APID: Agile Protein Interaction DataAnalyzer (original) (raw)
Abstract
Agile Protein Interaction DataAnalyzer (APID) is an interactive bioinformatics web tool developed to integrate and analyze in a unified and comparative platform main currently known information about protein–protein interactions demonstrated by specific small-scale or large-scale experimental methods. At present, the application includes information coming from five main source databases enclosing an unified sever to explore >35 000 different proteins and 111 000 different proven interactions. The web includes search tools to query and browse upon the data, allowing selection of the interaction pairs based in calculated parameters that weight and qualify the reliability of each given protein interaction. Such parameters are for the ‘proteins’: connectivity, cluster coefficient, Gene Ontology (GO) functional environment, GO environment enrichment; and for the ‘interactions’: number of methods, GO overlapping, iPfam domain–domain interaction. APID also includes a graphic interactive tool to visualize selected sub-networks and to navigate on them or along the whole interaction network. The application is available open access at http://bioinfow.dep.usal.es/apid/.
INTRODUCTION
Genome-wide and proteome-wide technologies on modern biochemistry and molecular biology provide vast and quickly increasing amounts of biological data that need to be stored, compared and organized using comprehensive and dynamic open access computational tools. One of the most productive areas is the one of protein–protein interactions and interactome data (1). The data about the interaction of two or more proteins come either from small-scale experimental work or from large-scale experimental methods. Both kind of data are being included in biological databases focus on protein interaction and several bioinformatic initiatives have been undertaken to this purpose [see reviews (1–3)]. However, several studies in recent years have reported comparative assessments of large-scale and high-throughput protein–protein interaction data (4,5) indicating that data quality is a critical problem in these datasets, that many times include a high proportion of false positive interactions due to low accuracy of the methods. Some bioinformatic and computational work has been done to assess the reliability of high-throughput observations and to gain confidence in the data (6–9). However, we consider that more efforts based on validated experimental information are essential to improve the quality of the protein–protein interaction data and therefore to improve the biological information that can be inferred from the interactome networks.
At present time, there are several major protein interaction databases [Biomolecular Interaction Network Database (BIND), Database of Interacting Proteins (DIP), InAct] (10–12) that are collecting the increasing amount of biological data produced in this area. Data about the interactions of two or more proteins are stored in many published scientific papers and the databases extract and integrate such information. However, each database has its own extraction, curation and storage protocols, and not all of them explore the same scientific papers. In fact, we have observed that the intersection and overlap between these source databases is small, and therefore in many cases their information is complementary and can be unified to increase and improve our knowledge about interactome networks. At the same time, the existence of several experimental evidences about many protein–protein interactions, reported by different literature references, allows to increase the number of methods that validate any given interaction. We consider that an integrative effort is essential to draw more clear maps about the protein interaction network and to explore sub-networks for specific proteins or protein families.
Keeping the key critical needs described above, i.e. (i) better assessing the quality of the protein–protein interaction data and (ii) more comprehensive integration of main currently known protein–protein interactions; we have developed an interactive bioinformatics web tool to integrate and analyze in a common and comparative platform main known protein interactomes. This web tool can be very helpful for the research on a specific protein or protein family, because it includes some score parameters that weight the reliability and functional meaning of the interactions.
METHODS
Agile Protein Interaction DataAnalyzer (APID) design tries to be as simple and light as possible keeping the minimal information to provide a correct and easy access to all included data sets. This design follows the software engineering methodology named ‘agile’ (13), that embraces software development using lightweight and adaptable methods. In this way, agile methods demand the idea of evolutionary design and seek to assume changes, allowing them to occur along all the live cycle of a product. Changes are controlled and easy to implement and the attitude of the designer is to enable them. APID has been designed following this strategy to achieve the purpose of a useful and active integration of the protein–protein interaction source databases included.
