Jasmonate signaling: a conserved mechanism of hormone sensing (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 1.
Published in final edited form as: Curr Opin Plant Biol. 2008 Jun 24;11(4):428–435. doi: 10.1016/j.pbi.2008.05.004
Summary
The lipid-derived hormone jasmonate (JA) regulates diverse aspects of plant immunity and development. Among the central components of the JA signaling cascade are the E3 ubiquitin ligase SCFCOI1 and JAZ proteins that repress transcription of JA-responsive genes. Recent studies provide evidence that amino acid-conjugated forms of JA initiate signal transduction upon formation of a COI1-JA-JAZ ternary complex in which JAZs are ubiquitinated and subsequently degraded. Coronatine, a virulence factor produced by the plant pathogen Pseudomonas syringae, is a potent agonist of this hormone receptor system. Coronatine-induced targeting of JAZs to COI1 obstructs host immune responses to P. syrinage, providing a striking example of how pathogens exploit hormone signaling pathways in the host to promote disease. These findings, together with homology between COI1 and the auxin receptor, TIR1, extend the paradigm of F-box proteins as intracellular sensors of small molecules, and suggest a common evolutionary origin of the auxin and JA response pathways.
Introduction
The plant hormone jasmonate (JA) regulates diverse aspects of plant growth, development, and immunity. This lipid-derived signal and its bioactive derivatives (collectively referred to as JAs) play a critical role in controlling defense responses to an extraordinary range of biotic aggressors, most notably arthropod herbivores and necrotrophic pathogens [1–3]. Other processes that depend on JA signaling include responses to UV radiation and ozone and, depending on the plant species, male and female reproductive development [4]. In general, JA promotes defense and reproduction while inhibiting growth-related processes such as cell division and photosynthesis. These contrasting activities of JA imply a broader role for the hormone in regulating the balance between growth- and defense-related processes, thereby optimizing plant fitness in rapidly changing environments.
A combination of genetic, molecular, and biochemical analyses indicates that the core signal transduction chain linking JA synthesis to hormone-induced changes in gene expression consists of a quartet of interacting players: a JA signal, the SCF-type E3 ubiquitin ligase SCFCOI1, JAsmonate ZIM-domain (JAZ) repressor proteins that are targeted by SCFCOI1 for degradation by the ubiquitin/26S proteasome pathway, and transcription factors (TFs) that positively regulate the expression of JA-responsive genes (Fig. 1). Several major advances in the identification of these players, and the way in which they harmonize, have been reported in the past year. Here, we review this progress and highlight knowledge gaps that remain to be filled. Although we keep to the general theme of biotic interactions by discussing these topics in the context of plant immunity, a common mechanism of JA action likely accounts for most JA-signaled processes, including developmental responses. Readers are referred to several recent review articles for additional information on JA signaling and other aspects of JA biology [4–11].
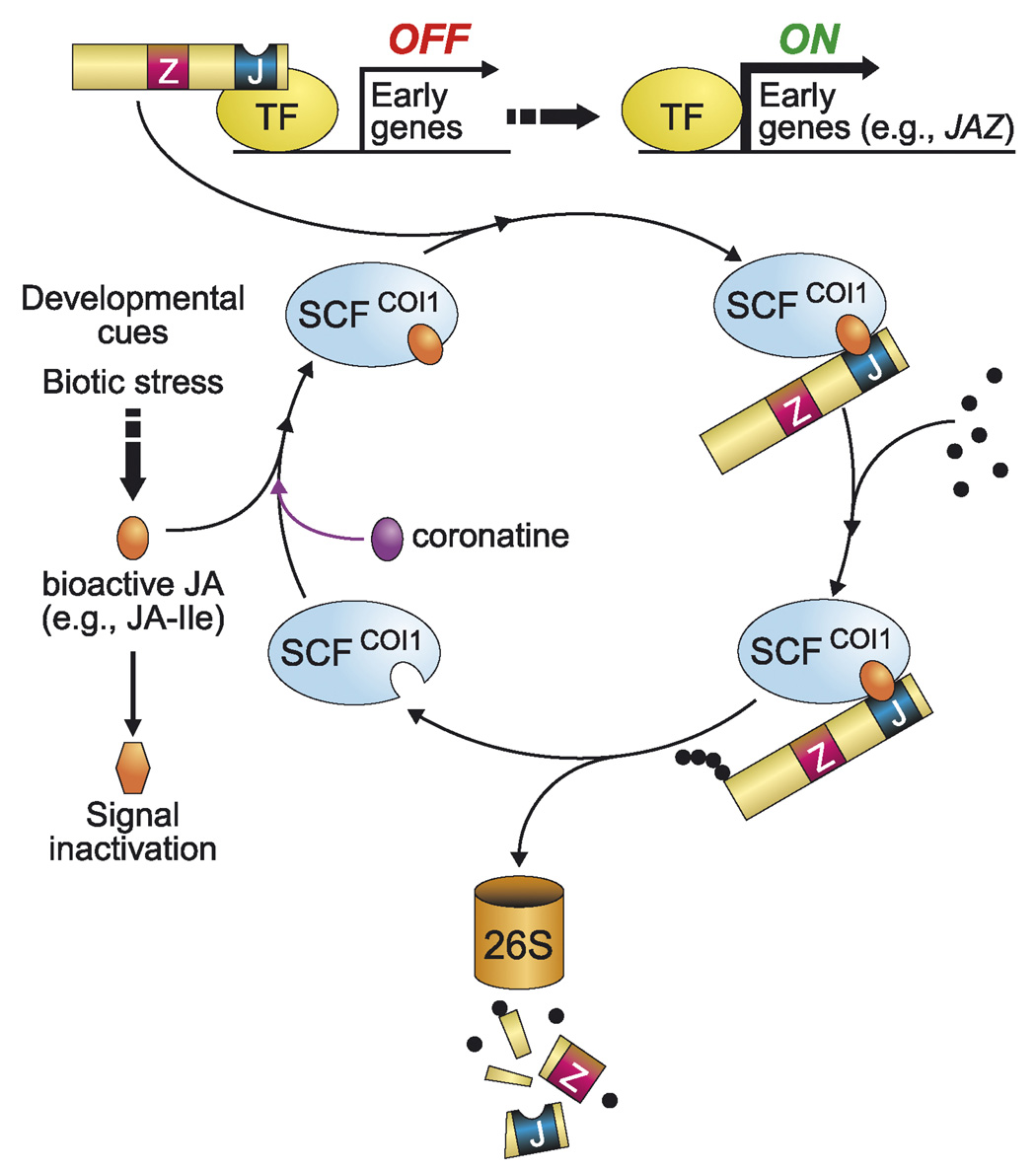
Figure 1. Model of JA signal transduction.
