Direct dynamin–actin interactions regulate the actin cytoskeleton (original) (raw)
Direct dynamin–actin interactions regulate the actin cytoskeleton
The GTPase dynamin regulates clathrin-dependent endocytosis and has a second, ill-defined, function in the regulation of the actin cytoskeleton. This study reports a direct interaction between dynamin and short actin filaments, which drives dynamin self-assembly, displacement of actin-capping proteins and actin polymerization.
Keywords: actin, cytoskeleton, dynamin
Abstract
The large GTPase dynamin assembles into higher order structures that are thought to promote endocytosis. Dynamin also regulates the actin cytoskeleton through an unknown, GTPase-dependent mechanism. Here, we identify a highly conserved site in dynamin that binds directly to actin filaments and aligns them into bundles. Point mutations in the actin-binding domain cause aberrant membrane ruffling and defective actin stress fibre formation in cells. Short actin filaments promote dynamin assembly into higher order structures, which in turn efficiently release the actin-capping protein (CP) gelsolin from barbed actin ends in vitro, allowing for elongation of actin filaments. Together, our results support a model in which assembled dynamin, generated through interactions with short actin filaments, promotes actin polymerization via displacement of actin-CPs.
Introduction
Dynamic cellular properties such as migration and division depend on the actin cytoskeleton, a dense meshwork of protein polymers that undergoes rapid cycles of assembly and disassembly and that is controlled by a large number of actin-associated proteins (reviewed in Pollard and Cooper, 2009). One of the proteins implicated in regulation of cell motility is the GTPase dynamin (reviewed in Schafer, 2004), best known for its essential role in clathrin-coated vesicle formation during endocytosis (reviewed in Mettlen et al, 2009). In addition to its role in endocytosis, dynamin co-localizes with actin filaments, often in locations where membranes undergo remodelling such as cortical ruffles and podosomes. Dynamin binds numerous actin-regulating or -binding proteins such as profilin, Nck and cortactin (reviewed in Schafer, 2004). These interactions are mediated through dynamin's C-terminal proline, arginine-rich (PRD) domain and the Src homology 3 (SH3) domains of actin-binding proteins. It is through these interactions that dynamin is thought to link the plasma membrane to actin and thereby assemble actin filaments on membranes that are undergoing remodelling (Orth and McNiven, 2003). According to this model, dynamin's regulation of the actin cytoskeleton is tightly connected to its membrane localization and its ability to bind actin-binding proteins.
Several lines of evidence suggest an alternative model in which dynamin has a more direct role in regulation of the actin cytoskeleton. Thus, expression of a dynamin mutant that cannot bind GTP, dynK44A, but can still bind SH3-domain containing proteins, significantly reduced formation of F-actin comets generated either by Listeria or on vesicles by overexpression of type I phosphatidylinositol 5-phosphate kinase (Lee and De Camilli, 2002; Orth et al, 2002). Interestingly, dynamin was not only concentrated at the actin–vesicle–membrane interface, but also extended along the length of the comet tails. Thus, in addition to nucleotide-independent, PRD-dependent interactions with actin-binding proteins (McNiven et al, 2000), these data raised the possibility of dynamin GTPase-dependent regulation of actin. Supporting this connection are studies showing a role for dynamin's GTPase activity in podosome structure and function (Bruzzaniti et al, 2005). Podosomes are highly dynamic actin-containing adhesion structures found in osteoclasts, macrophages and Rous sarcoma virus-transformed fibroblasts. While expression of dynK44A decreased osteoclast resorption and migration, overexpression of dynWT increased these processes (Bruzzaniti et al, 2005). Recent work by Mooren et al (2009) attempted to explain the role of dynamin's GTPase cycle in regulation of the actin cytoskeleton. They showed that in the presence of dynamin, GTP led to remodelling of actin filaments in vitro via the actin-binding protein cortactin. This suggested that a GTP hydrolysis-induced conformational change within dynamin is transduced to cortactin, which in turn alters orientation of the actin filaments. However, as interactions between dynamin's PRD and cortactin's SH3 domain do not require GTP binding or hydrolysis by dynamin, it is uncertain how such a conformational change within dynamin might be transmitted to cortactin.
In this study, we identify a direct interaction between dynamin and actin, which is mediated by a conserved domain in dynamin. Expression of dynamin mutants with impaired affinity for F-actin in podocytes decreased the number of actin stress fibres and disrupted the cortical actin cytoskeleton. In contrast, expression of a dynamin mutant with increased affinity for F-actin enhanced stress fibre formation. Importantly, actin binding was closely linked to the assembly status of dynamin. Thus, short actin filaments stimulated dynamin self-assembly via direct dynamin–actin interactions in vitro. Importantly, assembled dynamin displaced the capping protein (CP) gelsolin (Gsn) from barbed ends, which in turn promoted elongation of actin filaments. The effect was Gsn specific as dynamin could not displace another mammalian CP. Together, the data suggest that short actin filaments, generated by actin severing and CPs, lead to dynamin self-assembly. Dynamin oligomers in turn promote de-capping of barbed ends, allowing for actin filament elongation. The experiments reveal an intricate functional interplay between the actin cytoskeleton and the assembly status of dynamin.