All the work has been developed in Java programming language (http://java.sun.com/), and a J2EE architecture has been used to built the web interface and the applet graphic tool described below. For the parsing of source data we have used SAX and DOM Java programs to extract the information from the XML files, and JDBC programs to insert the processed data in the server. After the parsing efforts we still found problems to unify all the source data, being the main obstacle the heterogeneous and multiple protein identifiers given by the different sources, that many times cause false disjunction and incoherence in the data. To solve it we used the proteins sequences as the most unique and biological meaningful ‘protein code’, that allowed a good unification using algorithm BLAST2 (14) to find in UniProt each protein given by the source databases. Once a protein was recognized based on sequence alignment, we linked to it a univocal UniProt code. Together with the protein univocal code to obtain a coherent and uniform data, we also had to reach coherence about the experimental method or methods that validate any given interaction. The identification of the method also allows to find the existing consensus or agreement between the different databases for any given interaction. In this way, we have obtained a protocol able to store and unify protein interaction databases in a clear uniform structure, maintaining the integrity of the data and correcting some existing failures found in the original files.
Following the described strategy, the data unification has been done based on three key reference identifiers (IDs): (i) UniProt ID (i.e. UniProt accession number), to allow a specific identification of each protein and a direct link to its sequence and to the rest of the curated protein information included in UniProt (15); (ii) PSI-MI ID, to unify the experimental methods used in different publications to a common terminology developed by PSI-MI (16) (i.e. to a controlled vocabulary with standard identifiers); (iii) PubMed ID (PMID), to link each interaction validated by a given experimental method to a specific PubMed literature reference, and also to assign experimental method identifiers to the PubMed publications that describe each method. These main key identifiers constitute a simple information core that makes APID an agile tool to access and search through the interactomes.
At present, APID integrates data coming from five main source databases: BIND (10), DIP (11), HPRD (Human Protein Reference Database) (17), IntAct (Database system and analysis tools for protein interaction data) (12) and MINT (Molecular Interactions Database) (18). The data included in APID coming from these source databases correspond only to protein–protein interactions (i.e. not interactions of proteins with other ligands like DNA and the like) and the interactions have to be experimentally validated with a PubMed reference given. At the same time, as indicated above each protein has to be identified by its sequence and its UniProt code. In all cases, the web tool includes for each interaction links to the original files of the source databases, and to the PubMed references that validated each interaction. Finally, each protein includes links to the corresponding UniProt file and to other related databases [like InterPro, Pfam, Gene Ontology (GO), Ensembl, NCBI Gene].
PROGRAM DESCRIPTION
Workflow
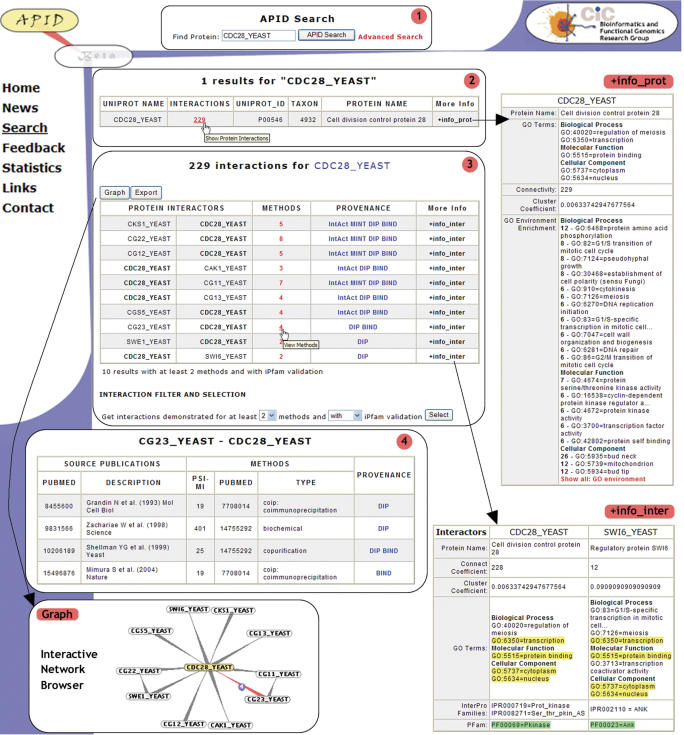
To illustrate the workflow and the different tools included in APID web server, we present in Figure 1 a schematic description of the steps usually given for a query. Each box included in the figure corresponds to a web window. Starting with box 1 a protein name, protein identifier, protein description or part of it is inserted in the general ‘APID search’ tool. As an example, CDC28 from yeast (‘CDC28_YEAST’) is the starting query. In box 2 the figure shows the result given by the search for ‘CDC28_YEAST’ that is a UniProt entry name. A simple table with only one row is presented because only one protein is found. This table includes six columns with information about the protein: the UniProt entry name, the number of interactions, the UniProt ID number, the taxon (NCBI Taxonomy ID), the protein name or description and a link with more information about the protein. Clicking on the link ‘+info_prot’ a new window with more detailed information about the query protein is displayed, including links to other referred biomolecular databases. The ‘+info_prot’ file also includes some calculated parameters about