In plant cells containing low JA levels, JA-responsive genes are repressed (OFF) by JAZ proteins (denoted with their ZIM and Jas motifs) that restrain the activity of transcription factors (TF; e.g., MYC2) involved in the expression of early response genes. The transition from the repressed to the active (ON) state of gene expression is initiated by developmental or environmental cues (e.g., biotic stress) that increase the accumulation of bioactive JAs (orange oval; see Box 1). COI1 is an F-box protein that determines the substrate specificity of the SCF-type E3 ubiquitin ligase, SCFCOI1 (blue oval). Bioactive JAs, such as JA-Ile, are proposed to bind to the LRR domain of COI1. Interaction of JAZs with ligand-bound COI1 leads to the formation of a COI1-ligand-JAZ ternary complex in which JAZs are polyubiquitinated (filled black circles) and subsequently degraded by the 26S proteasome (26S). Signaling is attenuated by metabolism of bioactive JAs to inactive forms of the hormone (orange hexagon), as well as by JA-induced de novo JAZ synthesis. Some plant pathogenic strains of Pseudomonas syringae produce a virulence factor called coronatine (purple oval) that is structurally similar to JA-Ile (Box 1). High-affinity binding of coronatine to COI1-JAZ complexes promotes proteolytic destruction of JAZs. Direct binding of JA-Ile or COR to COI1 in the absence of JAZ has not yet been demonstrated, and thus it is possible that COI1 and JAZ function as coreceptors. The model predicts that coi1 mutants fail to respond to JA and coronatine because JAZ proteins are not degraded in the presence of these signals.
All that JAZ: orchestrating four-part harmony
A decade ago, identification of CORONATINE-INSENSITIVE 1 (COI1) as an F-box protein led to the idea that negative regulators of JA signaling are subject to ubiquitin-dependent turnover in response to a JA signal [12,13]. Subsequent biochemical and genetic studies showed that COI1 associates with other proteins of the SCF complex, including ASK1, RBX1, and CUL1, and that these components are important for JA-signaled responses [14–16]. A major breakthrough in understanding how COI1 transduces the JA signal occurred in 2007, when JAZ proteins were identified as SCFCOI1 substrates that negatively regulate the response pathway [17**,18**,19*]. JAZs belong to a larger family of TIFY proteins that is defined by the highly conserved signature sequence TIF[F/Y]XG located in the so-called ZIM domain [20]. The TIFY family includes ZIM and ZIM-like proteins that contain a zinc-finger DNA-binding domain [21], as well as PEAPOD (PPD) proteins that regulate leaf development [22]. Although JAZs and PPDs do not contain a known DNA-binding domain, empirical evidence indicates that at least some JAZs accumulate in the nucleus [17**,18**,19*]. A distinguishing feature of JAZ proteins is the highly conserved Jas motif (SLX2FX2KRX2RX5PY) located near the C-terminus (Fig. 2; Fig. S1, Supplemental Material). PPD proteins, which have not been directly implicated in JA signaling, contain a modified Jas motif that lacks several JAZ-specific residues.
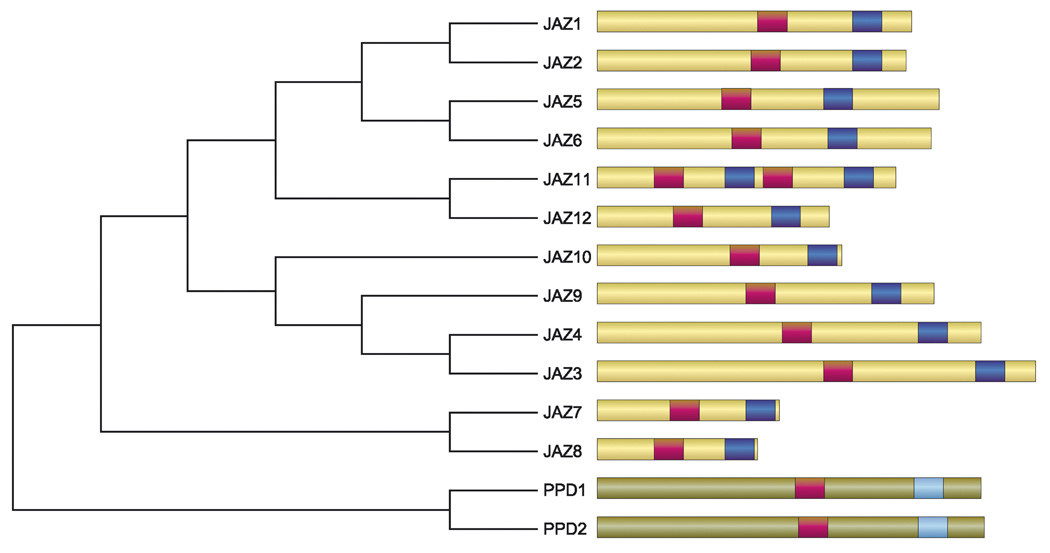
Figure 2. The JAZ protein family in Arabidopsis.
