The ORF7b Protein of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Is Expressed in Virus-Infected Cells and Incorporated into SARS-CoV Particles (original) (raw)
Abstract
Coronavirus replication is facilitated by a number of highly conserved viral proteins. The viruses also encode accessory genes, which are virus group specific and believed to play roles in virus replication and pathogenesis in vivo. Of the eight putative accessory proteins encoded by the severe acute respiratory distress syndrome associated coronavirus (SARS-CoV), only two—open reading frame 3a (ORF3a) and ORF7a—have been identified in virus-infected cells to date. The ORF7b protein is a putative viral accessory protein encoded on subgenomic (sg) RNA 7. The ORF7b initiation codon overlaps the ORF7a stop codon in a −1 shifted ORF. We demonstrate that the ORF7b protein is expressed in virus-infected cell lysates and from a cDNA encoding the gene 7 coding region, indicating that the sgRNA7 is bicistronic. The translation of ORF7b appears to be mediated by ribosome leaky scanning, and the protein has biochemical properties consistent with that of an integral membrane protein. ORF7b localizes to the Golgi compartment and is incorporated into SARS-CoV particles. We therefore conclude that the ORF7b protein is not only an accessory protein but a structural component of the SARS-CoV virion.
In 2002, a severe acute respiratory distress syndrome (SARS) of unknown etiology originated in China and subsequently spread to 32 countries on five continents, resulting in more than 8,000 suspected cases and a mortality rate of almost 11% (5). By March of 2003, the causative agent was identified as a novel coronavirus, SARS-CoV (13, 30, 51, 83), and the viral genome was sequenced and annotated (40, 62, 80a).
The SARS-CoV genome consists of genes characteristic of coronaviruses, including a bicistronic replicase (1a and 1b), nucleocapsid (N), envelope (E), membrane (M), and spike (S) (Fig. 1). These genes are essential for virus replication (4a). Coronaviruses and other members of the Nidovirales often contain additional genes that have no homologues in other, closely related viruses. These genes often are not required for viral replication in vitro but do contribute to viral fitness and pathogenesis in vivo (reviewed in reference 4a), hence their designation as accessory or group-specific genes. The SARS-CoV genome contains eight group-specific open reading frames (ORFs) (40, 62, 80a) that are conserved between human and animal isolates of SARS-CoV and in closely related coronaviruses (8, 32, 35). To date, the only group-specific ORFs documented to be expressed in SARS-CoV-infected cells are ORF3a and ORF7a (7, 14a, 33, 47, 79, 87a). ORF3a is incorporated into SARS-CoV particles and virus-like particles (25, 68). The ORF7a protein is expressed in SARS-CoV-infected cells (6, 7, 47), localizes to the Golgi compartment (28, 47) and, when overexpressed, induces apoptosis and inhibits host cell protein synthesis (7, 28, 77, 87a). The X-ray crystal and nuclear magnetic resonance structures of ORF7a have been determined (17, 47), and the protein has been found in SARS-CoV particles and virus-like particles (21).
FIG. 1.
SARS-CoV genomic organization and expression of the accessory protein ORF7b. (A) Genomic organization and sgRNAs of SARS-CoV. The ∼29-kb genome is transcribed into nine 3′-coterminal subgenomic RNAs in virus-infected cells. The sgRNAs encode replicase proteins (1a and 1b, white boxes) structural proteins (S, E, M, and N, black boxes), as well as eight proteins of unknown function (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b, gray boxes). The ORF7a and ORF7b proteins are encoded by sgRNA 7, with the ORF7a stop codon and the ORF7b initiation codon overlapping. The nucleotide and amino acid sequences for the ORF7a/ORF7b overlap region are shown. (B) Amino acid sequence of the ORF7b protein. The ORF7b protein is 44 amino acids in length with a single transmembrane domain (underlined) predicted by TMPred. (C) Vero cells were either mock infected or infected with SARS-CoV at an MOI of 5.0 for 18 h. The expression of ORF7b, N, and β-actin was determined by Western blotting. The band at ∼75 kDa is an unidentified cellular protein detected by the ORF7b sera. (D) Vero cells were either mock infected or infected with SARS-CoV at an MOI of 5.0 for 18 h. The cells were then processed for flow cytometry using antibodies specific for the ORF7a (horizontal axis) or ORF7b (vertical axis) proteins.
The SARS-CoV accessory genes are dispensable for virus replication in transformed cell lines, primary cell cultures, and the murine model of virus infection (69, 85a, 85b). However, the murine model for SARS-CoV infection does not show significant morbidity or mortality after infection. Therefore, it may not be an appropriate model for studying SARS-CoV pathogenesis and accessory gene function (75).
The SARS-CoV ORF7b protein is predicted to be a 44-amino-acid, highly hydrophobic protein with no identified sequence homology to other viral or cellular proteins. Given the highly hydrophobic nature of ORF7b, it has been hypothesized that ORF7b is a transmembrane protein and possibly a viral structural protein (1). Although the expression of ORF7b in virus-infected cells has not been documented to date, ORF7b-specific antibodies have been detected in SARS patient convalescent-phase serum (16b). Whereas the ORF7b coding region is highly conserved among sequenced SARS-CoV isolates, serial passage of SARS-CoV in Vero cells resulted in a 29-nucleotide deletion in ORF7b, demonstrating that the putative ORF7b protein is not essential for viral replication in vitro (80a).
We provide the first biochemical evidence for ORF7b expression in SARS-CoV-infected cells. We demonstrate that ORF7b localizes to the Golgi complex, is an integral membrane protein, is translated from the gene 7 mRNA via ribosomal leaky scanning, is associated with intracellular virus particles, and is present in purified virus particles.
MATERIALS AND METHODS
Cells.
Vero (ATCC CRL-1586) and 293T (76) cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum (Atlanta Biologicals), 1 mM glutamine, 1 mM sodium pyruvate (Invitrogen), 100 U of penicillin (Invitrogen)/ml, and 100 μg of streptomycin (Invitrogen)/ml. Cells were incubated in a 5% CO2 humidified incubator at 37°C.
Viruses.
All infections were performed in a biosafety level 3 laboratory using institution-approved procedures. SARS-CoV stocks (Urbani strain, courtesy of the Centers for Disease Control and Prevention) were generated by infecting Vero cells with SARS-CoV at a multiplicity of infection (MOI) of 0.01, followed by incubation at 37°C (47). Infected-cell supernatants were harvested 48 h postinfection, clarified by centrifugation at 1,000 × g, divided into aliquots, and stored at −70°C.
Infectious virus titers were determined by plaque assay or 50% tissue culture infectious dose (TCID50). PFU per ml were determined by serially diluting SARS-CoV in 10-fold increments and then incubating 250 μl of each dilution with Vero cells in six-well plates for 1 h at room temperature in quadruplicate. The inoculum was replaced with warm Dulbecco modified Eagle medium containing 10% fetal bovine serum, 1 mM glutamine, 1 mM sodium pyruvate, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 1% Ultrapure agarose (Invitrogen). When virus plaques were visible (usually after 4 days at 37°C), 1 ml of 1% formaldehyde in phosphate-buffered saline (PBS) was added to each well. One hour later, the formaldehyde and agarose was removed, and virus plaques were visualized by staining with naphthol blue-black. The TCID50 per ml was determined by serially diluting SARS-CoV in 10-fold increments and then adding 100 μl of each dilution to Vero cells in a 96-well plate in sextuplicate. The cells were incubated at 37°C for 4 days, visualized by staining with naphthol blue-black, and scored visually for cytopathic effect. A Reed and Muench calculation was used to determine TCID50 per ml (59).
To generate infected cells and infected cell lysates, Vero cells were incubated in 250 μl of medium containing SARS-CoV at various MOIs for 1 h at 37°C. The medium was aspirated, fresh medium was added, and cells were incubated for designated time periods at 37°C. Cells were lysed in 1% sodium dodecyl sulfate (SDS) or fixed with 2% paraformaldehyde in PBS.
For virion purification, Vero cells were infected at an MOI of 0.1 and incubated at 37°C for 48 h. The infected cells were lysed with 1% SDS. The infected-cell supernatant was collected and centrifuged at 1,000 × g for 10 min to remove cells and cellular debris.
Ultracentrifugation.
The virus-containing supernatant was collected, and virus was inactivated by either (i) the addition of an equal volume of 4% paraformaldehyde and incubation at 4°C overnight or (ii) the addition of 0.05% β-propiolactone and incubation at 4°C overnight, followed by incubation at 37°C for 2 h to allow β-propiolactone breakdown into nontoxic by-products (58, 71). The inactivated virions were pelleted by centrifugation over a 20% sucrose cushion at 100,000 × g for 1 h in an SW41 ultracentrifuge rotor. The pellet was resuspended in 2× Laemmli SDS loading buffer for Western blotting, in PBS for analysis by electron microscopy, or in NTE (100 mM Nacl, 10 mM Tris-HCl [pH 7.4], and 1 mM EDTA) for analysis by sucrose gradient ultracentrifugation. For sucrose gradient ultracentrifugation, the resuspended virus was loaded onto a continuous 20 to 60% sucrose gradient in NTE and centrifuged at 100,000 × g for 18 h in an SW41 ultracentrifuge rotor (50, 88). Then, 1-ml fractions were collected from the top of the centrifuge tube, and the virus was concentrated from each fraction by addition of 11 ml of NTE, followed by centrifugation at 100,000 × g for 2 h in an SW41 rotor. The resulting pellets were resuspended in 100 μl of 2× Laemmli SDS sample buffer and loaded onto SDS-15% polyacrylamide gel electrophoresis (PAGE) gels.