Results
Dynamin binds to filamentous actin
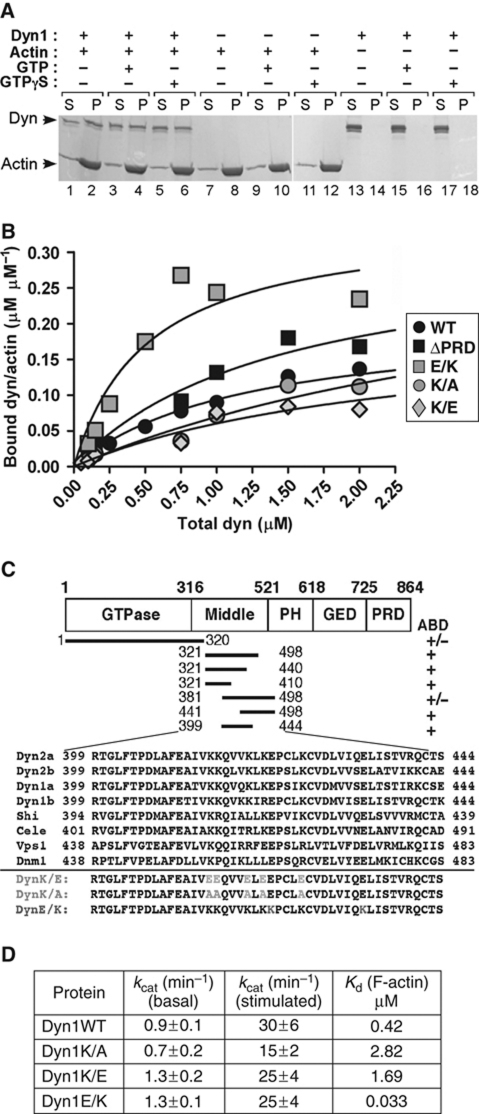
To revisit the question of how dynamin regulates the actin cytoskeleton, we tested whether dynamin binds to F-actin using a co-sedimentation assay. We used the neuronal isoform, dynamin 1 (dyn1), which has a lower propensity to spontaneously oligomerize (Warnock et al, 1997). At physiological ionic strength, dyn1 was soluble and thus found in the supernatant after ultracentrifugation (Figure 1A, lanes 13, 15 and 17). However, in the presence of F-actin, dynamin was found in the pellet (Figure 1A, compare lanes 2 and 14; Supplementary Figure S1A) independently of GTP or GTPγS (Figure 1A, compare lanes 2, 4 and 6). Recombinant dyn1 and isolated actin did not contain cortactin, an actin-binding protein known to interact with dynamin (Supplementary Figure S1B), suggesting that interactions between dynamin and actin are direct. The binding constant (_K_d) of dynamin for actin was ∼0.4 μM (Figure 1B, black circles); which is comparable with the _K_ds of known actin-binding proteins such as α-actinin 4 or cortactin (van Rossum et al, 2003; Weins et al, 2005). Based on a Scatchard analysis (Supplementary Figure S1C; saturation-binding curve), one dynamin tetramer bound 4–6 actin subunits of the filament.
Figure 1.
Direct dynamin–actin interactions are mediated by dynamin's middle domain. (A) Dyn1 co-sediments with F-actin. Representative Coomassie blue-stained gel of supernatants (S) and pellets (P) after centrifugation at 150 000 g in the presence and absence of 100 μM nucleotides as indicated in the figure; 1 μM dynamin was incubated with 5 μM rabbit skeletal muscle actin. (B) Actin-binding analysis of dynamin (wild-type, ΔPRD and actin-binding site mutants) to F-actin. Increasing concentrations of dyn1 were added to 2.5 μM F-actin. After centrifugation at 150 000 g, proteins were separated on SDS–PAGE and bands were analysed using densitometry. (C) Top, schematic diagram of dynamin domains and fragments tested in the actin co-sedimentation assay using IVT proteins. The presence or absence of an actin-binding domain (ABD) is indicated. Bottom, amino-acid sequence alignment of dyn2 (splice variants a and b), dyn1 (splice variants a and b), Drosophila dynamin (Shi), Caenorhabditis elegans dynamin (Cele), and yeast dynamin (Vps1). Dnm1 is a dynamin family member involved in mitochondrial morphogenesis. (D) Dynamin mutants with altered affinity for actin exhibit wild-type GTPase activities. Kinetic parameters were determined performing the GTPase assays using 0.2 μM dynamin and when indicated 80 μM lipids.
