Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn disease susceptibility (original) (raw)
. Author manuscript; available in PMC: 2009 Jan 19.
Published in final edited form as: Nat Genet. 2007 Jun 6;39(7):830–832. doi: 10.1038/ng2061
Abstract
A genome-wide association scan in Crohn disease by the Wellcome Trust Case Control Consortium1 detected strong association at 6 novel loci. We tested 37 SNPs from these and other loci for association in an independent case control sample. Replication was obtained for the IRGM gene on chromosome 5q33.1 which induces autophagy (replication P = 6.6 × 10−4, combined P = 2.1 × 10−10), and for 9 other loci including NKX2-3 and gene deserts on chromosomes 1q and 5p13.
Crohn disease (CD) is a common form of chronic inflammatory bowel disease. Established CD susceptibility genes NOD2 (CARD15), IL23R and _ATG16L1_ 2-5 showed strong evidence of association in the Wellcome Trust Case Control Consortium (WTCCC) genome-wide scan of 1748 CD cases and 2938 controls genotyped using the Affymetrix 500K chip. Six other loci also showed highly significant association. Although satisfying stringent statistical thresholds for significance (P < 5 × 10−7), replication in independent panels represents a key validation step.
We followed up 37 SNPs from 31 distinct loci associated at P < 10−5 on initial analysis of the WTCCC dataset. Support for some of these markers diminished in the final WTCCC analysis after extensive data filtering1. Two markers were selected for each locus where low LD between associated SNPs in areas of unbroken LD suggested distinct causal variants. SNPs were genotyped in a new panel of 1182 Caucasian CD cases using TaqMan assays (Supplementary Methods and Supplementary Table 1 online). Concordance with Affymetrix data was 99.7%, based on genotyping 96 WTCCC cases on both platforms. To target SNPs for replication testing and limit unnecessary genotyping, preliminary comparison was made between allele frequencies in the new CD panel and 5746 non-autoimmune WTCCC cases (bipolar disorder, coronary artery disease and hypertension). The latter represent a powerful resource of population allele frequencies1 assuming no overlap with CD aetiology. Twelve SNPs showed a difference in allele frequency between these two groups (Table 1), while for 25 other markers the allele frequencies converged (Supplementary Table 2). To exclude possible bias introduced by this approach, the 12 markers implicated in this preliminary comparison were then genotyped in 2024 independent population controls from the 1958 British Birth Cohort to test formally for association. Results are presented in Table 1, with odds ratios estimated from replication samples only.
Table 1. 12 SNPs genotyped in 1182 Crohn disease cases and 2024 controls.
Allele frequencies and association results for 12 SNPs genotyped in 1182 cases from the replication Crohn's disease panel (RCD) compared to frequencies for these markers in the original WTCCC Crohn Disease samples (WTCD), WTCCC Unaffected control samples (WTU1), WTCCC Unaffected case samples (WTU2) (from coronary artery disease, hypertension and bipolar disorder cohorts)1 and 2024 replication unaffected samples (RU) from the 1958 British Birth Cohort. Quoted p-values are for the final, clean WTCCC dataset.1 Some SNPs with p > 10−5 in this table showed stronger evidence in the preliminary data from which we fast-tracked them for follow-up. Odds ratios are estimated from the replication samples. Regions are shown in alternating rows of white and grey (adjacent rows of the same colour are from the same region).