The phylogeny includes JAZ and PEAPOD (PPD) proteins, which are members of the larger family of TIFY proteins that is defined by the TIF[F/Y]XG motif in the ZIM domain (red bar). JAZs are distinguished from other TIFY proteins by the presence of the C-terminally located Jas motif, SLX2FX2KRX2RX5PY (dark blue bar). PPD proteins contain a modified Jas motif (light blue bar) that lacks several invariant residues of the motif (Fig. S1, Supplemental material). The relative size of each protein and the position of the conserved motifs are drawn to scale.
The JAZ family in Arabidopsis consists of 12 members that generally cluster into sequence-related pairs (Fig. 2). Transgenic silencing of JAZ10 (also called JAS1), which lacks a closely related JAZ paralog, results in hypersensitivity to JA, thus confirming a negative regulatory function for this protein [19*]. The lack of obvious JA-related phenotypes in other jaz mutants suggests that different family members have overlapping or redundant functions in JA signaling [17**]. JAZ genes are present in angiosperms and gymnosperms but not in species outside the plant kingdom, including photosynthetic aquatic eukaryotes. Interestingly, putative JAZ orthologs are present in the liverwort Marchantia polymorpha [20] and in the genome of the moss Physcomitrella patens (Fig. S1, Supplemental Material). Although the ability of P. patens to synthesize or respond to JA has not been reported in the literature, it is noteworthy that this bryophyte contains genes for the JA biosynthetic enzymes allene oxide synthase/cyclase (I Feussner, personal communication), as well as putative JA-conjugating enzymes (Box 1)[23]. These observations raise the possibility that the JA pathway evolved in early land plants as an adaptation to the plethora of biotic and abiotic stress conditions that were encountered upon invasion of land by plants.
Bioactive and non-bioactive JAs.
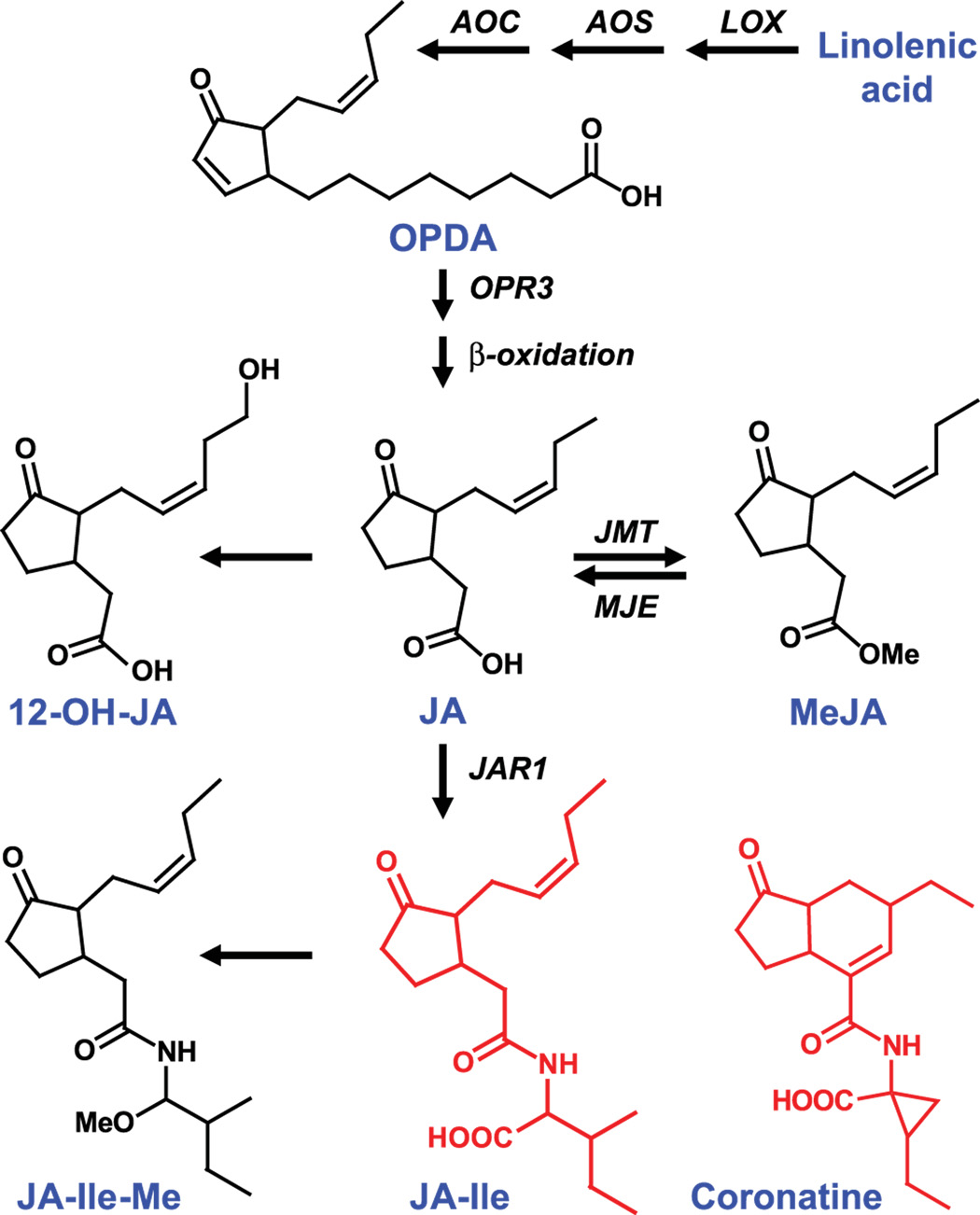
JA synthesis is initiated in the chloroplast, where linolenic acid is converted to 12-oxo-phytodienoic acid (OPDA) by the sequential action of lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC). Following transport to the peroxisome, OPDA is reduced to OPC-8:0 (not shown) by OPDA reductase3 (OPR3) and subjected to three cycles of β-oxidation to yield (+)-7-iso-JA (JA). JA is further metabolized to various inactive and bioactive derivatives. Only a subset of these compounds is shown. Jasmonate carboxyl methyltransferase (JMT) converts JA to the volatile compound MeJA. The reverse reaction is catalyzed by MeJA esterase (MJE). Conjugation of JA to Ile by JAR1 produces JA-Ile. JA-Ile can be methylated to produce JA-Ile-Me [59], the biological role of which is not known. Coronatine is a phytotoxic virulence factor produced by pathovars of Pseudomonas syringae.
Bioactive JAs can be defined as JA derivatives that directly promote COI1 interaction with one or more JAZ proteins to activate gene expression. Non-bioactive JAs are either precursors or deactivated forms of bioactive JAs. 12-hydroxy-JA (12-OH-JA) is an example of a deactivated form of JA [60]. Several criteria can be used to determine whether a particular compound is a bioactive signal. These criteria include: i) the compound is synthesized in plant cells; ii) the exogenous compound elicits a physiological response in wild-type but not coi1 mutant plants; iii) depletion of the compound in plant tissues by genetic or pharmacological means impairs physiological responses that depend on JA; and iv) the compound directly promotes COI1-JAZ interaction. Although OPDA, JA (i.e., jasmonic acid), MeJA, and JA-Ile satisfy the first three criteria, only JA-Ile has been shown to fulfill all four criteria.