Plasmids.
SARS-CoV viral RNA (Urbani strain, GenBank accession no. AY278741) was purified from infected cell supernatant using the QIAamp viral RNA purification kit (QIAGEN) according to the manufacturer's protocol. A cDNA encoding the entire gene 7 (7ab) was amplified by reverse transcription (RT)-PCR from the purified viral RNA using Superscript One-Step RT-PCR mix (Invitrogen) and oligonucleotide primers (primer sequences available upon request). Based on the Urbani strain sequence with the ORF7a initiation codon at positions 27273 to 27275, nucleotides 27268 through 27772 were isolated by PCR, cloned into the eukaryotic expression plasmid pCAGGS (48), and sequenced. This construct was subsequently used as a template for cloning all remaining ORF7ab and ORF7b constructs. The ORF7b coding region spanning nucleotides 27638 to 27772 (Urbani sequence) was isolated by PCR and cloned into the pCAGGS plasmid. PCR was then used to add the myc epitope sequence to the C terminus of the ORF7b cDNA (ORF7b-myc). The cDNA constructs encoding ORF7a and ORF7a fused to green fluorescent protein (ORF7a-GFP) were described previously (47).
For bacterial expression, full-length ORF7b was isolated by PCR and cloned into the pET28a bacterial expression vector (Invitrogen), resulting in an ORF7b construct containing an N-terminal His6 epitope tag (His6-ORF7b).
The ORF7ab constructs detailed in Fig. 2A were generated by using overlap PCR and encode the following mutations: 7ab 7a Kozak, nucleotides 27268 to 27272 (CGAAC) to GCCACC and nucleotides 27276 to 27277 (AA) to GC; 7ab ATG, nucleotide 27443 (T) to A; 7ab 7a Stop, nucleotide 27276 (A) to T and nucleotides 27291 to 27292 (CT) to TA;7ab 7a Stop KO, nucleotides 27641 to 27642 (AA) to GC; 7ab 7a Start KO, nucleotide 27275 (G) to C; and 7ab 7b Start KO, nucleotide 27640 (G) to A.
FIG. 2.
The ORF7b protein is translated via ribosome leaky scanning. (A). Schematic diagram of the cDNA constructs used to assess the translation of ORF7b. Gene 7 constructs included wild-type sequence (7ab), an optimal translation initiation consensus sequence at the ORF7a initiation codon (7ab 7a Kozak), a point mutation within the ORF7a coding region creating an out-of-frame ATG that does not alter the ORF7a amino acid sequence (7ab ATG), two point mutations within the ORF7a ORF resulting in two in-frame stop codons (7ab 7a Stop), a point mutation eliminating the ORF7a stop codon (7ab 7a Stop KO), a point mutation eliminating the ORF7a initiation codon (7ab 7a Start KO), and a point mutation eliminating the ORF7b initiation codon (7ab 7b Start KO). All ATG sequences are boxed, and stop codons are italicized. (B) 293T cells were transfected with plasmids encoding the indicated cDNAs and analyzed for ORF7a (horizontal axis) and ORF7b (vertical axis) expression by flow cytometry at 18 h posttransfection. (C) 293T cells were transfected with plasmids encoding the indicated cDNAs, lysed 18 h posttransfection, and analyzed for ORF7b and β-actin expression by Western blotting. (D) The ORF7b protein band intensities from panel C were quantified and normalized to β-actin expression by using phosphorimager analysis. The quantified data are from five independent experiments.
A plasmid (pEGFP-C1 vector; Clontech) containing the cDNA for GFP fused to the N terminus of Rab11 (GFP-Rab11) was provided by Phil Stahl, Washington University, St. Louis, MO. The expression vector for the influenza A virus M2 cDNA (pCAGGS M2) was described previously (42, 43). PCR was used to add the myc epitope sequence to the C terminus of the M2 ORF, generating the pCAGGS M2-Myc plasmid. The S15-GFP cDNA contains the GFP ORF fused to the first 15 amino acids of Src protein tyrosine kinase and has been described previously (60a).
Antiserum to ORF7b.
The pET28a ORF7b plasmid was transformed into BL21 cells and grown in 1 liter of Luria broth at 37°C to an optical density of 0.7 at 600 nm. The cells were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 25°C, collected by centrifugation at 7,000 × g, resuspended in denaturing lysis buffer (6 M guanidine HCl, 20 mM NaPO4, 500 mM NaCl [pH 7.8]), and sonicated on ice (550 Sonic Dismembranator). The His6-ORF7b protein was then purified using a ProBond column per manufacturer recommendations (Invitrogen). Purified His6-ORF7b protein was used to immunize New Zealand White rabbits using Ribi adjuvant (Covance, Pennsylvania).
Transient transfection of mammalian cells.
293T cells (1.5 × 105) were plated into a 9.5-cm2 tissue culture dish (Corning) and allowed to attach for 24 h at 37°C. Approximately 1 μg of DNA was mixed with 2 μl of LT-1 transfection reagent (Mirus) and incubated with the cells for 18 h at 37°C. For confocal microscopy, 7.5 × 104 Vero cells were plated into a 9.5-cm2 dish (Corning) containing glass coverslips and allowed to attach for 24 h at 37°C. Approximately 0.5 μg of DNA was mixed with 2 μl of LT-1 transfection reagent (Mirus) and incubated with the cells for 18 h at 37°C.
FACS.
Fluorescence-activated cell sorting (FACS) was performed as described previously (47). Cells were incubated with the anti-ORF7a murine monoclonal antibody (MAb) 2E11 (1:1,000 dilution) or anti-ORF7b rabbit serum (1:1,000 dilution), followed by goat anti-mouse immunoglobulin G (IgG; 1:500 dilution, Alexa Fluor 488 labeled; Molecular Probes) or goat anti-rabbit IgG (1:500 dilution, AlexaFluor 647 labeled; Molecular Probes) secondary antibody. All antibody dilutions were made in blocking buffer (PBS with 3% normal goat serum [NGS], 0.5% bovine serum albumin, and 0.1% saponin), and wash buffer consisted of PBS with 0.1% saponin. The cells were analyzed with a FACSCalibur dual-laser flow cytometer (Becton Dickinson), and data were collected by using CellQuest software.
Indirect immunofluorescence analysis and confocal microscopy.
For indirect immunofluorescence analysis, cells were washed three times with PBS, fixed with 2% paraformaldehyde in PBS for 10 min, washed three times with PBS, and then blocked in PBS containing 3% NGS (Sigma) and 0.5% bovine serum albumin for 10 min. The cells were then incubated with antibody diluted in blocking buffer. Primary antibodies were used as follows: rabbit anti-ORF7b polyclonal serum (1:1,000 dilution), anti-ORF7a mouse MAb 2E11 (1:1,000 dilution) (47), anti-influenza M2 mouse MAb 14C2 (1:1,000 dilution) (42, 43), anti-GM130 mouse MAb (1:100 dilution; BD Biosciences), anti-Golgin-97 mouse MAb (1:100 dilution; Molecular Probes), goat anti-calnexin IgG (1:100 dilution; Santa Cruz Biotechnology), rabbit anti-ERGIC53 IgG (1:100 dilution, Sigma), anti-CD63 mouse MAb (1:1,000 dilution, clone H5C6; BD Biosciences Pharmingen), anti-c-Myc mouse MAb (1:50 dilution, supernatant from hybridoma 9E10, ATCC CRL-1729). Cells were then washed three times with PBS and incubated with secondary antibody (goat anti-mouse IgG Alexa Fluor 488, goat anti-mouse IgG Alexa Fluor 594, goat anti-rabbit IgG Alexa Fluor 488, or goat anti-rabbit IgG Alexa Fluor 594; Molecular Probes) as appropriate (1:500 dilution). Nuclei were counterstained concurrently with TO-PRO-3 iodide (Molecular Probes). Where appropriate, cells were permeabilized by including 0.1% saponin in all antibody dilutions and wash buffers. For plasma membrane permeabilization, Vero cells were placed on ice at 18 h posttransfection and washed with KHM buffer (110 mM potassium acetate, 20 mM HEPES [pH 7.2], 2 mM magnesium acetate). Cells were then permeabilized in KHM buffer containing 25 μg of digitonin/ml on ice for 5 min, washed three times with KHM buffer, and fixed with 2% paraformaldehyde in PBS on ice for 10 min (10, 54). Coverslips were mounted onto microscope slides by using Prolong Antifade Gold (Molecular Probes) and visualized on a Zeiss LSM 510 confocal microscope. Colocalization was quantified by using Volocity image analysis software (Improvision).
SDS-PAGE and Western blotting.
Transfected or infected cells were lysed in 1% SDS in water and mixed at a 1:1 ratio with Laemmli 2× SDS-PAGE sample buffer with or without 200 mM dithiothreitol (DTT) and boiled for 5 min (50). Purified virions were resuspended in 2× SDS-PAGE sample buffer. Samples were loaded onto either 15% polyacrylamide gels or 17.5% polyacrylamide gels with 4 M urea (Mini Trans-Blot; Bio-Rad).