We next mapped the actin-binding domain (ABD) within dynamin. Importantly, dynamin lacking its PRD domain (dynΔPRD) bound F-actin as efficiently as dyn1WT with a _K_d of ∼0.3 μM (Figure 1B; Supplementary Figure S1D). Thus, dynamin–actin interactions are not mediated through SH3 containing actin regulators. Dynamin's GTPase effector domain (GED) did not bind F-actin (data not shown), suggesting that the actin-binding site was situated within the GTPase, middle or PH domains. Using in vitro transcription/translation (IVT), we generated several dynamin fragments and thereby mapped the actin-binding site to a region between amino acids 399 and 444 (Supplementary Figures S1E–G). As predicted for an ABD (Van Troys et al, 1996), this region contains several positively charged amino acids, and these are conserved from yeast to mammals (Figure 1C). Interestingly, this region is alternatively spliced within different mammalian dynamin isoforms (called variant a and b).
Site-directed mutagenesis was performed on conserved, charged residues within the ABD of dyn1b to generate putative ‘loss-of-function' mutants, dyn1K/E and dyn1K/A, and a putative ‘gain-of-function' mutant, dyn1E/K (Figure 1C). As predicted, the affinities of dyn1K/E and dyn1K/A for actin were reduced (_K_d of 1.7 and 2.8 μM, respectively), whereas the affinity of dyn1E/K for actin was increased (_K_d=0.03 μM) (Figure 1B; Supplementary Figures S1H and I). All three mutant proteins exhibited wild-type basal rates of GTP hydrolysis (Figure 1D), indicating proper folding. In addition, all three mutants supported lipid-stimulated GTP hydrolysis (Figure 1D), with dyn1K/A being partially impaired because of its reduced self-assembly (Supplementary Figure S1J, lane 3). In agreement with their normal GTPase activities, dyn1K/E and dyn1E/K supported endocytosis of transferrin (Supplementary Figures S2A and B). Expression of these mutant proteins did not change the overall level (Supplementary Figure S2C), or GTP-bound forms of known Rho-family GTPases (Supplementary Figure S2D) in cells. Together, these data suggest that the actin-binding mutants of dynamin are properly folded and that they do not detectably affect clathrin-mediated endocytosis or signalling by Cdc42, Rac1 or RhoA.
Direct dynamin–actin interactions regulate the actin cytoskeleton
To explore the function of direct dynamin–actin interactions in cells, we used cultured podocytes. Podocytes are specialized cells of the glomerulus containing a highly organized actin cytoskeleton that is necessary for their function as part of the glomerular filtration apparatus in the kidney (Saleem et al, 2008). Changes in podocyte morphology and attachment to the glomerular basement membrane, which are driven by reorganization of the actin cytoskeleton, are directly linked to chronic kidney diseases (Oh et al, 2004). The actin cytoskeleton in conditionally immortalized mouse podocytes supports podocyte adhesion, motility and morphogenesis, and like many other cell types can be roughly divided into three domains: (1) a lamellipodial actin network, (2) filopodial actin bundles and (3) contractile actin stress fibres (Figure 2A, panel 1).
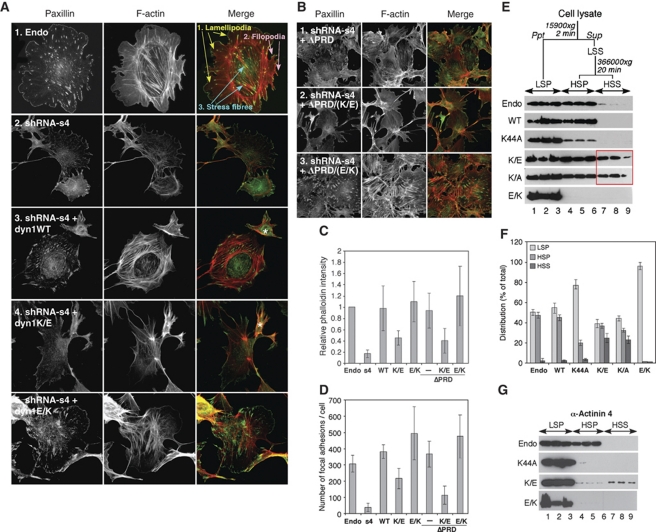
Figure 2.