In cells containing low JA levels, JAZs restrain the activity of TFs that positively regulate early JA-response genes (Fig. 1). This aspect of the model is based upon the key finding [18**] that Arabidopsis JAZ3 (also known as JAI3) interacts directly with MYC2 (also known as JIN1), a basic helix-loop-helix TF that serves a well-established role in JA signaling [18**,24–28]. The JAZ3-MYC2 interaction involves sequences located in the N- and C-terminal regions of MYC2 and JAZ3, respectively [18**]. Yeast two-hybrid studies showed that MYC2 also interacts with JAZ1 and JAZ9 and that these interactions, unlike COI1-JAZ partnering (see below), occur in both the presence and absence of exogenous JA [29*]. It remains to be determined how JAZs inhibit MYC2 activity. The fact that myc2 mutants are defective in some but not all (e.g., male fertility) JA responses predicts the existence of a larger JAZ-TF interactome.
Paradoxically, JAZ genes themselves are rapidly induced in response to JA treatment and environmental stress conditions that stimulate JA production (Fig. 1) [17**,18**,19*,20,30*]. Based on the function of JAZs as negative regulators, JA-induced JAZ expression appears to constitute a negative feedback loop in which newly synthesized JAZs desensitize or dampen the strength of the response by obstructing TF activity. The ability of MYC2 to bind a G-box motif in the promoter of JAZ genes, together with the fact that _JAZ_s are constitutively expressed in MYC2-overproducing plants, provides additional evidence for autoregulation of JA responses [18**,27]. Rapid expression of JAZ genes in response to increased endogenous JA content is consistent with the idea that early JA-response genes are expressed upon hormone-induced release of TFs from JAZ restraint [30*]. Experiments with the protein synthesis inhibitor cycloheximide further suggest that JAZ gene expression is activated upon COI1-dependent turnover of labile repressors, presumably JAZs [30*].
A COI weapon of JAZ destruction
The switch between restrained and active states of JA-responsive gene expression is triggered by hormone-induced proteolysis of JAZs via the SCFCOI1/ubiquitin/26S proteasome pathway. This transition is initiated in response to biotic stress or other cues that result in JA accumulation (Fig. 1). Two key pieces of experimental evidence support this view. First, JA stimulates turnover of JAZ-reporter fusion proteins in planta by a mechanism that requires COI1 and the 26S proteasome [17**,18**]. Second, bioactive JAs promote direct interaction between COI1 and JAZ proteins (see below). These findings support a scenario in which a JA signal triggers SCFCOI1/ubiquitin-mediated destruction of JAZs, which allows TFs (e.g., MYC2) to engage in transcription of early response genes. This simple hormone-induced switch provides tight temporal coupling between increased JA levels and changes in gene expression. For example, rapid (<5 minutes) JA accumulation in response to tissue damage occurs coincidently with the expression of early JA-response genes [30*]. The speed of this response may be advantageous for protection against attackers that betray their presence to the host by triggering JA synthesis in injured or infected tissue.
