concentrated midlog culture supernatants of indicated strains. Black arrow indicates position of Hcp. (B) Extracellular secretion of epitope-tagged substrates. V. cholerae strains V52 and SP120 (V52Δ_vasK_) maintaining a plasmid that allows arabinose-induced expression of tagged Hcp-2 and VgrG-2 were grown under inducing conditions. Cells and filtered supernatants were left untreated or incubated with either 0.1 mg/ml proteinase K (P.K) in the presence or absence of 1% SDS. Protease inhibitor PMSF was used to stop proteolysis after 20 min, and extracts were separated on a SDS/PAGE for immunoblotting with vesicular stomatitis virus glycoprotein antisera. The quality of pellet and supernatant fractionation was determined by localizing periplasmic β-lactamase (bla).
Analysis of culture supernatant fluids of V52 by electronspray ionization liquid chromatography tandem MS identified additional proteins in supernatants of V52 and _Dictyostelium_-attenuated mutants (see Table 3, which is published as supporting information on the PNAS web site). Strains carrying mutations in vasA, vasH, and vasK were still able to secrete proteins with hydrophobic amino-terminal signal sequences, namely chitinase, neuraminidase, PrtV protease, and HlyA hemolysin (see Fig. 5, which is published as supporting information on the PNAS web site). These proteins are known to be secreted by type I and type II secretion pathways. Critically, V52 secreted four proteins that could not be detected in supernatants of N16961, namely Hcp, VgrG-1, VgrG-2, and VgrG-3. These four proteins lack identifiable hydrophobic amino-terminal signal sequences (data not shown). In contrast, _Dictyostelium_-attenuated mutants SP17, SP44, SP83, SP95, and SP109 also showed undetectable levels of Hcp, VgrG-1, VgrG-2, and VgrG-3 in their culture supernatant fluids. In fact, in our initial mutant screen, we isolated two independent _Dictyostelium_-attenuated mutants, SP7 and SP83, that each carry a transposon insertion in the VgrG-2 gene. These results strongly suggest that VgrG-2 is also an essential component in the pathway leading to cytotoxicity of Dictyostelium amoebae. In conclusion, the V. cholerae VAS pathway does not appear to be required for secretion of any protein with hydrophobic amino terminus signal sequences, but is essential for secretion of Hcp, VgrG-1, VgrG-2, and VgrG-3, all of which lack such signal sequences.
We did not detect the predicted protein products of vasA and vasK in the culture supernatant of V52. These data suggest that these protein products are not secreted but are nonetheless required for the function of a secretion pathway that transports Hcp, VgrG-1, VgrG-2, and VgrG-3 to the exterior of bacterial cells. If this model is correct, epitope-tagged versions of Hcp-2 and VgrG-2 should be secreted by wild-type V52 but not by an isogenic vasK mutant. To follow VasK-mediated secretion, we introduced a plasmid that allows arabinose-induced expression of Hcp-2 and VgrG-2 tagged with a vesicular stomatitis virus glycoprotein epitope at their C termini into wild-type V52 and the isogenic vasK deletion mutant SP120. Only wild-type V52 was able to secrete these two tagged proteins into the culture supernatant (Fig. 3_B_). Both V52 and the vasK mutant SP120 produced equal amounts of Hcp-2 and VgrG-2 that accumulated inside the bacteria cells as evidenced by their resistance to proteolytic degradation when cells were treated with proteinase K (Fig. 3_B_). Thus, proteins Hcp-2 and VgrG-2 rely on VasK for their extracellular secretion.
VAS Genes Are Highly Regulated. Although all V. cholerae strains analyzed so far by microarray analysis carry DNA corresponding to VAS, hcp, and vgrG genes (2), our current data suggest that, unlike V52, most O1 and O139 strains, like N16961, are permissive for Dictyostelium predation under the in vitro conditions examined so far. This discrepancy may be explained by the results of our microarray analysis that show that the hcp genes are highly expressed in V52 compared with N16961 under in vitro conditions that stimulate VAS-dependent Hcp secretion by V52. In addition, N16961 is unable to secrete Hcp into culture fluids even when this gene is expressed via a heterologous promoter (data not shown). These experiments suggest that some V. cholerae strains with VAS gene clusters are unable to use the VAS secretion pathway perhaps because their Vas, Hcp, or VgrG genes are not properly regulated. This hypothesis is supported by the fact that VAS genes are tightly regulated in vivo. For example, vasK has been reported to be an _in vivo_-induced gene in a rabbit model for cholera (18), whereas its homolog in Salmonella enterica, sciS (19), is _in vivo_-induced in macrophages. Other VAS-related gene products have been identified as antigens recognized by catfish infected with Edwardsiella ictaluri, suggesting they, too, are _in vivo_-induced (20). Thus, transcriptional regulation in vivo may be a common characteristic of VAS genes and their homologues.