Direct dynamin–actin interactions are essential for organization of the actin cytoskeleton in podocytes. (A, B) Podocytes were first infected with adenoviruses expressing different dyn1 constructs as indicated; 18 h post-infection, cells were infected with lentivurus expressing shRNA construct S4, and dyn2 was downregulated for 3 days, after which cells were examined via immunofluorescence. Focal adhesions and F-actin were visualized with anti-paxillin antibodies and rhodamin phalloidin, respectively. In (A), white asterisk marks cells that were not infected with adenoviruses expressing dyn1 constructs. (C, D) Expression of dyn1WT, dyn1E/K, dyn1ΔPRD/EK, but not dyn1K/E or dyn1ΔPRD/KE promotes formation of stress fibres and focal adhesions in podocytes. Bar graphs depicting total F-actin (C) and the number of focal adhesions (D). Data represent measurements of >50 cells and are plotted as ±s.d. (_n_=3). (E) Effects of expression of dynamin mutants on the partitioning of actin into Triton-X 100 soluble and insoluble fractions. (Top) Flow chart of experiment, (bottom) western blot analysis of actin distribution among low-speed pellet (LSP), high-speed pellet (HSP), and high-speed supernatant (HSS) fractions in three separate experiments. (F) Bar graphs depicting the partitioning of actin in (E). The actin distribution in each fraction was expressed as a percentage of the total. Values shown are the mean ± s.d. (_n_=3) shown in (E). (G) Fractionation of α-actinin 4 by differential centrifugation in podocye lysates expressing different dynamin mutants. The result from three separate experiments are shown.
Downregulation of the ubiquitously expressed dynamin 2 (dyn2) using lentivirus-based short hairpin RNAs was monitored using RT–PCR (Supplementary Figure S3A) and western blot analysis (Supplementary Figure S3B). Three different shRNAs downregulated dyn2 protein levels by 70–90% (Supplementary Figure S3B), resulting in similar alterations in cell morphology with cells becoming smaller, less round and eventually detaching from the coverslip (Supplementary Figure S3C). There was a dramatic loss of F-actin and the number of focal adhesions (FAs) in dynamin-deficient cells (Figure 2A, panel 2 and 2C, 2D for quantification). These data are in agreement with the global alteration in the actin cytoskeleton observed after dyn2 was downregulated in U2OS cells (Mooren et al, 2009). The data are also in agreement with our previous finding that downregulation of dynamin by CatL processing resulted in loss of stress fibres (Sever et al, 2007).
We next examined whether expression of dyn1WT or our novel ABD mutants of dynamin could rescue the dyn2-depletion phenotype. Of note, >70% loss of dyn2 resulted in significant loss of cells from the coverslips (Supplementary Figure S3C and data not shown). Thus, all rescue experiments were performed under viral infection conditions in which downregulation of dynamin was ∼70% (Supplementary Figures S3D and E). Expression of the neuronal isoform of dynamin, dyn1WT, restored normal actin organization in dyn2-deficient cells (Figure 2A, panel 3; Supplementary Figure S3F). Quantification of the data showed that dyn1WT restored levels of F-actin (Figure 2C) and FAs (Figure 2D) to wild-type levels. In contrast, expression of ‘loss-of-function' dyn1K/E failed to rescue the depletion of dyn2 (Figure 2A, panel 4; Supplementary Figure S3F). Whereas dyn1K/E induced formation of FAs and stress fibres above levels seen in cells lacking dynamin (Figure 2C and D), it mostly induced formation of focal complexes and some disorganized actin cables within the cell body. Expression of ‘gain-of-function' dyn1E/K rescued formation of FAs within the cell body (Figure 2A, panel 5, 2D; Supplementary Figure S3F), as well as formation of stress fibres (Figure 2C). While expression of dyn1E/K clearly restored formation of stress fibres within the cell body (Figure 2A), it seems that these structures are also somewhat disorganized. As ABD mutants exhibit wild-type GTPase activity (Figure 1D) and do not affect clathrin-mediated endocytosis or signalling by Cdc42, Rac1 and RhoA (Supplementary Figure S2), our data indicate that direct dynamin–actin interactions are essential for actin polymerization, which drives formation of stress fibres as well as cortical actin cytoskeleton in podocytes.
Interactions between dynamin and SH3-domain containing proteins are not essential for stress fibre formation in podocytes
Until now, the role of dynamin in regulation of the actin cytoskeleton has been viewed through its interactions with actin-binding/regulatory proteins such as cortactin. As these interactions are mediated by dynamin's PRD domain, we examined effects of ABD mutants in the context of dynamin mutants that lack the PRD, dyn1ΔPRD. Unexpectedly, expression of dyn1ΔPRD complemented dyn2 knockdown to a similar extent as dyn1WT, as seen by the restoration of stress fibres and FAs within podocytes (Figure 2B, panel 1, 2C, 2D; Supplementary Figure S3F). In contrast, expression of the double mutant, dyn1ΔPRD/KE, which is impaired in actin binding, failed to restore stress fibres and FAs (Figure 2B, panel 2), whereas dyn1ΔPRD/EK behaved like dyn1WT (Figure 2B, panel 3). Together, these results further support the conclusion that the effects of dynamin on the actin cytoskeleton occurred through direct dynamin–actin interactions.