Truncated JAZ proteins (referred to here as JAZΔJas) lacking the Jas motif are stabilized against JA-induced turnover and confer a variety of dominant JA-insensitive phenotypes [17**,18**,29*,30*]. Overexpression of a splice variant (JAS1.3) of JAZ10/JAS1 that lacks 7 amino acids of the Jas motif also results in reduced sensitivity to JA [19*], suggesting that naturally occurring JAZΔJas proteins modulate JA responsiveness in vivo. These observations are consistent with the hypothesis that the Jas motif functions as a “dergron” that mediates JAZ interaction with COI1. This idea is supported by COI1-JAZ interaction studies in tomato [31**] and, in particular, by site-directed mutagenesis studies showing that the Jas motif is required for hormone-dependent binding of several JAZs to COI1 [29*]. A simple model to explain the dominant action of JAZΔJas proteins is that they fail to interact with SCFCOI1 and, as a consequence, accumulate and repress the activity of TFs involved in JA responses. Interestingly, however, Chini et al. [18**] reported that JAZ3ΔJas interacts with COI1 in a hormone-independent manner. Based on the fact JAZ3ΔJas does not interact with MYC2, these workers proposed an alternative “poison complex” model in which JA-independent binding of JAZ3ΔJas to SCFCOI1 obstructs the interaction of endogenous JAZs with the E3 ligase complex, thereby allowing endogenous JAZs to repress TF activity. Additional work is needed to determine the precise mechanism(s) by which JAZΔJas proteins reduce JA responsiveness.
Bioactive JAs: activation by conjugation
Newly synthesized jasmonic acid is subject to various enzymatic modifications that give rise to a plethora of JA derivatives [3,6,8]. An important challenge in the field of JA signal transduction is to identify the spectrum of JAs that directly promote COI1-JAZ interactions (Box 1). Cell-free and yeast-based assays showed that COI1 binding to certain JAZs is stimulated by jasmonoyl-isoleucine (JA-Ile) and structurally related JA-amino acid conjugates (e.g., JA-Val) [17**,29*,31**]. In light of increasing evidence that non-conjugated JAs are active per se as signals [32–37], it is noteworthy that jasmonic acid, MeJA, and OPDA were not active in these COI1-JAZ interaction assays. Although these findings support the view that JA-Ile (and perhaps related conjugates) is the primary signal for COI1-dependent responses, the possibility that non-conjugated JAs promote COI1 binding to other JAZ isoforms cannot be excluded. It is also conceivable that non-conjugated JAs regulate gene expression through as yet unidentified COI1 substrates [5].
Initial insight into the importance of JA-Ile as a bioactive signal came from studies of the Arabidopsis jar1 mutant that is defective in conjugating JA to Ile [10]. As a member of the GH3 family of carboxylic acid-conjugating enzymes, JAR1 exhibits high substrate specificity for jasmonic acid and Ile [36,38,39]. Phenotypes associated with jar1 mutants demonstrate that JA-Ile is important for plant protection against necrotrophic soil pathogens [40], lepidopteran insects [41], and various abiotic stresses as well [42]. JAR1 activity, however, is not required for all COI1-dependent responses [30*,34,36]. The persistence of certain JA-mediated responses in jar1 null mutants is consistent with the view that non-conjugated JAs (e.g., jasmonic acid) can promote COI1-JAZ interactions in vivo. An alternative explanation is that the jar1 mutants used in these studies produce sufficient amounts of JA-Ile to trigger COI1-mediated JAZ degradation and subsequent expression of JA-responsive genes. This idea is supported by the fact that flowers and wounded leaves of Arabidopsis jar1 mutants contain significant levels of JA-Ile [30*,36].
Coronatine (COR) is a phytotoxin produced by plant pathogenic strains of Pseudomonas syringae [43]. COR suppresses host immune responses by activating the JA signaling pathway in a COI1-dependent manner [26,44–47]. Indeed, COI1 derives its name from an Arabidopsis locus that, when mutated, results in insensitivity to COR and JA [44]. The structural similarity of COR to JA-Ile (Box 1) led to suggestion that the toxin acts as a molecular mimic of JA-Ile [10,38,48]. This hypothesis was validated by studies showing that COR triggers COI1-JAZ interaction in yeast two-hybrid and in vitro pull-down assays [31**,49]. Remarkably, COR is at least 100-fold more active than JA-Ile in promoting COI1-JAZ binding in vitro [31**]. COR-mediated JAZ degradation by P. syringae provides a compelling example of how pathogens exploit hormone signaling pathways in the host to promote infection (Fig. 1).
The jasmonate receptor: here come those TIRs again
The emerging view of JA signaling (Fig. 1) bears striking similarity to the mechanism of auxin action. It now appears that the functions of COI1, JAZ, and MYC2 in JA signaling are analogous to the core components of the auxin signaling pathway, namely the F-box protein TIR1 (and TIRl-like proteins), Aux/IAA repressor proteins, and auxin response factors, respectively. Sequence homology between COI1 and TIR1 implies a conserved role for these F-box proteins in signal transduction and, moreover, a common evolutionary origin of the two hormone response pathways. The lack of sequence similarity between JAZ and Aux/IAA proteins suggests that the interaction of these substrates with their respective F-box proteins may have evolved independently.