| SNP | Chr | Region (Mb) | Gene | WTCD | RCD | WTU1 | WTU2 | RU | WTCCC p | Rep p | Combined p | OR (95%CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12035082 | 1q24 | 169.53 - 169.67 | 0.438 | 0.429 | 0.389 | 0.396 | 0.398 | 2.15×10−6 | 0.01692 | 2.07×10−7 | 1.14 (1.02 - 1.27) | |
| rs10801047 | 1q31 | 188.17 - 188.32 | 0.09 | 0.108 | 0.067 | 0.072 | 0.076 | 8.15×10−5 | 4.78×10−5 | 2.83×10−8 | 1.47 (1.22 - 1.76) | |
| rs9858542 | 3p21 | 49.3 - 49.87 | Many | 0.331 | 0.324 | 0.282 | 0.289 | 0.291 | 7.71×10−7 | 0.01058 | 4.91×10−8 | 1.17 (1.04 - 1.31) |
| rs17234657 | 5p13 | 40.32 - 40.66 | 0.181 | 0.151 | 0.125 | 0.132 | 0.132 | 2.13×10−13 | 0.05305 | 3.80×10−12 | 1.16 (1.00 - 1.35) | |
| rs9292777 | 5p13 | 40.32 - 40.66 | 0.32 | 0.332 | 0.394 | 0.391 | 0.4 | 1.89×10−12 | 2.88×10−7 | 3.20×10−18 | 1.34 (1.20 - 1.50) | |
| rs10077785 | 5q23 | 131.40 - 131.90 | IBD5 | 0.192 | 0.202 | 0.234 | 0.241 | 0.232 | 1.81×10−6 | 0.008086 | 7.53×10−8 | 1.19 (1.05 - 1.36) |
| rs13361189 | 5q33 | 150.15 - 150.31 | IRGM | 0.098 | 0.108 | 0.067 | 0.079 | 0.081 | 4.38×10−8 | 0.000664 | 2.09×10−10 | 1.38 (1.15 - 1.66) |
| rs4958847 | 5q33 | 150.15 - 150.31 | IRGM | 0.147 | 0.158 | 0.113 | 0.123 | 0.125 | 2.41×10−6 | 0.00031 | 3.77×10−9 | 1.36 (1.17 - 1.59) |
| rs6887695 | 5q33 | 158.61 - 158.76 | IL12B | 0.343 | 0.369 | 0.318 | 0.314 | 0.318 | 0.009871 | 8.42×10−5 | 9.21×10−6 | 1.26 (1.12- 1.41) |
| rs10883365 | 10q24 | 101.26 - 101.32 | NKX2-3 | 0.537 | 0.523 | 0.477 | 0.489 | 0.482 | 1.41×10−8 | 0.00373 | 3.71×10−10 | 1.18 (1.05 - 1.32) |
| rs2542151 | 18p11 | 12.76 - 12.91 | PTPN2 | 0.208 | 0.196 | 0.163 | 0.168 | 0.175 | 4.56×10−8 | 0.0479 | 3.16×10−8 | 1.15 (1.00 - 1.32) |
| rs2836754 | 21q22 | 39.21 - 39.24 | 0.399 | 0.387 | 0.353 | 0.35 | 0.354 | 1.07×10−5 | 0.01224 | 5.48×10−7 | 1.15 (1.03 - 1.28) |
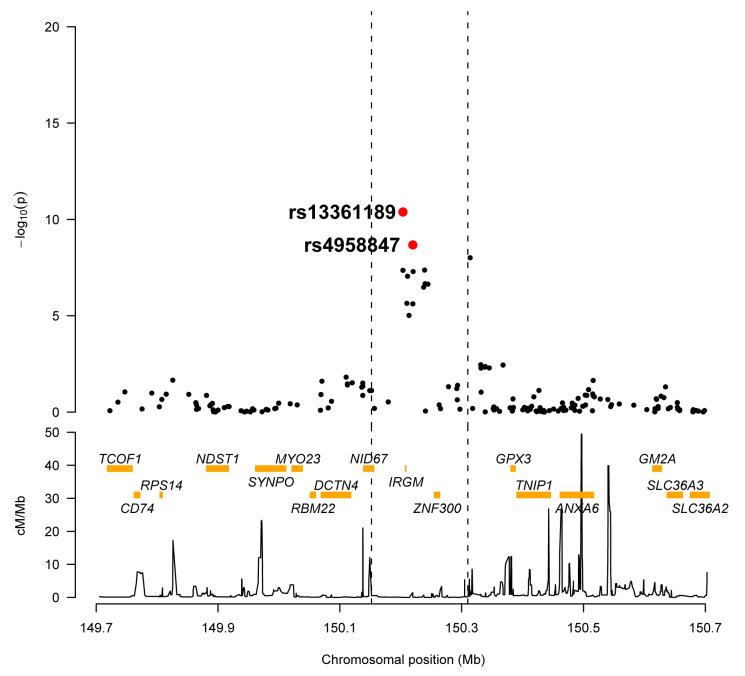
Among novel CD loci achieving genome-wide significance in the WTCCC scan1 the strongest replication adjacent to a known gene was for SNPs rs13361189 and rs4958847 (Prep=6.6×10−4 and 3.1×10−4 respectively) immediately flanking the IRGM gene [MIM 608212] on chromosome 5q33.1. Association in the combined panels of 2930 CD cases and 4962 controls was highly significant (Pcomb= 2.1×10−10 and 3.8×10−9 respectively) (Figure 1). The long coding exon of the human IRGM gene encodes a 20 kD protein of 181 amino acid residues.6,7 Since none of the associated SNPs are known to be functional, we re-sequenced this coding exon of IRGM and the 4 small putative downstream exons in 48 CD cases homozygous or heterozygous for risk alleles (Supplementary Methods). We detected two novel non-synonymous sequence variants, c.51G>C (p.Glu17Asp) and c.281C>A (p.Thr94Lys) and an exonic synonymous SNP, c.313T>C (rs10065172, p.L105). These SNPs were then genotyped in 769 unselected CD cases from our study and 705 controls. Only the silent 313T>C variant was associated with CD (P=0.008), and was in near perfect LD with SNP rs13361189 (r2=0.91). Sequencing of the IRGM coding region in a further 100 unselected CD cases and 100 controls did not detect additional variants. These results strongly suggest that the causal variant(s) do not change the amino acid sequence of the IRGM protein. They may lie in regulatory sequences, in LD with the associated SNPs, and affect IRGM expression; alternatively, the exonic (c.313T>C) SNP itself might affect transcript splicing or the rate of translation of the protein. Analysis of IRGM expression by PCR screening against uncloned cDNA showed transcription in several tissues, including colon, small bowel, peripheral blood leukocytes and the U937 monocytic cell line (data not shown).