The importance of direct dynamin–actin interactions in actin regulation is not unique to podocytes. Thus, it has been shown that expression of dyn1ΔPRD can induce stress fibre formation in rat hepatocytes called clone 9 cells (Supplementary Figure S4A, panel 1; McNiven et al, 2000). Consistent with our experiments in podocytes, dyn1ΔPRD/KE failed to induce stress fibre formation in clone 9 cells, whereas dyn1ΔPRD/EK retained this activity (Supplementary Figure S4A). In fact, expression of dyn1E/K induced stress fibre formation, whereas expression of dyn1K/E had the opposite effect (Supplementary Figure S4B). Together, these data suggest that in clone 9 cells, interactions between dynamin's PRD and SH3-domain containing proteins are not essential for formation of stress fibres, but that direct dynamin–actin interactions are essential.
Direct dynamin–actin interactions regulate actin polymerization
To further examine the role of dynamin–actin interactions in regulation of actin cytoskeleton, we overexpressed ABD mutants in podocytes. If dynamin–actin interactions are essential for actin polymerization, then overexpression of ABD mutants might be predicted to function as dominant negative or dominant positive with respect to actin polymerization. As shown previously (Sever et al, 2007), expression of dyn1K44A, a mutant that cannot bind GTP, dramatically altered organization of the actin cytoskeleton in podocytes, whereas expression of dyn1WT had no effect (Supplementary Figures S5A–C). Expression of ‘loss-of-function' dyn1K/E resulted in elongated podocytes whose cytoplasm collapsed into a narrow space around the nucleus (Supplementary Figure S5A), which correlated with a statistically significant loss of stress fibres and FAs (Supplementary Figures S5B and C). DynK/E was also generated in the context of dyn2, and the cells exhibited the same phenotypes as dyn1K/E (compare Supplementary Figure S5A, row 4 and Supplementary Figure S5D, row 3), showing that the effects were not isoform specific. In contrast, expression of the ‘gain-of-function' dyn1E/K exhibited a statistically significant increase in the number of FAs and stress fibres (Supplementary Figures S5B and C). Further supporting a role for dynamin–actin interactions in these phenotypes, both ABD mutants were able to interact with cortactin, an SH3-domain containing protein, to the same extent as the wild-type enzyme (Supplementary Figures S5E and F). Together, these data further show a role for direct dynamin–actin interactions in regulation of the actin cytoskeleton.
As a complementary means to examine dynamin-mediated regulation of actin polymerization, we performed subcellular fractionation using differential centrifugation (Watts and Howard, 1992). In this assay, higher order bundles and networks of F-actin are isolated in the low-speed fraction in the presence of Triton-X 100 (low-speed pellet, LSP in Figure 2E). The soluble fraction is recovered and re-centrifuged at high speed to separate non-crosslinked F-actin (high-speed pellet, HSP) from soluble G-actin monomers (high-speed supernatant, HSS). Wild-type mouse podocytes contained almost identical amounts of actin in the LSP (bundles) and HSP (F-actin) fractions and almost no actin in HSS (G-actin; Figure 2E, Endo; see Figure 2F for quantification), and this distribution was maintained in cells overexpressing dyn1WT (Figure 2E). Expression of dyn1K44A shifted F-actin into the LSP fraction (Figure 2E) consistent with formation of hyper-bundled cortical actin (Supplementary Figure S5A). In contrast, expression of ‘loss-of-function' dyn1K/E and dyn1K/A resulted in a significant increase in the amount of G-actin in HSS, while decreasing the levels of crosslinked F-actin in LSP and HSP (Figure 2E). These data are in agreement with the overall loss of stress fibres observed by immunofluorescence microscopy (Supplementary Figures S5A and B). Cells expressing the ‘gain-of-function' dyn1E/K exhibited a dramatic increase in F-actin pelleting into the LSP, and a concomitant decrease of F-actin in the HSP fraction (Figure 2E), consistent with increased stress fibre formation (Supplementary Figures S5A and B). A known podocyte actin bundling protein, α-actinin 4, co-fractionated with actin in cells expressing the different dynamin mutants (Figure 2G); thus, similar to the effects on actin, dyn1K44A and dyn1E/K expression shifted α-actinin 4 into the LSP, whereas dyn1K/E expression resulted in an increase of α-actinin 4 into the HSS fraction (Figure 2G, lanes 7–9). Together, these data show that dynamin–actin interactions have a major function in regulating actin polymerization in podocytes. Our data suggest that dynamin–actin interactions regulate both the formation of stress fibres and the cortical actin cytoskeleton in podocytes.