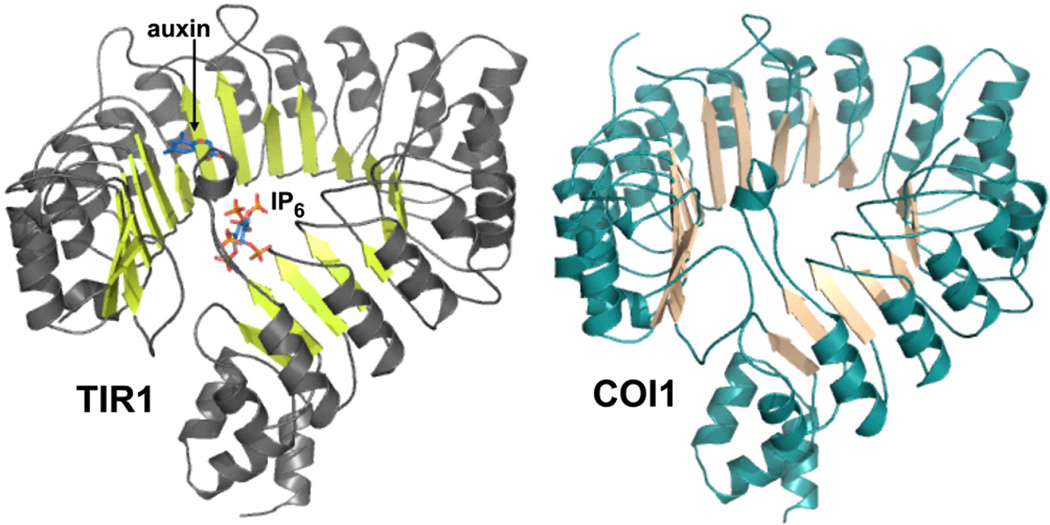
Elegant biochemical and crystallography studies have shown TIR1 functions as an auxin receptor [50**,51–53]. This work revealed that auxin binding to the TIR1 LRR promotes substrate recruitment, not allosterically, but rather by creating a hydrophobic pocket into which the Aux/IAA protein fits [50**]. Emerging evidence indicates that COI1 may play a similar role in JA perception. Homology modeling, for example, predicts that the COI1 LRR adopts the hallmark horseshoe-like shape of the TIR1 LRR (Fig. 3). An inositol-hexakisphosphate (IP6) cofactor positioned at the center of the TIR1-LRR solenoid lies in close proximity to the auxin-binding site. Although it remains to be determined whether COI1 possesses this cofactor, sequence and structural alignments indicate that amino acid residues in TIR1 involved in coordinating IP6 are conserved in COI1 [50**]. Residues that comprise the hormone-binding site on TIR1 are less well conserved in COI1, as might be expected from the structural differences in the respective hormones. Nevertheless, many of the amino acid substitutions within the putative ligand-binding pocket of COI1 retain the hydrophobic character of the auxin binding pocket (L Katsir and GA Howe, unpublished).
Figure 3. Homology model of COI1.
The x-ray crystal structure of TIR1 (a) [50**] was used as a template to construct a homology model of COI1 (b) with the program SWISS-MODEL. Images were rendered using PyMol software. The positions of the IP6 co-factor and auxin in the TIR structure are indicated.
Biochemical analysis of COI1-JAZ interactions in tomato indicate that COI1 plays a direct role in hormone binding [31**]. This study showed that radiolabled COR binds specifically and with high affinity (_K_d ~20 nM) to COI1-JAZ complexes. Detection of a COI1-COR-JAZ complex is consistent with the idea that COR, like auxin [50**], is a “molecular glue” that stabilizes the interaction between F-box proteins and their cognate substrates (Fig. 1). The ability of JA-Ile to compete with COR for binding indicates that both compounds are recognized by the same receptor. In addressing the question of whether the receptor is COI1, JAZ, or a COI1-JAZ complex, Katsir et al. [31**] showed that COR binding requires COI1, and that JAZ alone does not bind this ligand with high affinity. Yeast two-hybrid analyses have further shown that hormone-induced COI1-JAZ interaction does not require plant proteins other than COI1 and JAZ [17**,29*]. These collective findings support the conclusion that COI1 is an essential component of an intracellular receptor for JA-Ile and that COR is a potent agonist of this receptor (Fig. 1).
Conclusions and future perspectives
Major strides toward elucidating the molecular mechanism of JA action have been reported in the past year. These advances include the discovery of JAZ proteins as substrates for SCFCOI1 and the identification of COI1 as a component of the JA perception machinery. This work extends the paradigm [50**] of F-box proteins as intracellular receptors for small molecules that regulate fundamental processes in plants. This new view of JA signal transduction frames several key questions that remain to be answered. For example, how is the intracellular level of bioactive JAs controlled in response to developmental and environmental cues? Recent studies on lipases that control the release of JA precursors in leaves [54*] and flowers [55*] provide important advances in this direction. Another unanswered question is whether JA-Ile promotes the degradation of all JAZs, or whether other JA derivatives (e.g., non-conjugated JAs) also perform this function. In vitro and yeast-based COI1-JAZ interaction assays, together with mutants that completely lack JA-Ile, will help to resolve this issue. Because it remains to be determined whether COI1 binds JA-Ile/COR in the absence of JAZ, it will also be important to discriminate between models in which hormone binding is mediated by COI1 alone (Fig. 1) or an alternative model in which COI1 and JAZ act as co-receptors.