For Western blotting, separated polypeptides were transferred onto polyvinylidene difluoride membranes (Millipore) and blocked in PBS containing 0.3% Tween 20 and 5% nonfat dry milk (block buffer). Membranes were incubated with the following antibodies diluted in block buffer: rabbit anti-ORF7b serum (1:1,000 dilution), rabbit anti-SARS-CoV nsp8 protein (1:2,000 dilution) (56), anti-SARS-CoV N mouse MAb 87-A1 (1:1,000 dilution) (64), anti-SARS mouse hyperimmune serum (1:1,000 dilution, kindly provided by CDC), anti-β-actin mouse MAb (1:7,500 dilution; Abcam), anti-influenza A M2 mouse MAb 14C2 (1:2,000 dilution), anti-West Nile virus (WNV) NS1 mouse MAb 3NS1 (1:1,000) (9), or rabbit anti-GFP sera (1:2,500 dilution; Molecular Probes). Primary antibodies were detected by using species-specific IgG secondary antibodies coupled to horseradish peroxidase (Jackson Laboratories). The blots were soaked in chemiluminescent reagent (ECL Plus Pico; Amersham Biosciences) and imaged by using either chemiluminescence and exposure to X-ray film (Molecular Technologies) or chemifluorescence followed by phosphorimager analysis (FujiFilm FLA-5000) to quantify the signal intensity.
Membrane association assay.
293T cells were transfected with plasmids encoding the cDNAs for influenza A virus M2, SARS-CoV GFP-N, GFP-Rab11, or ORF7b. At 18 h posttransfection, cells were collected and centrifuged at 300 × g. The cell pellet was resuspended in hypotonic lysis buffer (10 mM Tris [pH 7.4], 10 mM KCl, 5 mM MgCl2, and a protease inhibitor cocktail) and incubated on ice for 30 min. Cells were lysed with 20 strokes of a Dounce homogenizer. Nuclei and intact cells were removed by centrifugation at 1,000 × g for 10 min. Postnuclear supernatants were separated into four aliquots and centrifuged at 100,000 × g for 2 h in a TLA 45 rotor (Beckman) at 4°C to separate membrane and cytosolic fractions. The high-speed supernatant was collected, and the high-speed pellet (HSP) containing the membrane fraction was resuspended in 250 μl of 1× NTE buffer (0.5 M NaCl, 10 mM Tris-HCl [pH 7.4], 5 mM EDTA). To dissociate peripheral membrane proteins (52), HSP suspensions were mixed with an equal volume of 8 M urea, 4 M KCl, or 100 mM carbonate buffer (pH 11.3) and incubated on ice for 30 min. HSP suspensions were then centrifuged at 100,000 × g for 2 h in a TLA 45 rotor at 4°C, the supernatant was collected, and the pellet resuspended in 2× Laemmli SDS-PAGE loading buffer.
Electron microscopy.
At 12 h postinfection SARS-CoV-infected cells were fixed with 4% paraformaldehyde-0.1% glutaraldehyde on ice for 60 min as previously described (38, 38a) and processed further for cryofixation and cryosectioning. Ultrathin cryosections were excised from the fixed material and immunolabeled with anti-ORF7b rabbit serum and/or an anti-N MAb. Antibody staining was visualized with protein A conjugated to 10- or 5-nm gold particles (Utrecht University, Utrecht, The Netherlands), as described previously (38, 38a). The immunolabeled sections were viewed on a JOEL 1010 transmission electron microscope and images were captured on a MegaView III side-mounted charge-coupled device camera (Soft Imaging Systems) and processed for publication in Adobe Photoshop.
RESULTS
Expression of ORF7b in SARS-CoV-infected cells.
The SARS-CoV genome encodes nine RNA species that serve as mRNAs for all of the proteins encoded by the virus (Fig. 1A). The 5′ end of each subgenomic RNA (sgRNA) is defined by the presence of a transcription regulatory sequence and the existence of the nine sgRNAs has been confirmed experimentally (70, 80a, 85b). Several of the sgRNAs are predicted to encode more than one protein (Fig. 1A). In some cases (sgRNAs 3 and 9) the two ORFs overlap significantly, while in others (sgRNAs 1, 7, and 8) the overlap is minimal. In all cases, the second ORF encoded by the sgRNA is in an unfavorable location for translation since it is located downstream of at least one initiation codon.
Although the expression of several SARS-CoV group-specific ORFs has been confirmed in tissue culture or in SARS-CoV-infected tissue samples (6, 7, 14a, 47, 78, 86), there have been no reports regarding the expression of the ORF7b protein. The ORF7b protein is encoded by an ORF beginning 365 nucleotides from the transcription regulatory sequence of sgRNA7. The putative start codon for ORF7b overlaps but is out of frame with the stop codon for ORF7a (Fig. 1A). SARS-CoV ORF7b is predicted to be a 44-amino-acid protein containing a hydrophobic central region that could span the membrane bilayer (Fig. 1B). Of the 130 SARS-CoV isolates that have been completely sequenced to date, 118 isolates have identical ORF7b amino acid sequences, 1 isolate has a single amino acid change in ORF7b, and 11 isolates have insertions or deletions that severely disrupt the ORF7b coding region (data not shown). The ORF7b proteins encoded by the eight bat coronavirus isolates sequenced to date contain only two to four amino acid changes from the sequence present in the majority of SARS-CoV isolates, suggesting a selective pressure to maintain the ORF7b ORF (32, 35, 55).
Sera from rabbits immunized with a bacterially expressed His6-ORF7b fusion protein recognized a protein band that migrated slightly faster than the 10-kDa molecular mass marker and was found in SARS-CoV-infected Vero cell lysates but not in mock-infected cell lysates (Fig. 1C). The expression of ORF7b was also confirmed by flow cytometry. Antibodies specific for the ORF7a (47) or ORF7b proteins reacted with SARS-CoV-infected cells but not with mock-infected cells (Fig. 1D). Most of the SARS-CoV-infected cells reacted with both ORF7a- and ORF7b-specific antibodies, indicating that both proteins were expressed in the same cells. These data indicate that the ORF7b protein is expressed in SARS-CoV-infected cells.
Expression of ORF7b from cDNA.
To confirm that the ORF7b protein can be expressed from an mRNA corresponding to the SARS-CoV sgRNA7, the sequences encoding both ORF7a and ORF7b (7ab) were amplified by reverse transcriptase PCR and cloned into the eukaryotic expression vector pCAGGS. Plasmids containing the coding regions for ORF7a or ORF7b were also constructed (Fig. 2A). The plasmids were transfected into 293T cells, and the expression of ORF7a or ORF7b was quantified by using flow cytometry. Antibodies specific for the ORF7a protein reacted with cells transfected with the cDNA encoding ORF7a or ORF7ab (Fig. 2B). Antibodies directed against the ORF7b protein recognized cells transfected with cDNAs encoding ORF7b or the ORF7ab, indicating the ORF7b protein could in fact be translated from an mRNA containing the entire gene 7 coding region, despite its position downstream of the ORF7a ORF (Fig. 2B). Neither antibody recognized mock-transfected cells.
The entire gene 7 coding region contains only three ATG codons. The first is predicted to initiate translation of ORF7a, and the second is predicted to serve as the initiation codon for the ORF7b protein (Fig. 2A). A third ATG resides immediately 3′ to the predicted ORF7b initiation codon, but translation initiation at this site would result in a peptide of only four amino acid residues. To confirm that the predicted ORF7a and ORF7b initiation codons were functional, the codons were mutated in the 7ab cDNA (7ab 7a Start KO and 7ab 7b Start KO, Fig. 2A), and protein expression after transfection into 293T cells was quantified by flow cytometry (Fig. 2B). Mutating the upstream ATG to ATC resulted in a loss of ORF7a expression, whereas mutating the downstream ATG to ATA resulted in a loss of ORF7b expression, confirming that these ATG codons were in fact the initiation codons for ORF7a and ORF7b (Fig. 1B).
Leaky scanning translation of ORF7b.
The downstream ORF in multicistronic mRNAs can be translated via a variety of mechanisms (reviewed in reference 29). Since the upstream ATG in the sgRNA 7 cDNA is in a poor translational or Kozak context (29), it seemed reasonable to surmise that the translation of ORF7b from sgRNA 7 occurred via leaky scanning. To test this hypothesis, the plasmid containing the entire gene 7 coding region was mutated to (i) place the ORF7a ATG in an ideal Kozak context (7ab 7a Kozak), (ii) introduce an additional ATG into the ORF7a ORF (7ab ATG), (iii) insert a stop codon into the ORF7a ORF (7ab 7a Stop), (iv) eliminate the ORF7a stop codon (7ab 7a Stop KO), and (v) eliminate the ORF7a initiation codon (7ab 7a Start KO) (Fig. 2A). The plasmids were transfected into 293T cells, and the cells were lysed 18 h posttransfection. The expression of ORF7b and β-actin was detected by Western blotting (Fig. 2C) and quantified by phosphorimager analysis (Fig. 2D). The ORF7b protein was expressed at a >2-fold higher amount from a plasmid encoding only the ORF7b cDNA compared to that from a plasmid expressing the full 7ab coding region, indicating that the ORF7b expression from the 7ab cDNA was inefficient. If ORF7b translation occurred by ribosome leaky scanning, then changing the context of the ORF7a initiation codon to an optimal Kozak sequence (Fig. 2A) should result in fewer ribosomes scanning past the ORF7a initiation codon, thereby reducing the amount of ORF7b expression. The expression of ORF7b from the 7ab 7a Kozak construct was significantly reduced (Fig. 2C), suggesting that leaky scanning translation could be responsible for the expression of ORF7b. The presence of additional ATG codons upstream of the ORF7b initiation codon, particularly ones that are in an optimized Kozak context, should result in reduced ORF7b expression since ribosomes that have scanned past the ORF7a initiation codon would encounter and engage at these initiation codons and not scan further down the mRNA (29). When an out-of-frame ATG codon in a near-optimal Kozak context was introduced into the ORF7a coding region, the expression of ORF7b was reduced (Fig. 2C, 7ab ATG), further supporting leaky scanning as the mechanism of ORF7b translation. The introduction of two premature stop codons into the ORF7a coding region (Fig. 2C, 7ab 7a Stop) or the elimination of the ORF7a stop codon (Fig. 2C, 7ab 7a Stop KO) had little effect on the expression of ORF7b. Eliminating the ORF7a start codon resulted in an ca. 50% increase in ORF7b expression (Fig. 2C, 7ab 7a Start KO). Eliminating the ORF7b initiation codon resulted in a complete loss of ORF7b expression (Fig. 2C, 7ab 7b Start KO). Taken together, the data in Fig. 2 indicate that the gene 7 mRNA is bicistronic and the translation of ORF7b from this mRNA occurs via ribosome leaky scanning.