As the coordinated interplay between the cortical actin cytoskeleton and stress fibres underlies cell motility, we examined whether the dynamin–actin interaction influences podocyte motility. Podocyte motility was examined using a scratch wound-healing assay (Asanuma et al, 2006). As predicted from actin cytoskeleton organization, overexpression of dyn1WT had no effect on podocyte motility, in contrast to diminished podocyte motility in cells expressing dyn1K44A (Supplementary Figures S5G and H for quantification). Importantly, motility was also impaired in cells expressing any of the three ABD mutants of dynamin (Supplementary Figure S5G). Thus, both ‘loss-of-function' and ‘gain-of-function' ABD dynamin mutants act as dominant negatives with respect to podocyte motility. The loss of motility in cells expressing dyn1E/K can be explained by the increase in the number of FAs within the cell body (Supplementary Figure S5C). Together, these data show that direct dynamin–actin interactions control global organization of the actin cytoskeleton by regulating actin polymerization. To our knowledge, dynamin is the only GTPase known to directly regulate global organization of the actin cytoskeleton in cells.
Dynamin bundles filamentous actin
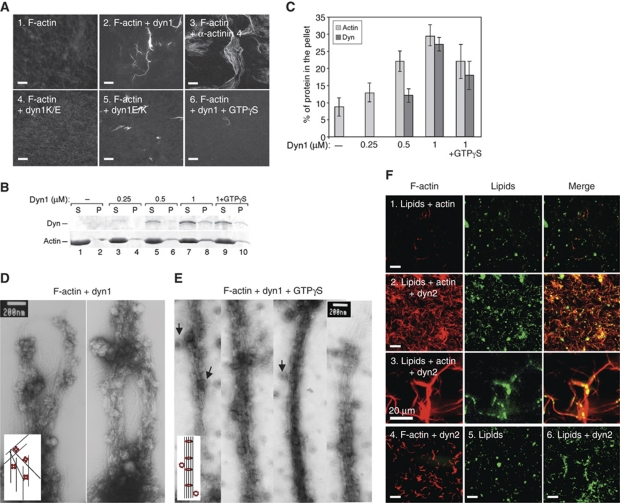
We next investigated the molecular mechanism by which dynamin–actin interactions influence the organization of the actin cytoskeleton. First, we examined whether dynamin alters F-actin organization. Figure 3A shows that dyn1 bundles F-actin in vitro, as visualized by fluorescence microscopy of actin after FITC-phalloidin staining (Figure 3A, panel 2). Importantly, dyn1E/K also bundled F-actin (panel 5), whereas dyn1K/E did not (panel 4), indicating that bundling is mediated by direct dynamin–actin interactions mediated by the dynamin ABD. Although dynamin can bundle F-actin, it does so less effectively than α-actinin 4, a known actin bundling protein (Figure 3A, panel 3). Given dynamin's ability to oligomerize into higher order structures such as rings, we tested whether dynamin rings also bind and bundle F-actin. Dynamin was induced to oligomerize into rings by GTPγS, a non-hydrolysable GTP analogue. Although bundles were detected, the presence of GTPγS diminished the extent of bundling (Figure 3A, compare panels 2 and 6). Co-sedimentation at low speed was used to independently measure the ability of dynamin to crosslink F-actin into bundles. In the absence of dyn1, <10% of F-actin sedimented during centrifugation (Figure 3B, lane 2, and 3C for quantification). Addition of 1 μM dyn1 increased the amount of F-actin found in the pellet to ∼35% of the total (Figure 3B, lane 8), but this was slightly reduced in the presence of GTPγS (Figure 3B, lane 10). Therefore, unassembled dynamin tetramers and, to a lesser extend, dynamin rings can bundle actin filaments.
Figure 3.