Hcp Mediates Translocation of VgrG-1 and VgrG-2. We have identified two other non-O1, non-O139 V. cholerae strains, SCE223 and SCE226, that are virulent for Dictyostelium and express and secrete Hcp under in vitro conditions (see Supporting Text, and Fig. 6, which are published as supporting information on the PNAS web site). As in V52, in-frame vasK deletions in these two strains rendered them sensitive to predation by Dictyostelium and blocked Hcp export (data not shown). Thus, expression and secretion of Hcp by a VAS pathway correlates with Dictyostelium virulence for other strains of V. cholerae besides V52. Because Hcp appears to be a central component of the VAS pathway, we asked whether Hcp was essential for the secretion of VgrG-1 and VgrG-2. When a plasmid that allows arabinose-induced expression of Hcp was introduced into a V52 mutant strain with deletions in both Hcp genes, VgrG-1 and VgrG-2 could be detected in culture supernatants by MS only when Hcp expression was induced. Thus, Hcp is both secreted by the VAS pathway and required for the extracellular secretion of other proteins like VgrG-1 and VgrG-2.
V. cholerae Uses the VAS Pathway to Mediate Virulence Toward J774 Macrophages. By analogy to T3SS and T4SS (21, 22), the extracellular secretion of Hcp, VgrG-1, and VgrG-2 may actually reflect a more complex process that involves the translocation of these proteins by V. cholerae into eukaryotic target cells. Recently, Sheahan et al. (23) reported that VgrG-1 and the V. cholerae RtxA toxin share a subdomain that mediates actin covalent crosslinking and cytotoxicity when expressed in the cytosol of mammalian cells. The extracellular cytotoxin RtxA, however, is not required for Dictyostelium virulence, because a rtxA mutant of V52 still kills amoebae (data not shown). Thus, the actin-crosslinking activity of VgrG-1 suggests that this protein might be a cytotoxic effector transported into mammalian target cells by the VAS secretion pathway.
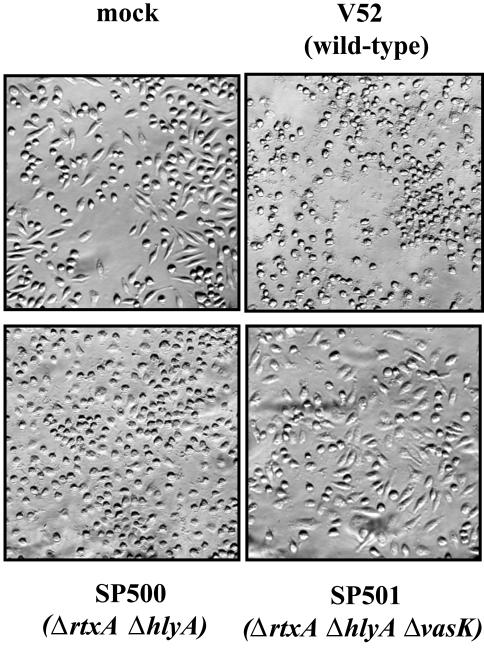
We asked whether VAS-mediated secretion was associated with _V. cholerae_-mediated cytotoxicity toward a mammalian macrophage cell line. Because several different V. cholerae toxins, including RtxA and HlyA, can disrupt mammalian cell structures (24, 25), we examined the effect of a vasK mutation in the context of mutations in these other two factors. As shown in Fig. 4, the morphology of J774 macrophages is disrupted within 2 h of exposure to live V52. Mutant SP501 disrupted in rtxA, hlyA, and vasK lost all detectable cytotoxicity toward J774 cells, whereas its parent strain SP500, disrupted in only rtxA and hlyA, retained this property. Interestingly, media supernatants from wells infected with SP500 showed no cell rounding activity when added to wells containing uninfected J774 cells, suggesting bacterial-macrophage cell-cell contact is a requisite for SP500 cytotoxicity (data not shown). In conclusion, vasK and the VAS-dependent secretion pathway contribute significantly to the cytotoxicity that V. cholerae displays toward this mammalian macrophage cell line in a cell-contact-dependent manner.