ORF7b is an integral membrane protein.
The ORF7b amino acids 9 to 28 are highly hydrophobic (Fig. 1B) and predicted to be a transmembrane domain by the TMPred software (http://www.ch.embnet.org/software/TMPRED_form.html). In order to experimentally determine whether ORF7b is an integral membrane protein, 293T cells were transfected with plasmids expressing GFP fused to the N terminus of the cellular GTPase Rab11 (GFP-Rab11, a protein that is found in the cytosol and bound to membranes) (26), GFP fused to the N terminus of SARS-CoV nucleocapsid protein (GFP-SARS N, a cytosolic protein) (64), the influenza M2 protein (an integral membrane protein) (31), or the ORF7b protein. At 18 h posttransfection, cells were lysed in hypotonic lysis buffer, and the membrane and cytosolic fractions were separated by high-speed centrifugation. The resulting pellet (containing the cellular membranes) was resuspended in PBS and treated with urea, alkaline pH, or KCl—conditions known to extract peripheral membrane proteins but not integral membrane proteins from membranes (16, 52). The ORF7b protein pelleted with cellular membranes after hypotonic lysis, as did the influenza A virus M2 protein (Fig. 3). As expected, the cytosolic protein GFP-SARS N was not present in the pellet, whereas the GFP-Rab11 was present in both fractions, indicating that these centrifugation conditions did not result in pelleting of a significant amount of cytosolic proteins. Treatment of the pelleted membranes with urea, carbonate, or KCl did not dissociate ORF7b or M2 from membranes, indicating that both of these proteins were integral membrane proteins. The GFP-Rab11 protein was removed from membranes with urea and carbonate, indicating these conditions could strip peripheral membrane proteins from membranes. KCl treatment did not dissociate GFP-Rab11 from membranes, presumably because the interactions that tether GFP-Rab11 to membranes are not ionic interactions, and therefore are not disrupted by high amounts of KCl (72). These results strongly suggest the ORF7b protein is an integral membrane protein.
FIG. 3.
The ORF7b protein is an integral membrane protein. 293T cells were transfected with plasmids encoding the cDNAs for GFP-Rab11, SARS-CoV GFP-N, M2 from influenza A virus, or SARS-CoV ORF7b. At 18 h posttransfection, cells were collected, lysed in hypotonic lysis buffer, and homogenized. The lysate was separated into pellet (P) and supernatant (S) fractions by centrifugation at 100,000 × g. Pellets containing cellular membranes were subsequently treated with 4 M urea, 50 mM Na2CO3 (pH 11.3), or 2 M KCl to disrupt peripheral membrane proteins, and the samples were centrifuged at 100,000 × g. The resultant pellet (P) and supernatant (S) were analyzed by Western blotting with rabbit anti-GFP polyclonal sera, anti-M2 MAb, or rabbit anti-ORF7b polyclonal sera.
The ORF7b protein encodes cysteine residues at amino acids 12 and 41 (Fig. 1B). The cysteine at amino acid 41 is present downstream of the predicted transmembrane domain, whereas the cysteine at amino acid 12 lies within the predicted transmembrane domain. To determine whether the ORF7b protein can form disulfide bond-linked oligomers, plasmids expressing either the influenza A virus M2 protein or the SARS-CoV ORF7b protein were transfected into 293T cells. At 18 h posttransfection, the cells were lysed in 1% Triton X-100, polypeptides were separated by SDS-PAGE under either reducing or nonreducing conditions, and the proteins were detected by Western blotting. As shown in Fig. 4A, the M2 protein forms characteristic dimers (d) and tetramers (t) under nonreducing conditions (20) and exists as a monomer (m) under reducing conditions. In contrast, no change in migration was detected when the ORF7b protein was analyzed under reducing or nonreducing conditions (Fig. 4B), indicating the protein does not form disulfide-linked oligomers.
FIG. 4.
The ORF7b protein does not form disulfide-linked oligomers. 293T cells were transfected with either control plasmid (mock) or plasmids expressing the cDNAs for the influenza A virus M2 protein (A) or SARS-CoV ORF7b (B). The cells were lysed 18 h posttransfection, separated by SDS-PAGE under reducing (+DTT) or nonreducing (−DTT) conditions, and analyzed for protein expression by Western blotting. The M2 protein formed characteristic disulfide-linked dimers (d) and tetramers (t) under nonreducing conditions and monomers (m) in reducing conditions.
Subcellular localization of ORF7b.
Since the SARS-CoV ORF7b protein appears to be an integral membrane protein, we investigated its subcellular localization after cDNA expression or in virus-infected cells by confocal microscopy of both permeabilized and intact cells. As a positive control for surface expression, cells were transfected with a plasmid containing the cDNA for the influenza A virus M2 protein or a C-terminal myc epitope-tagged M2 protein. M2 is a type III integral membrane protein with an extracellular N terminus, an intracellular C terminus and no signal peptide (31, 87). Cell surface expression of the M2 protein was discernible in permeabilized or intact cells stained with the MAb 14C2 which recognizes the extracellular N terminus of the protein (Fig. 5A, a to d). The increased perinuclear staining observed in permeabilized cells represents the M2 protein that accumulates in the Golgi compartment as a result of intra-Golgi pH changes induced by the protein (65). The C-terminal myc epitope-tagged M2 protein was detected at the plasma membrane of permeabilized but not intact cells (Fig. 5A, e to h), which is consistent with the fact that the M2 C terminus is located in the cytoplasm and therefore not accessible to antibody in nonpermeabilized cells (87).
FIG. 5.
The ORF7b protein is not expressed on the cell surface of transfected cells. (A) Vero cells were transfected with plasmids encoding the cDNAs for the surface-expressed influenza A virus M2 protein (a to d), a C-terminal myc-epitope tagged M2 protein (e to h), ORF7a (i to l), ORF7b (m to p), or an ORF7b protein containing a C-terminal myc epitope tag (q to t). Transfected cells were fixed and permeabilized with 0.1% saponin (left columns) or stained with primary antibody at 4°C and fixed immediately poststaining (nonpermeabilized). Cells were incubated with antibodies specific for the indicated proteins, with the exception of the myc epitope-tagged proteins, which were detected with an anti-myc MAb. Only influenza virus M2 transfected cells stained with the 14C2 MAb (targeting the N-terminal extracellular domain of M2) demonstrated surface staining. (B) Vero cells were cotransfected with cDNAs expressing ORF7b and a GFP construct that localizes to the plasma membrane of transfected cells (S15-GFP). At 18 h posttransfection, the cells were immunostained with antibodies that recognize the ORF7b protein (red). The lack of colocalization between the proteins further suggests little or no ORF7b is present at the cell surface. All images were obtained with a ×63 oil immersion objective and represent a z-stack projection of 0.5-μm slices obtained by confocal microscopy. Nuclei were counterstained with TO-PRO-3 (blue).
When cells transfected with the ORF7b cDNA were immunostained with polyclonal antibodies raised to the entire ORF7b protein, the protein was detected in a concentrated perinuclear region with no significant staining associated with the plasma membrane (Fig. 5A, m to p). ORF7b was detected in permeabilized but not in intact cells. The expression pattern of ORF7b was similar to that observed with SARS-CoV ORF7a (Fig. 5A, i to l), which is not expressed at the plasma membrane but localizes primarily to the Golgi compartment (47).
Since the ORF7b epitopes that are recognized by the polyclonal ORF7b sera have not been mapped, it is not clear whether antibodies to both the N and the C termini of the protein are present in the polyclonal anti-ORF7b sera. Cells transfected with a cDNA encoding ORF7b containing a C-terminal myc epitope tag were immunostained to gain insight into the membrane topology of the protein. Antibodies that recognize the myc epitope-immunostained ORF7b in permeabilized but not in intact cells, indicating that the C terminus of ORF7b was not exposed to the extracellular media. In addition, the lack of a discernible plasma membrane staining pattern further supported the conclusion that ORF7b was most likely retained at an intracellular site. In order to unambiguously demonstrate that ORF7b targets to intracellular membranes, 293T cells were transfected with plasmids expressing the cDNAs of ORF7b and a short, acylated polypeptide fused to GFP (S15-GFP), a protein previously demonstrated to bind to the plasma membrane (60a). Cotransfected cells were analyzed by confocal microscopy (Fig. 5B, a to d). Little or no colocalization (lack of yellow staining in the merge image) between S15-GFP (green) and the ORF7b protein (red) was detected, indicating that ORF7b is not present at the plasma membrane of transfected cells.