Dynamin crosslinks actin filaments into bundles. (A) Actin bundling viewed with confocal microscopy of phalloidin staining of 5 μM F-actin in control sample lacking dynamin (1), in the presence of 1 μM dyn1WT (2), 1 μM α-actinin 4 (3), 1 μM dyn1K/E (4), 1 μM dyn1E/K (5) and 1 μM dyn1WT + 200 μM GTPγS (6). Pictures were taken 1 h after initiation of the reaction. Bar is 100 μm. (B) Effects of increasing dynamin concentrations on the partitioning of actin filaments after centrifugation for 20 min at 15 000 g. Only filaments crosslinked into the bundles sediment at this speed. To generate long filaments of similar lengths, actin was polymerized in the presence of gelsolin, a barbed end-binding protein at the ratio of G1:A1000. Where indicated, 200 μM GTPγS was added. Dynamin and F-actin were detected using Coomassie blue staining of the gels. (C) Quantification of F-actin and dynamin partitioning in (B). The actin and dynamin recovery in the pellet was expressed as a percentage of the total. Values shown are mean ± s.d. (_n_=3). (D, E) Electron micrographs of actin filaments generated after 1 h in the presence of 1 μM dyn1, 5 μM Gsn–F-actin (G1:A1000), and without (D), or with 100 μM of GTPγS (E). Arrowheads indicate dynamin rings attached to actin bundles. Cartoons depict possible mechanisms by which dynamin crosslinks long actin filaments into bundles. (F) Dynamin binds and crosslinks F-actin in the presence of lipids. Actin filaments and lipid vesicles were visualized with rhodamine phalloidin (red) and fluorescein-PE (green), respectively. Reactions contained 50 μM PC:PIP2 (90:10 mol:mol), 5 μM F-actin and 0.5 μM dyn2. Single actin filaments distributed on the coverslip surface are not detected in these images because the exposure time for collecting images of the actin filament bundles was short. Bar is 100 μm, except in panel 3 where it is 20 μm.
To visualize actin bundles generated by dynamin, negative staining and electron microscopy were performed. In agreement with the light microscopy, the addition of dyn1 to F-actin resulted in bundles that could be visualized in the electron microscope (Figure 3D). White ‘knobs' attached to actin filaments are recombinant dynamin. As observed by the IF, the EM analysis also showed that dynamin bundled F-actin in the presence of GTPγS (Figure 3E). In Figure 3E, black arrows point to the dynamin rings that associate with actin bundles in the presence of GTPγS. The filament-to-filament spacing in the presence of GTPγS was 17–20 nm and quite regular, showing that in contrast to unassembled dynamin, dynamin rings generate bundles with defined spacing (see model in Figure 3E). Therefore, both tetrameric and oligomeric dynamin can interact with and crosslink F-actin filaments into bundles and modify actin structure.
Dynamin is known to bind phospho-lipids. Thus, we tested whether dynamin might recruit actin filaments to membranes. Actin filaments and PIP2-containing lipid vesicles were visualized with rhodamine phalloidin and fluorescein phosphatidylethanolamine (PE), respectively (Schafer et al, 2002). As seen before (Schafer et al, 2002), addition of dyn2 induced aggregation of lipid vesicles and resulted in association of actin filaments with lipid aggregates (Figure 3F). In fact, addition of lipid vesicles increased dynamin's ability to bundle actin filaments into disordered and entangled networks of thin filament bundles. Together, these data show that dynamin can crosslink actin filaments into bundles even when associated with the membranes, and thus suggest that it can directly influence the organization of the actin filaments in the vicinity of the membrane.
Short actin filaments promote dynamin oligomerization
To fully explore the functional connection between actin and dynamin, we addressed whether actin also modulates the properties of dynamin. The GTPase activity of dynamin is tightly connected to its oligomerization cycle. In its basal state at high ionic strength, dynamin exhibits a dimer–tetramer equilibrium (Muhlberg et al, 1997). At low ionic strength, or in the presence of lipid vesicles (reviewed in Praefcke and McMahon, 2004), dynamin tetramers self-assemble into higher order structures such as rings and/or spirals. Dynamin self-assembly increases its GTPase activity because of activation of an internal GTPase-activating protein domain (Sever et al, 1999). GTP hydrolysis in turn drives dynamin disassembly (Warnock et al, 1996).