Fig. 4.
V. cholerae cytotoxicity toward J774 macrophages. J774 cells were infected for 2 h with V52 (wild type) and isogenic mutant SP500 (Δ_rtxA_, Δ_hlyA_) or mutant SP501 (Δ_rtxA_, Δ_hlyA_, Δ_vasK_). Cells were fixed with 3% paraformaldehyde to assess the morphology of infected cells.
Discussion
The data presented here provide strong evidence that the V. cholerae VAS genes of V. cholerae strain V52 encode a unique secretion apparatus that minimally transports proteins to the exterior of V. cholerae cells by a mechanism that does not require hydrophobic N-terminal signal sequences. We hypothesize that vasA, vasK, and probably other proteins encoded by the IAHP gene cluster are structural components of the VAS secretion apparatus because these proteins do not appear in the culture supernatant fluids. The fact that concentrated culture supernatants of V52 that contain Hcp, VgrG-1, VgrG-2, and VgrG-3 display no cytotoxicity for Dictyostelium amoebae further suggests that the likely function of the VAS-encoded secretion system is to transport one or more of these secreted proteins into the cytosol of eukaryotic target cells upon cell-cell contact. The actin-crosslinking activity of VgrG-1 (23) is consistent with the hypothesis that this protein might be an “effector” that is delivered into target cells, including mammalian target cells. However, it is also possible that Hcp, VgrG-1, VgrG-2, and VgrG-3 themselves are components of the bacterial secretion apparatus or components of a secreted “translocon” (26) that assembles in target cells membranes.
Bioinformatic analysis (6) previously found that vasK and vasF (Fig. 2) show a high degree of sequence conservation with L. pneumophila icmF and icmH (dotU) (13, 14), whereas an adjacent gene (VCA0116) has homology to the ClpB family of AAA proteins (27), which have recently been implicated in the ATP-dependent export of proteins by the T3SS pathway (28). These three genes, together with several other closely linked genes, constitute a region that Das and Chaudhuri (6) designated the IAHP gene cluster, which can be found in one or more copies in many pathogenic and commensal Gram-negative bacterial species, including Yersinia pestis, P. aeruginosa, E. coli O157, S. enterica, Agrobacterium tumefaciens, E. ictaluri, and R. leguminosarum (6, 8, 19, 20, 29).
In several bacterial species, mutations in genes belonging to IAHP clusters have been implicated in infection and virulence-related processes ranging from intracellular survival in macrophages to pathogenicity in fish or mice (19, 30-33). In several of these cases, the IAHP genes implicated have been found to be up-regulated in macrophages or during infection (19, 32, 34). Work in R. leguminosarium is frequently cited as evidence that genes in an IAHP cluster are involved in secretion of proteins (8, 29). However, the protein most clearly demonstrated to be secreted in an IAHP-dependent fashion in R. leguminosarium was a ribose binding protein that is synthesized with a typical N-terminal hydrophobic signal sequence (8). Complicating matters further, Das et al. (35) reported that an icmF (i.e., vasK) mutant of V. cholerae strain O395 showed a pleiotropic phenotype (i.e., was affected in motility, adherence, and conjugation recipient efficiency). However, because O395 is not virulent for Dictyostelium, we conclude that mutations in icmF/vasK probably affects these phenotypes in a nonspecific fashion. Evidence for this comes from 2D gel analysis that shows some secreted proteins with hydrophobic amino terminus secretion signals (e.g., protease, PrtV, and hemolysin, HlyA) are present in reduced abundance in the supernatant fluids of our vasK mutant; reduced secretion of HlyA does not, however, affect the hemolytic properties of a vasK mutant, which forms halos on blood plates comparable to wild-type V52 (Fig. 1_B_). Thus, mutations in icmF homologs may be pleiotropic and affect the cell architecture in a nonspecific fashion. In contrast, a vasH mutant shows no alteration in the levels of PrtV, HlyA, or other secreted proteins that carry hydrophobic amino-terminal signal sequences (see Fig. 5). Thus, the V. cholerae VAS pathway is only essential for secretion of Hcp, VgrG-1, VgrG-2, and VgrG-3, all of which lack such signal sequences. By analogy, we predict that genes associated with IAHP clusters in other Gram-negative species will encode a secretion system that exports a narrow subset of protein effectors by a mechanism that does not require its substrates to have N-terminal hydrophobic signal sequences.