Figure 1.
Associated region at the gene IRGM. Strength of association of Affymetrix 500K SNPs in the region,1 with combined p-values shown in red for replicated SNPs. Recombination map showing how the signals are clearly demarcated by recombination hotspots. Nearby genes are shown in yellow.
IRGM belongs to the p47 immunity-related GTPase family. LRG-47 (irgm1), its mouse homologue, critically controls intracellular pathogens by autophagy, and _LRG-47_−/− mice showed markedly increased susceptibility to Toxoplasma gondii and Listeria monocytogenes.8 Consistent with this, IRGM induces autophagy and thereby control of intracellular M. tuberculosis in human macrophages.7
Three other associations with genome-wide significance in the WTCCC scan produced strong evidence of replication (P≤0.01), two of which are novel. The strongest was SNP rs9292777 (Prep=2.9×10−7; Pcomb=3.2×10−18) which maps to a 1.2 Mb gene desert on chromosome 5p13.1 recently associated with CD.9 The most significant novel association was SNP rs10883365 (Prep=0.0037, Pcomb=3.7 × 10−10), which maps within the NKX2-3 gene (NK2 transcription factor related, locus 3) on chromosome 10q24.2. _Nkx2.3_-deficient mice develop splenic and gut-associated lymphoid tissue abnormalities with disordered segregation of T- and B- cells.10 The second novel locus at rs9858542 (Prep=0.010, Pcomb=4.9×10−8) on chromosome 3p21 is a 1 Mb region of high LD that contains over 20 genes, including MST1 (macrophage stimulating 1), encoding a protein which induces phagocytosis by resident peritoneal macrophages.
The modest evidence of replication for SNP rs2542151 (P = 0.048) at the PTPN2 locus (protein tyrosine phosphatase, non-receptor type 2) on chromosome 18p11 (Pcomb = 3.2 × 10−8) is of interest since PTPN2 encodes a T cell protein tyrosine phosphatase, a key negative regulator of inflammation and is also associated with Type 1 Diabetes.11 Allele frequencies for rs10761659 on chromosome 10q21, strongly associated in the WTCCC scan, were similar in replication CD cases and population controls (SOM table 2), but association in this intergenic region was recently detected in a North American whole-genome CD scan.12
Allele frequencies for most of the markers from the 25 other loci studied that did not achieve genome-wide significance in the WTCCC scan but had an initial P<10−5 converged with control frequencies in the CD replication panel (Supplementary Table 2). Five of these loci, however, provided evidence of replication, thus indicating that a systematic follow-up of all CD hits in the 10−5-10−7 range is likely to yield a number of additional CD susceptibility loci. Four of these loci are novel putative CD risk loci; intriguingly they include two gene deserts, both on chromosome 1q. SNP rs10801047 (Prep=4.8×10−5, Pcomb=2.8×10−8) is located in a gene desert of 1.7Mb on chromosome 1q31.2, and rs12035082 (Prep=0.017, Pcomb=2.1×10−7) maps to chromosome 1q24 (the association signal does not extend to nearest gene, TNFSF18). Although previous studies did not implicate IL12B (interleukin 12B),4,13 modest WTCCC association at rs6887695 from this locus replicated here (Prep=8.4×10−5, Pcomb=9.2×10−6) and warrants follow up, particularly given its association with psoriasis14 and the common association of IL23R with psoriasis and CD.4,14 SNP rs10077785 (Prep=0.008) tags the known IBD5 locus15 on chromosome 5q31 in which, despite extensive resequencing, disease-causing coding variants have yet to be confirmed. Finally, rs2836754 (Prep=0.012, Pcomb=5.5×10−7) maps to an intron in FLJ45139, a gene of unknown function on chromosome 21 that is expressed in bone marrow and colon.