The data in Fig. 5 indicate that the ORF7b protein does not localize to the plasma membrane after expression from cDNA. However, the data do not provide insights into the membrane topology of the protein because an intracellularly targeted protein will not have any epitopes exposed to the extracellular milieu. The ORF7b protein could be a type III integral membrane protein with its N terminus in the lumen and its C terminus exposed to the cytoplasm, or it could be a type II integral membrane protein with its N terminus exposed to the cytoplasm and its C terminus in the luminal space. To distinguish between these possibilities, selective plasma membrane permeabilization with digitonin was utilized. After transfection with cDNAs encoding SARS-CoV ORF7a-GFP, C-terminal myc-tagged M2 or C-terminal myc-tagged ORF7b, the cells were either completely permeabilized with saponin or selectively permeabilized with digitonin. To ensure the digitonin treatment was selectively permeabilizing the plasma membrane, ORF7a-GFP-expressing cells were immunostained with an antibody, 2E11, which recognizes the luminal domain of the protein (47). Whereas GFP staining was visible regardless of permeabilization, the 2E11 immunostaining was only visible after saponin permeabilization, indicating that the digitonin was not permeabilizing the Golgi membranes (Fig. 6A, D, E, and H). Immunostaining of cells expressing the C-terminally tagged M2 protein showed antibody staining with both detergent treatments, indicating the digitonin was in fact permeabilizing the plasma membrane (Fig. 6B and F). Immunostaining of the ORF7b-myc protein-expressing cells was detected under both conditions, clearly indicating the C terminus of ORF7b-myc was exposed to the cytoplasm (Fig. 6C and G). Unfortunately, introducing a myc epitope tag at the N terminus of ORF7b altered the intracellular distribution of the protein, preventing us from further characterizing the membrane orientation of ORF7b (data not shown).
FIG. 6.
The C terminus of ORF7b is exposed to the cytoplasm. Vero cells were transfected with plasmids expressing the cDNA for ORF7a-GFP (A, D, E, and H), M2-myc (B and F), or ORF7b-myc (C and G). At 18 h posttransfection, the cells were fixed with 2% paraformaldehyde, washed extensively, and permeabilized with PBS containing 0.1% saponin to disrupt all cellular membranes (A to D) or 25 μg of digitonin/ml to selectively permeabilize the plasma membrane (E to H). The cells were incubated with the 2E11 MAb, which recognizes the luminal domain of ORF7a (A, D, E, and H), or the 9E10 MAb, which recognizes the myc epitope tag (B, C, F, and G). The primary antibodies were detected using a goat anti-mouse IgG conjugated to Alexa Fluor 594 and mounted, and the cells were visualized by confocal microscopy using a ×63 oil immersion objective.
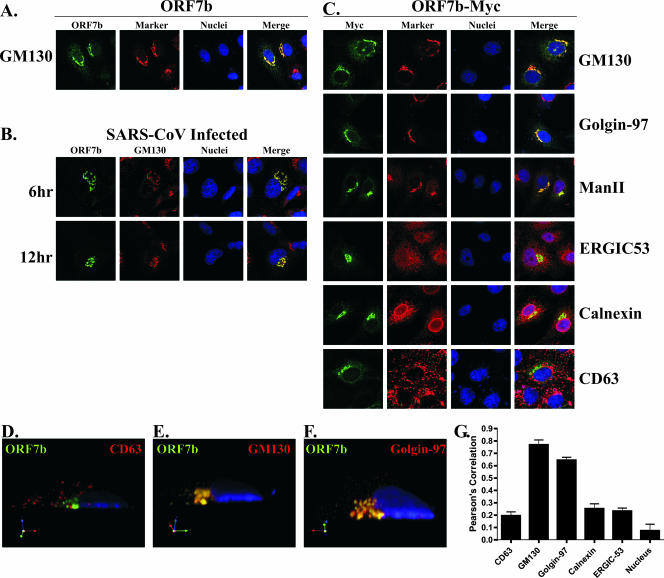
To determine the precise intracellular localization of ORF7b, Vero cells transfected with a plasmid expressing the cDNA for the ORF7b protein were analyzed by indirect immunofluorescence confocal microscopy. The ORF7b protein exhibited a juxtanuclear localization pattern that highly colocalized with the _cis_-Golgi marker GM130 (Fig. 7A). The localization of ORF7b was also analyzed in SARS-CoV-infected Vero cells at 6 and 12 h postinfection (Fig. 7B). The ORF7b protein colocalized with GM130 at both times postinfection, indicating that the intracellular localization of ORF7b was similar in cDNA-transfected and virus-infected cells. The Golgi appears expanded and distended in SARS-CoV-infected cells compared to those in mock-infected or cDNA-transfected cells. This is most likely due to an expansion of the Golgi in order to handle the large amount of viral glycoproteins transiting through or localizing to the Golgi.
FIG. 7.
The ORF7b protein localizes to the Golgi apparatus in cDNA transfected or SARS-CoV-infected cells. (A) Vero cells grown on coverslips were transfected with a plasmid expressing the ORF7b cDNA. At 18 h posttransfection, the cells were immunostained with antibodies against ORF7b (green) or the _cis_-Golgi marker protein GM130 (red) by using confocal microscopy. Nuclei were counterstained with TO-PRO-3 (blue). (B) Vero cells were plated onto coverslips and then infected at an MOI of 5.0 with SARS-CoV. At the indicated times postinfection, the cells were fixed, and colocalization of ORF7b (green) with the _cis_-Golgi marker GM130 (red) was assessed by confocal microscopy. (C) Vero cells grown on coverslips were transfected with a plasmid expressing the ORF7b protein containing a C-terminal myc-epitope tag and then assessed by confocal microscopy for colocalization with cellular proteins that reside in various subcellular compartments (red) including the _cis_-Golgi network (GM130), the _trans_-Golgi network (Golgin-97), _cis_- and _medial_-Golgi (α-mannosidase II), ER-to-Golgi intermediate compartment (ERGIC53), ER (Calnexin), and lysosomes/multivesicular body (CD63). The ORF7b and ORF7b-myc antigen (green) colocalized with all three Golgi markers. All images were obtained with a ×63 oil immersion objective lens and represent a z-stack projection of 0.7-μm slices obtained by confocal microscopy. (D to F) Three-dimensional confocal images were generated by using Volocity software (Improvision) and analyzed for colocalization between ORF7b (green) and GM130, Golgin-97, or CD63 (red). (G) Shown is a cross-section along the y axis of the cell. Nuclei were counterstained with TO-PRO-3 (blue). The extent of colocalization was quantified for five cells in each condition, and the measure of fluorophore colocalization is expressed as a Pearson correlation coefficient (39).
The subcellular localization of ORF7b was then compared to those of a number of cellular proteins. For these studies, the ORF7b-myc protein was used as rabbit and mouse antibodies recognizing the myc epitope are available. The ORF7b-myc protein colocalized with GM130 indicating the epitope-tagged protein displayed the same intracellular localization as the untagged protein (Fig. 7C). Interestingly, ORF7b-myc colocalized with two other Golgi markers, the _trans_-Golgi network marker Golgin-97 and the _cis/medial_-Golgi marker α-mannosidase II (Fig. 7C), indicating that ORF7b-myc was present throughout the Golgi stacks.
The ORF7b protein did not colocalize to a great extent with the cellular proteins calnexin (ER resident protein), ERGIC53 (ER-to-Golgi intermediate compartment protein), or CD63 (lysosome/multivesicular body protein) (Fig. 7C). The amount of colocalization of ORF7b-myc with CD63 (Fig. 7D), GM130 (Fig. 7E), or Golgin-97 (Fig. 7F) was quantified by reconstructing a three-dimensional image from the confocal images and calculating a Pearson coefficient of colocalization (Fig. 7G) (39). The ORF7b colocalization with GM130 (0.769 ± 0.068) and Golgin-97 (0.645 ± 0.040) was significantly higher than its colocalization with CD63 (0.195 ± 0.054), calnexin (0.252 ± 0.067), ERGIC53 (0.233 ± 0.048), or the nuclear counterstain TO-PRO-3 (0.074 ± 0.106). The data from Fig. 7 clearly indicate that the ORF7b protein localizes to the Golgi compartment in SARS-CoV-infected cells and after expression from cDNA.
ORF7b is incorporated into SARS-CoV virions.
The assembly and budding of coronavirus particles occurs at intracellular membranes (reviewed in reference 4a). While the precise intracellular location of SARS-CoV budding has yet to be determined, several other coronaviruses bud into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and the E and M proteins that drive the budding process localize to the ERGIC or Golgi (11a, 11c, 36, 45). Since ORF7b localized near the presumed site of SARS-CoV budding, we investigated whether ORF7b was incorporated into virus particles.