the protein interaction network (i.e. connectivity and cluster coefficient) and about the protein functional environment based on GO annotation (i.e. GO environment and GO environment enrichment). In this way, it can be seen that connectivity 229 corresponds to the number of proteins that interact with CDC28 from yeast. This is a big number of interactions that may include false positives. Clicking on 229 in box 2 a new window is displayed including a table with details about the 229 interactions that have been reported for CDC28_YEAST. This table (Figure 1, box 3) has five columns with information about: the interaction protein partners, the number of methods that validate each interaction, the provenance source databases (with links to them) and a final column with more information about the interaction: ‘+info_inter’. Clicking on any ‘+info_inter’ a new window with more detailed information about the corresponding interaction protein pair is displayed, including marks in yellow that show GO terms overlapping and marks in green that show iPfam doamin–domain interaction. This is the case shown for protein pair CDC28_YEAST and SWI6_YEAST. At the same time, as presented in box 3 a certain subset of protein interactions can be selected from the original 229 interaction using a filter that limitates the display to interaction pairs proven by two methods at least and that also show iPfam domain–domain interaction. Doing this the number of interaction partners for CDC28 is reduced to only 10 proteins (as seen in box 3). Finally, clicking on the number of methods APID displays another window with the information about all the methods that validate any given protein–protein interaction (e.g. CG23_YEAST – CDC28_YEAST in box 4), presenting for each method: (i) the publications that describe and prove the interaction, linking to PubMed by the pubmed accession number (PMID) and including a description about the publication (i.e. first author, year, journal); (ii) the type of method (i.e. name given by the controlled vocabulary), the PMID of the publication that explains such experimental technique or method, and the PSI-MI method identifier; (iii) the source databases that include these data.
Figure 1.
Schematic representation showing APID workflow example. Search query: ‘cdc28_yeast’ (box 1). Protein found with the text search (box 2) and its additional information (+info_prot). The found protein presents 229 protein partners and the filtered interactions (with iPfam validation and at least two methods) are show in next chart (box 3), that links to the graphical tool APIN where the corresponding interaction network can be visualized and explored in an interactive way. Each interaction also links to its additional information (+info_inter). The experimental methods that prove each interaction are indicated and the details about such methods for the protein pair CG23_YEAST and CDC28_YEAST are shown by clicking the corresponding number 4 (box 4). Each presented box corresponds to consecutive web pages in the APID website.
When any subset of protein interaction pairs is selected, as done in box 3, APID also includes a ‘Graph’ tool that opens a graphical interactive network browser, where the proteins are nodes and the interactions edges. This application tool visualizes dynamically the data, and allows interactive exploring and navigating along the network. The tool includes information about the proteins and the interactions that can be shown by opening windows with basic information and with links to the reference databases UniProt, PubMed and so on.
Statistics and overlap between source databases
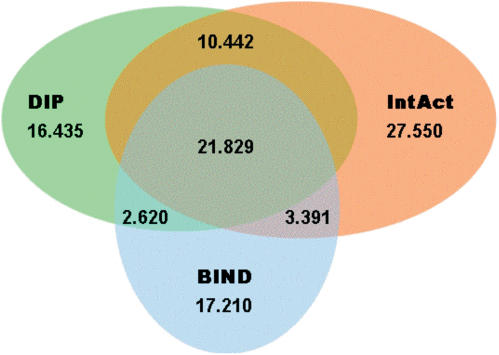
At present time (February 2006) the ‘statistics’ section in APID web tool shows that the application includes >35 000 proteins from several organism and >111 000 interactions. The ‘statistics’ web page also presents the proteins and interactions per organism, the number of methods that report the same interaction and the detail numbers for the overlap and different intersections between the five source databases used. A more simple and graphical analysis of the overlapping of only three protein interaction databases (BIND, DIP and IntAact) is presented in Figure 2, that shows a Venn diagram with the number of interactions for the multiple intersections between these three databases. It is worthy of note that 62% of the overall protein interactions included in BIND, DIP and IntAct are presented in only one of these databases, i.e. they are exclusive to one of the protein interaction resources.