To date, five distinct prokaryotic secretion mechanisms, classified as types I-V, have been described (36, 37). Because the VAS gene cluster represents an ensemble of genes that is novel in its linkage to a secretion pathway of proteins that lack N-terminal signal sequences, we propose to call this type of secretion system a type VI secretion system (T6SS). The genes of the IAHP gene cluster (6, 8, 19, 20, 29), together with other VAS, Hcp, and VgrG genes, likely encode the T6SS apparatus and several of its translocated effectors. Because so many pathogenic Gram-negative bacterial species carry VAS gene clusters, we predict that the primary function of the T6SS is to mediate extracellular export of virulence factors and their translocation into target eukaryotic cells. Because this transport will likely have deleterious effects on the host, the components of the T6SS constitute exciting candidates for the development of preventative or therapeutic vaccines and targets for antimicrobial drug development.
Materials and Methods
Strains and Culture Conditions. D. discoideum strain AX3 was used in all experiments. AX3 was grown in liquid HL/5 cultures or in lawns of K. aerogenes on SM/5 plates, as described by Sussman (38). V. cholerae O37 serogroup strain V52 and El Tor biotype strain N16961 were used in all experiments. E. coli strains DH5α-λpir and SM10λpir were used for cloning and mating, respectively. All bacterial strains were grown in Luria broth (LB). J774 cells were obtained from the American Type Culture Collection.
Transposon Library of V. cholerae Strain V52. Mariner transposon TnAraOut was introduced into V. cholerae by using DTH2129-2 (gift of D. T. Hung, Harvard Medical School), a derivative of suicide plasmid pNJ17 (7). E. coli BW20767 was used to mobilize DTH2129 by conjugation into streptomycin-resistant V. cholerae strain V52 by incubating donor and recipient at a 10:1 ratio on LB agar for 60 min at 37°C. Bacteria were collected, and dilutions were plated on LB agar containing 100 μg/ml kanamycin and 100 μg/ml streptomycin to select for V. cholerae clones carrying TnAraOut.
Isolation of _Dictyostelium_-Attenuated V. cholerae. Amoebae (5 × 106) were mixed with 1 × 103 TnAraOut mutants of V. cholerae strain V52 and plated onto SM/5 plates containing 100 μg/ml kanamycin. Plates were incubated at 22°C for 3 days and then scored for notched V. cholerae colonies formed by _Dictyostelium_-attenuated V. cholerae mutants. Bacteria were restreaked on SM/5 plates containing 5 μg/ml blasticidin to kill amoebae.
Plaque Assay. Bacteria were grown in LB for 16 h, pelleted by centrifugation, washed once, and resuspended in SorC (16.7 mM Na2H/KH2PO4/50 μM CaCl2, pH 6.0) at a final OD of 5.5 at 600 nm. D. discoideum cells from midlogarithmic cultures were collected by centrifugation, washed once with SorC (38), and added to the bacterial suspensions at a final concentration of 5 × 102 cells per ml suspension; 0.2 ml of this mixture was plated on SM/5 plates and allowed to dry under a sterile flow of air. Plates were incubated for 3-5 days and examined for plaques formed by Dictyostelium amoebae.
Plate Killing Assay. Bacterial strains were plated with D. discoideum on SM/5 plates as described for the plaque assay. After 24 h, bacterial lawns containing amoebae were collected and enumerated by plating with tetracycline-resistant E. coli B/r on SM/5 plates containing 30 μg/ml tetracycline. Plaques were counted 3 days later.
Secretion Assay. Hcp was isolated from midlog cultures of V. cholerae. Briefly, culture supernatants were sterilized by passing through a 0.2-μm filter (Millipore), and proteins were precipitated with trichloroacidic acid (TCA) and subjected to 4-12% gradient SDS/PAGE. Extracellular secretion of epitope-tagged substrates was determined by growing V. cholerae strains maintaining a plasmid with tagged Hcp-2 and VgrG-2 fused to the arabinose-inducible PBAD promoter in LB containing 0.1% arabinose. Midlog cultures were harvested and cells were isolated by centrifugation. Cells and 0.2-μm filtered supernatants were left untreated or incubated with either 0.1 mg/ml proteinase K in the presence or absence of 1% SDS. After a 20-min incubation at room temperature, protease inhibitor PMSF (final concentration of 1 mM) was added to all samples, and proteins were precipitated with TCA, solubilized in sample buffer, and separated on a SDS/PAGE for immunoblotting with vesicular stomatitis virus glycoprotein antisera. The quality of pellet and supernatant fractionation was determined by localizing periplasmic β-lactamase.