Supernatant from SARS-CoV-infected cells was inactivated with paraformaldehyde and then centrifuged over a 20% (wt/vol) sucrose cushion in order to partially purify virus particles. The pelleted virus particles were resuspended and analyzed by SDS-PAGE and Western blotting for the expression of viral and cellular proteins. SARS-CoV cell lysates contained the expected viral (nsp8, S, and ORF7b) and cellular (β-actin) proteins, whereas mock-infected cells contained only β-actin (Fig. 8). The partially purified virus pellet contained the SARS-CoV S and ORF7b proteins but did not possess a detectable signal for the nsp8 protein or the cellular protein β-actin (Fig. 8). The lack of β-actin in the SARS-CoV virus pellet indicated that there was not a significant amount of cellular contaminants in the preparation, and the lack of nsp8 protein indicated that not all SARS-CoV proteins could be packaged into virus particles.
FIG. 8.
The ORF7b protein is present in purified virus particles. (A) SARS-CoV particles were inactivated with paraformaldehyde, the supernatant was precleared by centrifugation at 1,000 × g and layered on a 20% sucrose cushion, and virus particles were pelleted at 100,000 × g. The virus pellet was resuspended in 2× Laemmli SDS-PAGE buffer, and protein expression was detected by Western blotting. The virus lysate contains no β-actin or nsp8 but is enriched for both S (the asterisk indicating a highly glycosylated virus associated form of S) and ORF7b. (B) The inactivated, pelleted virus particles were loaded onto a 20 to 60% continuous sucrose gradient and centrifuged for 18 h at 100,00 × g, and 1-ml fractions were collected from the top. The fractions were analyzed for the presence of SARS-CoV N or ORF7b by Western blotting. (C) The density of each fraction of the 20 to 60% sucrose gradient was determined by weighing 100 μl of the fraction.
The partially purified virus particle preparation was subjected to ultracentrifugation on a continuous 20 to 60% sucrose gradient for 18 h at 100,000 × g. The sucrose gradients were fractionated from the top, and each fraction was analyzed by Western blotting for SARS-CoV N or ORF7b protein. Fractions 8 to 10 contained the peak amounts of both SARS-CoV N and ORF7b proteins, indicating the particles containing either protein cosedimented (Fig. 8B). The density of fractions 8 to 10 was approximately 1.20 g/ml (Fig. 8C), which is consistent with the density of SARS-CoV particles (23, 25). Identical results were obtained with viruses inactivated with β-propiolactone (data not shown), suggesting the cosedimentation of ORF7b and N was not dependent on the method of virus inactivation. These data suggest the SARS-CoV ORF7b protein is incorporated into virus particles.
It has recently been reported that the SARS-CoV 3a protein is released from transiently transfected cells in membranous structures (22). Coronavirus E proteins can also be released from cDNA-transfected cells and are key components of the virus budding machinery (10, 21, 22, 46). In order to determine whether ORF7b could be secreted or associated with cellular vesicles that are secreted, we analyzed the supernatants of cells transfected with cDNA plasmids encoding ORF7b, influenza A virus M2 or WNV NS1. The influenza A virus M2 protein is an integral membrane protein that is not secreted or shed from transfected cells, while the WNV NS1 protein is secreted from infected or transfected cells in a glycosylated form (37). Western blotting of transfected cell lysates indicated that all of the proteins were expressed (Fig. 9). When transfected cell supernatants were analyzed, no ORF7b was detected, indicating the ORF7b protein is not secreted into the media of cDNA-transfected cells (Fig. 9). As expected, no influenza A virus M2 protein was present in the supernatants, whereas the WNV NS1 protein was readily detected.
FIG. 9.
The ORF7b protein is not secreted from transfected cells. 293T cells were transfected with plasmids encoding the cDNAs for the influenza A virus M2 protein, the WNV NS1 protein, or the SARS-CoV ORF7b protein. Mock samples were transfected with the plasmid vector containing no cDNA insert. At 18 h posttransfection, supernatants were collected and centrifuged at 1,000 × g to remove cellular debris, and an equal volume of 2× Laemmli SDS-PAGE buffer was added. Samples were analyzed by Western blotting for the indicated proteins. NS1 was present in cell lysates (l) and secreted (s) as a higher-molecular-weight glycosylated form, whereas the M2 and ORF7b proteins were present only in the cell lysates.
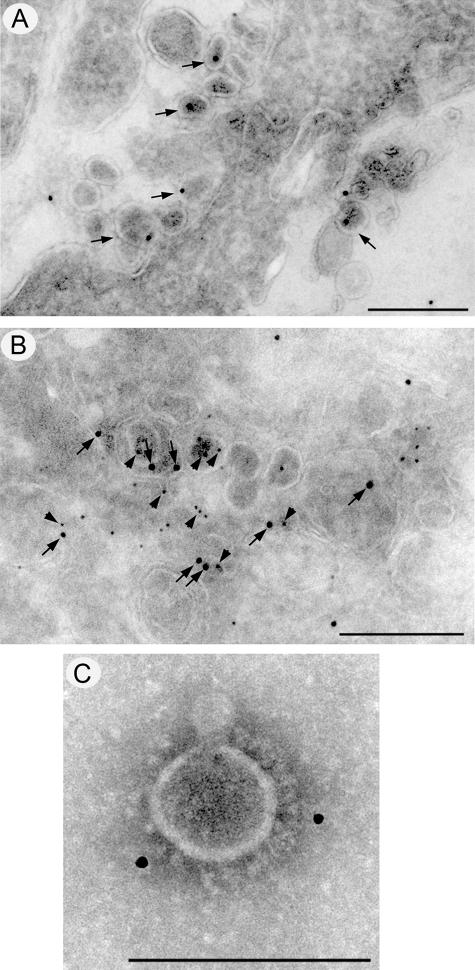
To confirm the association of ORF7b with virus particles, infected Vero cells were analyzed by immunogold labeling and electron microscopy (Fig. 10). At 12 (Fig. 10A) and 24 h (data not shown) postinfection, intracellular SARS-CoV particles were labeled strongly with anti-ORF7b antibodies. The association of ORF7b with SARS-CoV virions was further confirmed by dual-labeling experiments with anti-ORF7b (arrows) and anti-N (arrowheads) antibodies (Fig. 10B). Both antibodies labeled intracellular virions. A more direct association of the ORF7b protein with the SARS-CoV is shown in Fig. 10C, where purified SARS-CoV particles were decorated with anti-ORF7b antibodies, further indicating the ORF7b protein is packaged into SARS-CoV particles.
FIG. 10.
Immunogold staining for ORF7b in SARS-CoV particles. Cryosections from SARS-CoV-infected Vero cells were immunolabeled with antibodies raised against ORF7b (A and B) or the SARS-CoV N protein (B) and visualized with 10-nm or 5-nm protein A-gold, respectively. Localization of the ORF7b protein to intracellular virions is highlighted with arrows in panels A and B, and colocalization with N protein (arrowheads) is shown in panel B. (C) Immunolabeling of ORF7b on purified SARS-CoV particles. Magnification bars represent 200 nm in panels A and B and 100 nm in panel C.
DISCUSSION
Coronavirus accessory genes have been shown to play important roles in the in vivo replication and pathogenesis of a number of coronaviruses (11b, 49). However, there are a number of coronavirus accessory genes whose function has yet to be determined (4, 44, 85b). In this report, the expression of the SARS-CoV ORF7b protein was confirmed in SARS-CoV-infected cells and after expression of the gene 7 coding region from a eukaryotic expression plasmid. To date, the expression of the SARS-CoV ORF3a (14a, 22, 25, 68, 86) and ORF7a (47, 87a) accessory proteins has been demonstrated in infected cells, but both of those proteins are located near the 5′ end of their respective sgRNAs and are therefore in a favorable position for translation. Expression of the ORF3b, ORF6, ORF8a, ORF8b, and ORF9b proteins in SARS-CoV-infected cells has yet to be demonstrated, although sera from SARS-CoV-infected patients does contain antibodies to some of these proteins (16b). To our knowledge, the present study represents the first documentation of a SARS-CoV bicistronic RNA encoding an accessory protein. However, bicistronic RNAs have been identified in other coronaviruses, including all coronavirus replicase ORF1a/1b genes (1, 1a, 12), the bovine coronavirus N and I proteins (66, 67), the MHV ORF5a and ORF5b (80b), the IBV 5a and 5b proteins (4), the IBV 3a and 3b proteins (18), the TGEV 3a and 3b proteins (48a), and the TGEV 7a and 7b proteins (49). Although we cannot formally rule out the presence of a sgRNA whose transcript initiates at a noncanonical sequence allowing for monocistronic expression of ORF7b, it is clear from our studies of gene 7 expression from cDNA that the ORF7b protein can be translated in the context of a sgRNA 7-like transcript.
The biochemical and cell biological data suggest that the ORF7b protein is an integral membrane protein that localizes to the Golgi apparatus and has its C terminus exposed to the cytoplasm. The ORF7b protein is most likely a type III integral membrane protein; however, we were not able to raise antibodies to the N terminus of the protein, nor were we able to successfully epitope tag the N terminus of the protein without altering the intracellular localization of the protein. Introduction of potential N-linked glycosylation sites into the N terminus of the protein did not lead to the production of a glycosylated form of ORF7b, although the limited number of amino acids in the predicted luminal domain of ORF7b (nine amino acids) may preclude access to the glycosylation machinery (data not shown). It is interesting that the M2 protein of influenza A virus—a type III integral membrane protein—has a membrane-proximal sequence for N-linked glycosylation in its luminal domain that is not recognized by cellular glycosylation enzymes (87). For these reasons, formal proof of the ORF7b orientation in membranes must await the development of additional reagents.