One of the most important questions in JA biology is how the diversity of hormone-mediated developmental and defense-related responses is regulated in specific organs and cell types. Unlike the auxin signaling pathway in which hormone responses are controlled by TIR1 and several additional TIR1-like receptors [56], the strong JA-insensitive phenotype of coi1 null mutants [12,57] indicates that most JA responses are controlled by the activity of a single E3 ligase, SCFCOI1. It therefore seems likely that the specificity of COI1-JA-JAZ and JAZ-TF interactions plays a central role in determining which of the myriad JA responses are expressed in a given cell type. Likewise, the JAZ-TF interactome may comprise a regulatory node for integrating JA responses with other hormone signaling pathways [11,58]. Future progress in these areas will be facilitated by detailed phenotypic analysis of jaz mutants, elucidation of conserved functional domains in JAZs, and the identification of additional JAZ-interacting proteins. The application of X-ray crystallography approaches [50**] to study COI1 and its interacting partners may ultimately provide the best picture of how bioactive JAs mediate COI1-JAZ interaction and, perhaps, how F-box proteins evolved as hormone sensors.
Supplementary Material
01
Acknowledgments
We thank Ning Zheng for helpful comments on the manuscript, Ivo Feussner for sharing unpublished results, and Marlene Cameron for assistance in the preparation of figures. This work was supported by grants from the National Institutes of Health (R01GM57795) and the U.S. Department of Energy (DE-FG02-91ER20021).
Abbreviations
JA
jasmonate
MeJA
methyl jasmonate
JA-Ile
jasmonoyl-isoleucine
JAZ
Jasmonate ZIM-domain
OPDA
12-oxo-phytodienoic acid
COI1
CORONATINE-INSENSITIVE1
SCF
Skp/Cullin/F-box
JAR1
JASMONIC ACID RESISTANT1
PPD
PEAPOD
COR
coronatine
TF
transcription factor
IP6
inositol-hexakisphosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Leron Katsir, Email: katsirle@msu.edu.
Hoo Sun Chung, Email: chunghoo@msu.edu.
Abraham J.K. Koo, Email: koojeon1@msu.edu.
Gregg A. Howe, Email: howeg@msu.edu.
References and recommended reading
- 1.Howe G, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 2.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 3.Browse J, Howe GA. Update on jasmonate signaling: New weapons and a rapid response against insect attack. Plant Physiol. 2008;146:832–838. doi: 10.1104/pp.107.115683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browse J. Jasmonate: An oxylipin signal with many roles in plants. Vitam Horm. 2005;72:431–456. doi: 10.1016/S0083-6729(05)72012-4. [DOI] [PubMed] [Google Scholar]
- 5.Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 2008:301–318. doi: 10.1111/j.1469-8137.2007.02292.x. [DOI] [PubMed] [Google Scholar]
- 6.Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilmiller AL, Howe GA. Systemic signaling in the wound response. Curr Opin Plant Biol. 2005;8:369–377. doi: 10.1016/j.pbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Schaller F, Schaller A, Stintzi A. Biosynthesis and metabolism of jasmonates. J Plant Growth Regu. 2005;23:179–199. [Google Scholar]
- 9.Liechti R, Gfeller A, Farmer EE. Jasmonate signaling pathway. Sci STKE. 2006;2006:cm2. doi: 10.1126/stke.3222006cm2. [DOI] [PubMed] [Google Scholar]
- 10.Staswick PE. JAZing up jasmonate signaling. Trends Plant Sci. 2008;13:66–71. doi: 10.1016/j.tplants.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Kazan K, Manners JM. Jasmonate signaling: toward an integrated view. Plant Physiol. 2008;146:1459–1468. doi: 10.1104/pp.107.115717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 13.Creelman RA. Jasmonate perception: characterization of COI1 mutants provides the first clues. Trends Plant Sci. 1998;3:367–368. [Google Scholar]
- 14.Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32:457–466. doi: 10.1046/j.1365-313x.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 16.Lorenzo O, Solano R. Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol. 2005;8:532–540. doi: 10.1016/j.pbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- **17.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960.The authors use an expression profiling approach to identify the family of JAZ genes that encode negative regulators of the JA signaling pathway. JAZ proteins were shown to be targets of SCFCOI1 during the JA response. In vitro pull-down and yeast two-hybrid assays demonstrated that COI1 interaction with JAZ1 is mediated by JA-Ile but not by non-conjugated JAs including jasmonic acid, MeJA, and OPDA.
- **18.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006.This paper reports the use of a forward genetics approach to identify JAZ proteins as targets of SCFCOI1. JAZ3/JAI3 was shown to physically interact with the transcription factor MYC2, a positive regulator of JA signaling. The paper also presents evidence that MYC2 directly regulates the expression of JAZ genes.
- *19.Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708.An alternative splicing event that removes 7 amino acids from the Jas motif of JAS1 (JAZ10) confers a dominant JA-insensitive phenotype, implicating endogenous JAZΔJas proteins in the regulation of JA responses. The authors also report that loss of JAS1/JAZ10 function results in hypersensitivity to JA. This finding is consistent with a role for JAS1/JAZ10 as a negative regulator of the pathway.