Cell Rounding of J774 Macrophages. Bacterial midlog cultures grown in LB were washed with PBS and added to adherent J774 cells (multiplicity of infection ≈10) cultured in advanced DMEM (GIBCO) containing 10% FCS. Cells were infected for 2 h at which time supernatants were replaced with 3% paraformaldehyde to fix adherent cells. Saved supernatants were sterilized by centrifugation and treatment with 0.1 mg/ml gentamicin for 30 min at 37°C and transferred to wells containing uninfected J774 cells. Cell rounding was monitored with a Nikon Diaphot 200 inverted microscope equipped with computer interface.
Supplementary Material
Supporting Information
Acknowledgments
We thank Deborah Hung, Vincent Tam, Elizabeth Shaknovich, Vincent Lee, Ann Thanawastien, Arne Rietsch, David Raskin, Joseph Mougous, Vincent Lee, Richard Kessin, and Virginia Miller for helpful discussions; Meike Gummich for kind support; Matt Waldor (Tufts University, Boston) for strain V52; Ewen Cameron for characterization of the TnAraOut mutants; and Su Chiang for editing the manuscript. This work was supported by National Institutes of Health Grant AI-18045 (to J.J.M.), Ellison Medical Foundation Grant ID-T-0007-01 (to J.J.M.), and National Institute of Allergy and Infectious Diseases Contract N01-AI-30071 (to C. M. Fraser, The Institute for Genomic Research).
Author contributions: S.P., A.T.M., J.F.H., and J.J.M. designed research; S.P., A.T.M., D. Sturtevant, B.K., D. Sarracino, and W.C.N. performed research; S.P. contributed new reagents/analytic tools; S.P., A.T.M., D. Sturtevant, B.K., D. Sarracino, W.C.N., J.F.H., and J.J.M. analyzed data; and S.P. and J.J.M. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: Hcp, hemolysin-coregulated protein; IAHP, IcmF-associated homologous protein; LB, Luria broth; T3SS, type III secretion system; T4SS, type IV secretion system; VAS, virulence-associated secretion.
References
- 1.Zinnaker, Y. & Carpenter, C. C. (1972) Johns Hopkins Med. 131**,** 403-411. [PubMed] [Google Scholar]
- 2.Dziejman, M., Balon, E., Boyd, D., Fraser, C. M., Heidelberg, J. F. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99**,** 1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinert, M. & Heuner, K. (2005) Cell. Microbiol. 7**,** 307-314. [DOI] [PubMed] [Google Scholar]
- 4.Pukatzki, S., Kessin, R. H. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99**,** 3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vance, R. E., Rietsch, A. & Mekalanos, J. J. (2005) Infect. Immun. 73**,** 1706-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, S. & Chaudhuri, K. (2003) In Silico Biol. 3**,** 287-300. [PubMed] [Google Scholar]
- 7.Judson, N. & Mekalanos, J. J. (2000) Nat. Biotechnol. 18**,** 740-745. [DOI] [PubMed] [Google Scholar]
- 8.Bladergroen, M. R., Badelt, K. & Spaink, H. P. (2003) Mol. Plant Microbe. Interact. 16**,** 53-64. [DOI] [PubMed] [Google Scholar]
- 9.Williams, S. G., Varcoe, L. T., Attridge, S. R. & Manning, P. A. (1996) Infect. Immun. 64**,** 283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal, G., Purcell, M. & Shuman, H. A. (1998) Proc. Natl. Acad. Sci. USA 95**,** 1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marra, A., Blander, S. J., Horwitz, M. A. & Shuman, H. A. (1992) Proc. Natl. Acad. Sci. USA 89**,** 9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon, J. M., Rupper, A., Cardelli, J. A. & Isberg, R. R. (2000) Infect. Immun. 68**,** 2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sexton, J. A., Miller, J. L., Yoneda, A., Kehl-Fie, T. E. & Vogel, J. P. (2004) Infect. Immun. 72**,** 5983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.VanRheenen, S. M., Dumenil, G. & Isberg, R. R. (2004) Infect. Immun. 72**,** 5972-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal, G. & Shuman, H. A. (1998) Trends Microbiol. 6**,** 253-255. [DOI] [PubMed] [Google Scholar]