The ORF7b protein was found to be a structural component of SARS-CoV virions, making it the third accessory gene to be found in virions. The importance of virion incorporation to ORF7b protein function will be an important question to address in future studies. Both the ORF3a (25, 68) and the ORF7a (21) accessory proteins have been determined to be SARS-CoV structural proteins. This is not surprising, since all three of these proteins localize to the Golgi compartment and are therefore quite close to the presumed site of SARS-CoV budding. The incorporation of viral proteins into SARS-CoV particles is not well understood, and it will be important to determine whether specific virion incorporation signals are present in ORF7b or whether it is simply packaged into the SARS-CoV particles because it is present in high amounts near the virus budding site. The latter would be similar to the pseudotyping of retroviral cores with integral membrane proteins expressed at high levels (11).
As yet, no function has been ascribed to the ORF7b protein and a recombinant SARS-CoV encoding GFP in place of the gene 7 coding region is not attenuated in mice or in primary human airway epithelial cell cultures (69, 85b). Furthermore, a mouse hepatitis virus (strain JHM) encoding ORF7b as an extra gene displayed no change in virulence or replication compared to the parental virus (53). Since the mouse is a good model for in vivo replication of SARS-CoV but not for SARS-CoV pathogenesis, it is possible that the protein may be dispensable for replication in mice but not in other SARS-CoV animal models of infection such as hamsters (60), bats (32, 55), civit cats (84), ferrets (41, 80), cats (41), or nonhuman primates (34, 41a, 57, 63a, 82). Alternatively, the ORF7b protein may be essential for virus replication in the as-yet-unidentified natural host of the virus. Our studies of the translation of ORF7a and ORF7b provide insight into how to make recombinant SARS-CoVs that expresses only one of the two gene 7 products without altering the level of expression of the other gene product. This is an important concept, since it will allow one to attribute a replication phenotype to the absence of one gene product and not the over- or underexpression of the other.
The high degree of conservation between the SARS-CoV and bat coronavirus gene 7 sequences implies the gene 7 products are conserved or selected for in a natural infection. Elucidating a role for the gene 7 products in SARS-CoV infection will require a careful, methodical analysis of SARS-CoV replication, pathogenesis, and host immune response in an appropriate animal model. However, valuable insights into the cell biology of SARS-CoV gene expression can be gained through the study of the translation and cell biology of SARS-CoV accessory gene proteins.
Acknowledgments
We thank all of the members of the Pekosz laboratory for insightful discussions and comments. We acknowledge and thank Ying Fang, South Dakota State University, Brookings, SD (antibodies to SARS-CoV N protein); Mark Denison, Vanderbilt University, Nashville, TN (antibodies to SARS-CoV nsp8 protein); Michael Diamond, Washington University School of Medicine, St. Louis, MO (antibodies to WNV NS1 protein and the NS1 cDNA); Wayne Rodgers, Oklahoma Medical Research Foundation, Oklahoma City, OK (pS15-GFP plasmid); and Phil Stahl, Washington University School of Medicine, St. Louis, MO (cDNA for GFP-Rab11). We also acknowledge the Molecular Microbiology Imaging Facility for help with microscopy and Jason Ader for technical assistance.
This study was supported by the Markey Pathway (S.R.S), T32 HL07317 (S.R.S), and AI059328 (A.P.).
Footnotes
▿
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Astell, C. R., R. A. Holt, S. J. M. Jones, and M. A. Marra. 2005. Genome organization and structural aspects of the SARS-related virus, p. 101-128. In A. Schmidt, M. H. Wolff, and O. Weber (ed.), Coronaviruses with special emphasis on first insights concerning SARS. Birkhauser Verlag, Basel, Switzerland.
- 1a.Brierley, I., and F. J. Dos Ramos. 2006. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 119**:**29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Reference deleted.
- 4.Casais, R., M. Davies, D. Cavanagh, and P. Britton. 2005. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 79**:**8065-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Cavanagh, D. 2005. Coronaviridae: a review of coronaviruses and toroviruses, p. 1-54. In A. Schmidt, M. H. Wolff, and O. Weber (ed.), Coronaviruses with special emphasis on first insights concerning SARS. Birkhauser Verlag, Basel, Switzerland.
- 5.Centers for Disease Control and Prevention. 2003. Revised U.S. surveillance case definition for severe acute respiratory syndrome (SARS) and update on SARS cases-United States and worldwide, December 2003. Morb. Mortal. Wkly. Rep. 52**:**1202-1206. [PubMed] [Google Scholar]
- 6.Chan, W. S., C. Wu, S. C. Chow, T. Cheung, K. F. To, W. K. Leung, P. K. Chan, K. C. Lee, H. K. Ng, D. M. Au, and A. W. Lo. 2005. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Modern Pathol. 18**:**1432-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y. Y., B. Shuang, Y. X. Tan, M. J. Meng, P. Han, X. N. Mo, Q. S. Song, X. Y. Qiu, X. Luo, Q. N. Gan, X. Zhang, Y. Zheng, S. A. Liu, X. N. Wang, N. S. Zhong, and D. L. Ma. 2005. The protein X4 of severe acute respiratory syndrome-associated coronavirus is expressed on both virus-infected cells and lung tissue of severe acute respiratory syndrome patients and inhibits growth of BALB/c 3T3 cell line. Chin. Med. J. 118**:**267-274. [PubMed] [Google Scholar]
- 8.Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303**:**1666-1669. [DOI] [PubMed] [Google Scholar]
- 9.Chung, K. M., G. E. Nybakken, B. S. Thompson, M. J. Engle, A. Marri, D. H. Fremont, and M. S. Diamond. 2006. Antibodies against West Nile virus nonstructural protein NS1 prevent lethal infection through Fcγ receptor-dependent and -independent mechanisms. J. Virol. 80**:**1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74**:**4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronin, J., X. Y. Zhang, and J. Reiser. 2005. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5**:**387-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.de Haan, C. A., L. Kuo, P. S. Masters, H. Vennema, and P. J. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72**:**6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296**:**177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11c.de Haan, C. A. M., H. Vennema, and P. J. M. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74**:**4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dos Ramos, F., M. Carrasco, T. Doyle, and I. Brierley. 2004. Programmed −1 ribosomal frameshifting in the SARS coronavirus. Biochem. Soc. Trans. 32**:**1081-1083. [DOI] [PubMed] [Google Scholar]
- 13.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H.-R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. M. Fouchier, A. Berger, A.-M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J.-C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H.-D. Klenk, A. D. M. E. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med.:NEJMoa030747. [DOI] [PubMed]
- 14.Reference deleted.
- 14a.Fielding, B. C., Y.-J. Tan, S. Shuo, T. H. P. Tan, E.-E. Ooi, S. G. Lim, W. Hong, and P.-Y. Goh. 2004. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 78**:**7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Gilmore, R., and G. Blobel. 1983. Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell 35**:**677-685. [DOI] [PubMed] [Google Scholar]
- 16a.Greenough, T. C., A. Carville, J. Coderre, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, and K. Mansfield. 2005. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am. J. Pathol. 167**:**455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Guo, J. P., M. Petric, W. Campbell, and P. L. McGeer. 2004. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology 324**:**251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanel, K., T. Stangler, M. Stoldt, and D. Willbold. 2006. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin-like fold and suggests a binding activity to integrin I domains. J. Biomed. Sci. 13**:**281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgson, T., P. Britton, and D. Cavanagh. 2006. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 80**:**296-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Holsinger, L. J., and R. A. Lamb. 1991. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology 183**:**32-43. [DOI] [PubMed] [Google Scholar]
- 21.Huang, C., N. Ito, C.-T. K. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 80**:**7287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C., K. Narayanan, N. Ito, C. J. Peters, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 80**:**210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y., Z.-Y. Yang, W.-P. Kong, and G. J. Nabel. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78**:**12557-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 79**:**3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordens, I., M. Marsman, C. Kuijl, and J. Neefjes. 2005. Rab proteins, connecting transport, and vesicle fusion. Traffic 6**:**1070-1077. [DOI] [PubMed] [Google Scholar]
- 27.Reference deleted.