Figure 2.
Venn diagram with the number of interactions for the multiple intersections between BIND, DIP and IntAct.
Protein interaction network assessment
High-throughput experimental technologies used to prove protein–protein interactions of complete proteomes, using the two-hybrid system (19) or mass spectrometry (20), have highly increased the data included in the protein interaction databases. However little overlap between the high-throughput datasets (6) and frequent disagreement with small-scale experiments jeopardize high-throughput interactions confidence. Several efforts have been undertaken to tackle this problem (4,5), but some critical steps to solve it are to achieve more comprehensive and integrated resources of the interactomic data and to include certain calculated parameters that weight the reliability of a given interaction between two proteins. These steps are the ones followed by APID application that is an integrated repository of interactions and includes some tools to assess the interactions:
- Number of methods: number of experimentally validated methods that prove a protein–protein interaction, given the PubMed reference and link.
- GO overlapping: tool that shows the GO terms assigned to each protein pair and marks the ones that are common to both.
- iPfam domain–domain interaction: tool that identifies the Pfam domains of each protein pair and marks the ones that interact according to iPfam database.
At the same time, APID also infers data about proteins in the interaction network, since the web application measures graph parameters as the connectivity and the cluster coefficient of each node, and it also qualifies the functional environment around any given protein using GO:
- GO environment: tool that identifies and lists all the Gene Ontology (GO) terms that are assigned to the proteins directly interacting with a query protein.
- GO environment enrichment: tool that selects the most-represented and non-self GO terms assigned to the proteins interacting with a query protein.
The use of these quality parameters will allow to make functional predictions about the proteins based on the assumption that interacting proteins tend to have related functions or at least to be involved in common biological processes. Using protein neighbourhoods such biological processes can be explored and mapped on a more reliable interactome landscape.
Acknowledgments
We thank Alberto de Luís and Ángel Román for helpful discussions. We acknowledge the funding and support provided by the Spanish Ministerio de Sanidad y Consumo, ISCIII (research grant ref. PI030920), Junta de Castilla y Leon (research grant ref. SA104/03) and Fundación BBVA (Bioinformatics Grants Program). C.P. holds a research grant for PhD from Junta de Castilla y Leon (ref. BOCyL no. 119, EDU/777/2005).
Conflict of interest statement. None declared.
REFERENCES
- 1.Xenarios I., Eisenberg D. Protein interaction databases. Curr. Opin. Biotechnol. 2001;12:334–339. doi: 10.1016/s0958-1669(00)00224-x. [DOI] [PubMed] [Google Scholar]
- 2.Legrain P., Wojcik J., Gauthier J.M. Protein–protein interaction maps: a lead towards cellular functions. Trends Genet. 2001;17:346–352. doi: 10.1016/s0168-9525(01)02323-x. [DOI] [PubMed] [Google Scholar]
- 3.De Las Rivas J., De Luis A. Interactome data and databases: different types of protein interaction. Comp. Funct. Genom. 2004;5:173–178. doi: 10.1002/cfg.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader G.D., Hogue C.W. Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 5.von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 6.Deane C.M., Salwinski L., Xenarios I., Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Bader J.S. Greedily building protein networks with confidence. Bioinformatics. 2003;19:1869–1874. doi: 10.1093/bioinformatics/btg358. [DOI] [PubMed] [Google Scholar]
- 8.Iossifov I., Krauthammer M., Friedman C., Hatzivassiloglou V., Bader J.S., White K.P., Rzhetsky A. Probabilistic inference of molecular networks from noisy data sources. Bioinformatics. 2004;20:1205–1213. doi: 10.1093/bioinformatics/bth061. [DOI] [PubMed] [Google Scholar]
- 9.Bader J.S., Chaudhuri A., Rothberg J.M., Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 10.Alfarano C., Andrade C.E., Anthony K., Bahroos N., Bajec M., Bantoft K., Betel D., Bobechko B., Boutilier K., Burgess E., et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cockburn A. Agile Software Development. London, UK: Addison-Wesly Longman; 2002. [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 17.Peri S., Navarro J.D., Amanchy R., Kristiansen T.Z., Jonnalagadda C.K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T.K., Gronborg M., et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 19.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 20.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]