- 16.Kirby, J. E. & Isberg, R. R. (1998) Trends Microbiol. 6**,** 256-258. [DOI] [PubMed] [Google Scholar]
- 17.Dziejman, M., Serruto, D., Tam, V. C., Sturtevant, D., Diraphat, P., Faruque, S. M., Rahman, M. H., Heidelberg, J. F., Decker, J., Li, L., et al. (2005) Proc. Natl. Acad. Sci. USA 102**,** 3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, S., Chakrabortty, A., Banerjee, R., Roychoudhury, S. & Chaudhuri, K. (2000) FEMS Microbiol. Lett. 190**,** 87-91. [DOI] [PubMed] [Google Scholar]
- 19.Parsons, D. A. & Heffron, F. (2005) Infect. Immun. 73**,** 4338-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, M. M., Fernandez, D. L. & Thune, R. L. (2002) Dis. Aquat. Organ. 52**,** 93-107. [DOI] [PubMed] [Google Scholar]
- 21.Mota, L. J. & Cornelis, G. R. (2005) Ann. Med. 37**,** 234-249. [DOI] [PubMed] [Google Scholar]
- 22.Christie, P. J., Atmakuri, K., Krishnamoorthy, V., Jakubowski, S. & Cascales, E. (2005) Annu. Rev. Microbiol. 59**,** 451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheahan, K. L., Cordero, C. L. & Satchell, K. J. (2004) Proc. Natl. Acad. Sci. USA 101**,** 9798-9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fullner, K. J. & Mekalanos, J. J. (2000) EMBO J. 19**,** 5315-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa-Arredondo, P., Heuser, J. E., Akopyants, N. S., Morisaki, J. H., Giono-Cerezo, S., Enriquez-Rincon, F. & Berg, D. E. (2001) Infect. Immun. 69**,** 1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goure, J., Broz, P., Attree, O., Cornelis, G. R. & Attree, I. (2005) J. Infect. Dis. 192**,** 218-225. [DOI] [PubMed] [Google Scholar]
- 27.Squires, C. L., Pedersen, S., Ross, B. M. & Squires, C. (1991) J. Bacteriol. 173**,** 4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akeda, Y. & Galan, J. E. (2005) Nature 437**,** 911-915. [DOI] [PubMed] [Google Scholar]
- 29.Roest, H. P., Mulders, I. H., Spaink, H. P., Wijffelman, C. A. & Lugtenberg, B. J. (1997) Mol. Plant Microbe. Interact. 10**,** 938-941. [DOI] [PubMed] [Google Scholar]
- 30.Rao, P. S., Yamada, Y., Tan, Y. P. & Leung, K. Y. (2004) Mol. Microbiol. 53**,** 573-586. [DOI] [PubMed] [Google Scholar]
- 31.Folkesson, A., Lofdahl, S. & Normark, S. (2002) Res. Microbiol. 153**,** 537-545. [DOI] [PubMed] [Google Scholar]
- 32.Gray, C. G., Cowley, S. C., Cheung, K. K. & Nano, F. E. (2002) FEMS Microbiol. Lett. 215**,** 53-56. [DOI] [PubMed] [Google Scholar]
- 33.Nano, F. E., Zhang, N., Cowley, S. C., Klose, K. E., Cheung, K. K., Roberts, M. J., Ludu, J. S., Letendre, G. W., Meierovics, A. I., Stephens, G. & Elkins, K. L. (2004) J. Bacteriol. 186**,** 6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golovliov, I., Ericsson, M., Sandstrom, G., Tarnvik, A. & Sjostedt, A. (1997) Infect. Immun. 65**,** 2183-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das, S., Chakrabortty, A., Banerjee, R. & Chaudhuri, K. (2002) Biochem. Biophys. Res. Commun. 295**,** 922-928. [DOI] [PubMed] [Google Scholar]
- 36.Pugsley, A. P., Francetic, O., Driessen, A. J. & de Lorenzo, V. (2004) Mol. Microbiol. 52**,** 3-11. [DOI] [PubMed] [Google Scholar]
- 37.Desvaux, M., Parham, N. J. & Henderson, I. R. (2004) Curr. Issues Mol. Biol. 6**,** 111-124. [PubMed] [Google Scholar]
- 38.Sussman, M. (1987) Methods Cell. Biol. 28**,** 9-29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information