- 28.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, and P. Palese. 2006. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 80**:**785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak, M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196**:**947-950. [DOI] [PubMed] [Google Scholar]
- 30.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348**:**1953-1966. [DOI] [PubMed] [Google Scholar]
- 31.Lamb, R. A., S. L. Zebedee, and C. D. Richardson. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40**:**627-633. [DOI] [PubMed] [Google Scholar]
- 32.Lau, S. K. P., P. C. Y. Woo, K. S. M. Li, Y. Huang, H.-W. Tsoi, B. H. L. Wong, S. S. Y. Wong, S.-Y. Leung, K.-H. Chan, and K.-Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102**:**14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Law, P. T. W., C.-H. Wong, T. C. C. Au, C.-P. Chuck, S.-K. Kong, P. K. S. Chan, K.-F. To, A. W. I. Lo, J. Y. W. Chan, Y.-K. Suen, H. Y. E. Chan, K.-P. Fung, M. M. Y. Waye, J. J. Y. Sung, Y. M. D. Lo, and S. K. W. Tsui. 2005. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 86**:**1921-1930. [DOI] [PubMed] [Google Scholar]
- 34.Lawler, J. V., T. P. Endy, L. E. Hensley, A. Garrison, E. A. Fritz, M. Lesar, R. S. Baric, D. A. Kulesh, D. A. Norwood, L. P. Wasieloski, M. P. Ulrich, T. R. Slezak, E. Vitalis, J. W. Huggins, P. B. Jahrling, and J. Paragas. 18April2006. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 3**:**e149 [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L.-F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310**:**676-679. [DOI] [PubMed] [Google Scholar]
- 36.Lim, K. P., H. Y. Xu, and D. X. Liu. 2001. Physical interaction between the membrane (M) and envelope (E) proteins of the coronavirus avian infectious bronchitis virus (IBV). Adv. Exp. Med. Biol. 494**:**595-602. [DOI] [PubMed] [Google Scholar]
- 37.Macdonald, J., J. Tonry, R. A. Hall, B. Williams, G. Palacios, M. S. Ashok, O. Jabado, D. Clark, R. B. Tesh, T. Briese, and W. I. Lipkin. 2005. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 79**:**13924-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73**:**9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220**:**232-240. [DOI] [PubMed] [Google Scholar]
- 39.Manders, E. M. M., F. J. Verbeek, and J. A. Aten. 1993. Measurement of colocalization of objects in dual-colour confocal images. J. Microsc. 169**:**375-382. [DOI] [PubMed] [Google Scholar]
- 40.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300**:**1399-1404. [DOI] [PubMed] [Google Scholar]
- 41.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425**:**915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.McAuliffe, J., L. Vogel, A. Roberts, G. Fahle, S. Fischer, W.-J. Shieh, E. Butler, S. Zaki, M. St. Claire, B. Murphy, and K. Subbarao. 2004. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330**:**8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCown, M., M. S. Diamond, and A. Pekosz. 2003. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology 313**:**514-524. [DOI] [PubMed] [Google Scholar]
- 43.McCown, M. F., and A. Pekosz. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79**:**3595-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGoldrick, A., J. P. Lowings, and D. J. Paton. 1999. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 144**:**763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nal, B., C. Chan, F. Kien, L. Siu, J. Tse, K. Chu, J. Kam, I. Staropoli, B. Crescenzo-Chaigne, N. Escriou, S. van der Werf, K.-Y. Yuen, and R. Altmeyer. 2005. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M, and E. J. Gen. Virol. 86**:**1423-1434. [DOI] [PubMed] [Google Scholar]
- 46.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74**:**8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 13**:**75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108**:**193-199. [DOI] [PubMed] [Google Scholar]
- 48a.O'Connor, J. B., and D. A. Brian. 2000. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology 269**:**172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308**:**13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In Molecular virology: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 51.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361**:**1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pekosz, A., and R. A. Lamb. 1998. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc. Natl. Acad. Sci. USA 95**:**13233-13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A Severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79**:**11335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plutner, H., H. W. Davidson, J. Saraste, and W. E. Balch. 1992. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 119**:**1097-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poon, L. L. M., D. K. W. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. C. Leung, S. K. P. Lau, P. C. Y. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79**:**2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279**:**10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin, C., J. Wang, Q. Wei, M. She, W. A. Marasco, H. Jiang, X. Tu, H. Zhu, L. Ren, H. Gao, L. Guo, L. Huang, R. Yang, Z. Cong, Y. Wang, Y. Liu, Y. Sun, S. Duan, J. Qu, L. Chen, W. Tong, L. Ruan, P. Liu, H. Zhang, J. Zhang, D. Liu, Q. Liu, T. Hong, and W. He. 2005. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J. Pathol. 206**:**251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin, E., H. Shi, L. Tang, C. Wang, G. Chang, Z. Ding, K. Zhao, J. Wang, Z. Chen, M. Yu, B. Si, J. Liu, D. Wu, X. Cheng, B. Yang, W. Peng, Q. Meng, B. Liu, W. Han, X. Yin, H. Duan, D. Zhan, L. Tian, S. Li, J. Wu, G. Tan, Y. Li, Y. Li, Y. Liu, H. Liu, F. Lv, Y. Zhang, X. Kong, B. Fan, T. Jiang, S. Xu, X. Wang, C. Li, X. Wu, Y. Deng, M. Zhao, and Q. Zhu. 2006. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine 24**:**1028-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50% endpoints. Am. J. Hyg. 27**:**493-499. [Google Scholar]
- 60.Roberts, A., L. Vogel, J. Guarner, N. Hayes, B. Murphy, S. Zaki, and K. Subbarao. 2005. Severe acute respiratory syndrome coronavirus infection of Golden Syrian hamsters. J. Virol. 79**:**503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60a.Rodgers, W. 2002. Making membranes green: construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. BioTechniques 32**:**1044-1051. [DOI] [PubMed] [Google Scholar]
- 61.Reference deleted.
- 62.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M.-h. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300**:**1394-1399. [DOI] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 63a.Rowe, T., G. Gao, R. J. Hogan, R. G. Crystal, T. G. Voss, R. L. Grant, P. Bell, G. P. Kobinger, N. A. Wivel, and J. M. Wilson. 2004. Macaque model for severe acute respiratory syndrome. J. Virol. 78**:**11401-11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rowland, R. R. R., V. Chauhan, Y. Fang, A. Pekosz, M. Kerrigan, and M. D. Burton. 2005. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells. J. Virol. 79**:**11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakaguchi, T., G. P. Leser, and R. A. Lamb. 1996. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell Biol. 133**:**733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Senanayake, S., M. Hofmann, J. Maki, and D. Brian. 1992. The nucleocapsid protein gene of bovine coronavirus is bicistronic. J. Virol. 66**:**5277-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Senanayake, S. D., and D. A. Brian. 1997. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 48**:**101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330**:**286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79**:**15511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331**:**991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stadler, K., A. Roberts, S. Becker, L. Vogel, M. Eickmann, L. Kolesnikova, H. D. Klenk, B. Murphy, R. Rappuoli, S. Abrignani, and K. Subbarao. 2005. SARS vaccine protective in mice. Emerg. Infect. Dis. 11**:**1312-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stein, M.-P., J. Dong, and A. Wandinger-Ness. 2003. Rab proteins and endocytic trafficking: potential targets for therapeutic intervention. Adv. Drug Deliv. Rev. 55**:**1421-1437. [DOI] [PubMed] [Google Scholar]
- 73.Reference deleted.
- 74.Reference deleted.
- 75.Subbarao, K., and A. Roberts. 2006. Is there an ideal animal model for SARS? Trends Microbiol. 14**:**299-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 76**:**1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan, Y.-J., B. C. Fielding, P.-Y. Goh, S. Shen, T. H. P. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 78**:**14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan, Y.-J., E. Teng, S. Shen, T. H. P. Tan, P.-Y. Goh, B. C. Fielding, E.-E. Ooi, H.-C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78**:**6723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reference deleted.
- 80.ter Meulen, J., A. B. H. Bakker, E. N. van den Brink, G. J. Weverling, B. E. E. Martina, B. L. Haagmans, T. Kuiken, J. de Kruif, W. Preiser, and W. Spaan. 2004. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet 363**:**2139-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80a.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84**:**2305-2315. [DOI] [PubMed] [Google Scholar]
- 80b.Thiel, V., and S. Siddell. 1994. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 75**:**3041-3046. [DOI] [PubMed] [Google Scholar]
- 81.Reference deleted.
- 82.Reference deleted.
- 83.Wang, D., A. Urisman, Y. T. Liu, M. Springer, T. G. Ksiazek, D. D. Erdman, E. R. Mardis, M. Hickenbotham, V. Magrini, J. Eldred, J. P. Latreille, R. K. Wilson, D. Ganem, and J. L. DeRisi. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1**:**E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu, D., C. Tu, C. Xin, H. Xuan, Q. Meng, Y. Liu, Y. Yu, Y. Guan, Y. Jiang, X. Yin, G. Crameri, M. Wang, C. Li, S. Liu, M. Liao, L. Feng, H. Xiang, J. Sun, J. Chen, Y. Sun, S. Gu, N. Liu, D. Fu, B. T. Eaton, L.-F. Wang, and X. Kong. 2005. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 79**:**2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reference deleted.
- 85a.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100**:**12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85b.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79**:**14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuan, X., J. Li, Y. Shan, Z. Yang, Z. Zhao, B. Chen, Z. Yao, B. Dong, S. Wang, J. Chen, and Y. Cong. 2005. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 109**:**191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zebedee, S. L., C. D. Richardson, and R. A. Lamb. 1985. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J. Virol. 56**:**502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87a.Zeng, R., R. F. Yang, M. D. Shi, M. R. Jiang, Y. H. Xie, H. Q. Ruan, X. S. Jiang, L. Shi, H. Zhou, L. Zhang, X. D. Wu, Y. Lin, Y. Y. Ji, L. Xiong, Y. Jin, E. H. Dai, X. Y. Wang, B. Y. Si, J. Wang, H. X. Wang, C. E. Wang, Y. H. Gan, Y. C. Li, J. T. Cao, J. P. Zuo, S. F. Shan, E. Xie, S. H. Chen, Z. Q. Jiang, X. Zhang, Y. Wang, G. Pei, B. Sun, and J. R. Wu. 2004. Characterization of the 3a protein of SARS-associated coronavirus in infected Vero E6 cells and SARS patients. J. Mol. Biol. 341**:**271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, J., G. P. Leser, A. Pekosz, and R. A. Lamb. 2000. The cytoplasmic tails of the influenza virus spike glycoproteins are required for normal genome packaging. Virology 269**:**325-334. [DOI] [PubMed] [Google Scholar]
- 89.Reference deleted.