- 20.Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends Plant Sci. 2007;12:239–244. doi: 10.1016/j.tplants.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Shikata M, Takemura M, Yokota A, Kohchi T. Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci Biotechnol Biochem. 2003;67:2495–2497. doi: 10.1271/bbb.67.2495. [DOI] [PubMed] [Google Scholar]
- 22.White DWR. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:13238–13243. doi: 10.1073/pnas.0604349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terol J, Domingo C, Talon M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene. 2006;371:279–290. doi: 10.1016/j.gene.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurie-Berry N, Joardar V, Street IH, Kunkel BN. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact. 2006;19:789–800. doi: 10.1094/MPMI-19-0789. [DOI] [PubMed] [Google Scholar]
- 27.Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick PE, Browse J, Howe GA, He SY. The bacterial toxin coronatine and the plant hormone jasmonoyl isoleucine target the physical interaction between the Arabidopsis COI1 F-box protein and the Jas domain of JAZ repressor proteins. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03566.x. in press.This paper shows that the Jas motif mediates hormone-dependent interaction of COI1 with Arabidopsis JAZ1, JAZ3, and JAZ9. The results also demonstrate that coronatine can substitute for JA-Ile in promoting COI1-JAZ interactions, and the multiple JAZs interact with MYC2.
- *30.Chung HS, Koo AJK, Gao X, Jayany S, Thines B, Jones AD, Howe GA. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691.Gene expression and JA/JA-Ile measurements show that COI1-medaited transcriptional responses are activated within 5 minutes of tissue damage. Increased susceptibility of a JAZ1ΔJas-expressing line of Arabidopsis to Spodoptera exigua establishes a role for JAZs in anti-insect defense.
- **31.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105.This paper shows that tomato COI1 plays a direct role in binding JA-Ile and the bacterial virulence factor coronatine, thereby indicating the COI1 or a COI1-JAZ complex is a JA receptor. The authors also show that the Jas domain-containing C-terminal region of tomato JAZ3 is critical for hormone-dependent binding of JAZ3 to COI1.
- 32.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci U S A. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci U S A. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Allmann S, Wu J, Baldwin IT. Comparisons of LOX3- and JAR4/6-silenced plants reveal that JA and JA-AA conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter A, Mazars C, Maitrejean M, Hopke J, Ranjeva R, Boland W, Mithofer A. Structural requirements of jasmonates and synthetic analogues as inducers of Ca2+ signals in the nucleus and the cytosol of plant cells. Angew Chemie. 2007;46:4783–4785. doi: 10.1002/anie.200604989. [DOI] [PubMed] [Google Scholar]
- 36.Suza WP, Staswick PE. The role of JAR1 in jasmonoyl-L-isoleucine production in Arabidopsis wound response. Planta. 2008;227:1221–1232. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 37.Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y. Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSTIVE 1-dependent pathway. Plant Physiol. 2008 doi: 10.1104/pp.108.119321. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiryaki I, Staswick PE. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 2002;130:887–894. doi: 10.1104/pp.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staswick PE, Yuen GY, Lehman CC. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998;15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 41.Kang JH, Wang L, Giri A, Baldwin IT. Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell. 2006;18:3303–3320. doi: 10.1105/tpc.106.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell. 2000;12:1633–1646. doi: 10.1105/tpc.12.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 46.Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant Journal. 2005;42:201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 47.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157: H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 48.Krumm T, Bandemer K, Boland W. Induction of volatile biosynthesis in the lima bean (Phaseolus lunatus) by leucine- and isoleucine conjugates of 1-oxo- and 1-hydroxyindan-4-carboxylic acid: evidence for amino acid conjugates of jasmonic acid as intermediates in the octadecanoid signalling pathway. FEBS Lett. 1995;377:523–529. doi: 10.1016/0014-5793(95)01398-9. [DOI] [PubMed] [Google Scholar]
- 49.Melotto M, Underwood W, He SY. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol. 2008;46 doi: 10.1146/annurev.phyto.121107.104959. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731.The crystal structure of the F-box protein TIR1 reveals a novel mode of E3 ligase-substrate interaction in which auxin acts as a "molecular glue" to bind TIR1 to Aux/IAA substrates. Sequence homology between TIR1 and COI1 suggests that COI1 may play a similar role in JA signaling.
- 51.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 52.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13:1418–1422. doi: 10.1016/s0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- 53.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- *54.Hyun Y, Choi S, Hwang HJ, Yu J, Nam SJ, Ko J, Park JY, Seo YS, Kim EY, Ryu SB, et al. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev Cell. 2008;14:183–192. doi: 10.1016/j.devcel.2007.11.010.This paper identifies a lipase, called DONGLE, which is responsible for initiating JA biosynthesis in Arabidopsis leaves. DONGLE is a chloroplast-targeted galactolipase that works together with a closely lipase (DAD1) to release fatty acid precursors of JA in response to wounding.
- *55.Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467.This paper shows that the floral identity gene AGAMOUS controls JA responses during late-stage floral development by regulating the expression of the DAD1 lipase involved in JA synthesis. The results illustrate that JA-signaled processes are precisely regulated by developmental cues.
- 56.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Navarro L, Bari R, Achard P, Lison P, Nemri A, Harberd NP, Jones JD. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 59.Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C. Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 2000;24:113–126. doi: 10.1046/j.1365-313x.2000.00861.x. [DOI] [PubMed] [Google Scholar]
- 60.Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 2007:114–127. doi: 10.1111/j.1469-8137.2007.02252.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01