We performed ‘within-cases’ interaction analysis between known CD susceptibility variants and replicating SNPs. For CARD15, rs2066844, rs2066845 and rs2066847 were tested independently or in combination classifying individuals as homozygous wild type, heterozygous or compound heterozygote/rare-allele homozygote. No significant interactions were seen. Testing CD associated variants rs11805303 in IL23R and rs10210302 in ATG16L1 also showed no interactions. Thus risk loci in CD appear to act independently.
We examined six sub-phenotypes of CD and two modifiers for differential effects within cases at markers studied. Cases were classified as affected or unaffected for disease site (‘pure ileal’, ‘pure colorectal’, ‘perianal involvement’) and behaviour (‘inflammatory’, ‘stenosing’ and ‘penetrating’); and by age of onset and smoking status (Supplementary Table 3). Inflammatory behaviour appeared less strongly associated with rs10210302 (in ATG16L1, P=0.0003, Bonferroni P=0.010) than stenosing or penetrating behaviour. No further significant differential associations were seen, although ‘pure ileal’ tends to associate more strongly with most replicating SNPs than ‘pure colorectal’.
This study confirms all four previously unpublished CD loci which achieved genome-wide significance in the WTCCC scan1, and the recently reported 5p13 gene desert9, as well as implicating four additional novel risk loci. These data, combined with the association in the WTCCC scan and the North American study of an intergenic region on chromosome 10q211,12, underline the value of the whole-genome scan approach. Taken together, the genetic evidence regarding IRGM, ATG16L1 (converging on autophagy pathways), CARD15 and IL23R strongly implicates defects in the early immune response, particularly innate immune pathways and the handling of intracellular bacteria, in the pathogenesis of CD. Whether the latter is restricted to a single pathogen or, more likely, a class effect of sub-pathogenic bacteria relying on defective host innate immunity for survival awaits further investigation.
Supplementary Material
Supplementary Material
Acknowledgements
We acknowledge use of DNA from the 1958 British Birth Cohort collection (R. Jones, S. Ring, W. McArdle and M. Pembrey), funded by the Medical Research Council (grant G0000934) and The Wellcome Trust (grant 068545/Z/02). We also acknowledge the National Association for Colitis and Crohn's disease and the Wellcome Trust for supporting the case DNA collections, and UCB Pharma who supported this study with an unrestricted educational grant. We thank Dr Daniel Kelberman, all subjects who contributed samples, and consultants and nursing staff across the UK who helped with recruitment of study subjects.
References
- 1.Wellcome Trust Case Control Consortium Nature. 2007 in press. [Google Scholar]
- 2.Hugot JP, et al. Nature. 2001;411:299–603. [Google Scholar]
- 3.Ogura Y, et al. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 4.Duerr RH, et al. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hampe J, et al. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 6.Bekpen C, et al. Genome Biol. 2005;6:R92.1–92.18. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh SB, et al. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 8.Collazo CM, et al. J. Exp. Med. 2001;194:181–187. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libioulle C, et al. PLOS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pabst O, et al. EMBO J. 2000;19:2015–2023. doi: 10.1093/emboj/19.9.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd J, et al. Nat. Genet. 2007 this issue. [Google Scholar]
- 12.Rioux JD, et al. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwiers A, et al. Genes Immun. 2004;5:675–677. doi: 10.1038/sj.gene.6364131. [DOI] [PubMed] [Google Scholar]
- 14.Cargill M, et al. Am. J. Hum. Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rioux JD, et al. Nat. Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material