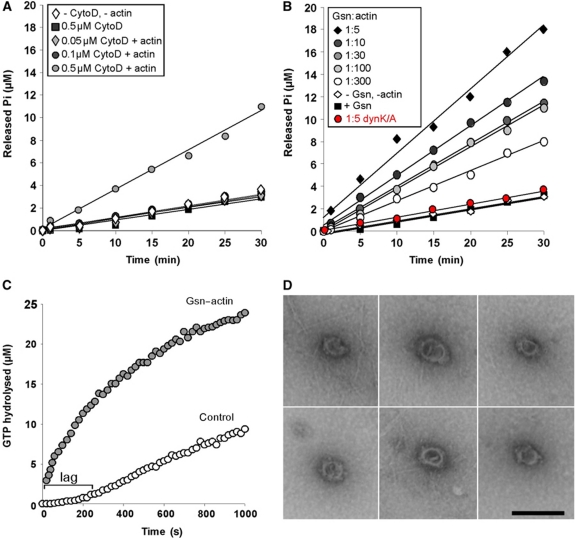
GTPase assays were performed in the presence and absence of actin. Addition of G-actin, G-actin treated with latranculin A (to ensure that actin remains monomeric at the salt conditions of the assay) or F-actin did not significantly alter the basal rate of dynamin GTP hydrolysis (data not shown). To determine whether actin filament length might affect the GTPase activity of dyn1, F-actin (2 μM final G-actin concentration) was sheared by five passages through a 27-gauge needle, followed immediately by the addition of different concentrations of cytochalasin D (cytoD), an inhibitor of actin polymerization. This shearing procedure generates short filaments that are ∼0.7 μm long (Supplementary Figure S6A), which are efficiently capped by 0.5 μM cytoD. As uncapped actin filaments can anneal through an end-to-end mechanism (Andrianantoandro et al, 2001), lower concentrations of cytoD are expected to generate actin filaments longer than 0.7 μm. As shown in Figure 4A, the shortest actin filaments generated by the addition of 0.5 μM cytoD stimulated dynamin's GTPase activity by ∼3.5-fold, whereas a lower concentration of cytoD or cytoD alone had no effect. These data suggest that short actin filaments can promote dynamin oligomerization into higher order structures, which in turn increases dynamin's GTPase activity.
Figure 4.
Short actin filaments promote dynamin oligomerization into ring-like structures. (A) Time course of GTP hydrolysis by 0.2 μM dyn1 incubated without or with 2.5 μM sheared F-actin treated with the indicated concentrations of Cyto D. (B) Time course of GTP hydrolysis by 0.2 μM dyn1WT or dyn1K/A (red circle) incubated with 2.5 μM Gsn or 2.5 μM Gsn-capped F-actin complexes generated with the indicated ratios of Gsn to actin. (C) A coupled assay of GTP hydrolysis by 0.2 μM dyn1 in the absence or presence of 2.5 μM Gsn-capped F-actin (G1:A100). The GTPase activity was measured at the indicated times. (D) Electron micrographs of dynamin rings formed in the presence of Gsn–F-actin (G1:A100). Bar is 100 nm. Note actin filaments around and adjacent to dynamin rings.
In cells, short actin filaments are generated as a result of F-actin cleavage and capping by severing proteins such as Gsn (Barkalow et al, 1996; Pollard and Borisy, 2003). Thus, we next tested whether dynamin's GTPase activity can be stimulated by the addition of short filaments generated in the presence of Gsn, a calcium-activated protein, and calcium (Yin et al, 1981). Under our experimental conditions, Gsn caps actin barbed ends, thus inhibiting actin polymerization. By varying the ratio of Gsn to actin (G:A), actin filaments of different lengths can be generated (Supplementary Figure S6B). Strikingly, Gsn-treated F-actin complexes increased dynamin's basal rate of GTP hydrolysis up to eight-fold, with the highest stimulation achieved by the shortest filaments (_k_cat∼8 min−1; Figure 4B, black diamonds). EM images revealed that actin filaments were 51±34 nm long for a G1:A5 ratio (which is equivalent to ∼19 monomers), whereas they were 3.2±1.7 μm long for a G1:A300 ration (which poorly stimulated dynamin). Together, these data show that filament lengths between 51±34 and 700 nm have the best ability to stimulate dynamin oligomerization into higher order structures.
The observed increase in GTPase activity was dependent on dynamin–actin interactions, as Gsn–F-actin complexes could neither activate the dyn1K/A mutant (Figure 4B, red circles), nor could addition of only Gsn (Figure 4B, black squares). As dynamin's GTPase activity is dependent on its oligomerization state, these findings suggest that actin filaments can promote dynamin oligomerization into rings in a length-dependent manner, with the shortest filaments being most potent. As an alternative approach to investigate the ability of short actin filaments to promote dynamin oligomerization into rings, GTP hydrolysis was monitored continuously in a coupled assay (Ingerman and Nunnari, 2005). Addition of Gsn–F-actin complexes to this assay overcame the kinetic lag that reflects dynamin assembly into rings (Figure 4C, grey circles).
To directly visualize dynamin's oligomerization status in the presence of Gsn–F-actin complexes, samples were negatively stained with uranyl acetate and observed in the electron microscope. In agreement with biochemical experiments, Gsn–F-actin complexes induced limited assembly into ring-like structures ∼40 nm in diameter (Figure 4D), similar to rings that self-assemble at low ionic strength (Hinshaw and Schmid, 1995). Oligomerization into single rings is consistent with the maximum eight-fold increase in the rate of dynamin's GTP hydrolysis, as assembly into spirals causes up to a 100-fold increase (Warnock et al, 1996). Together, these data suggest that short actin filaments can have a direct and profound effect on the oligomerization status of dynamin.
Dynamin rings induce elongation of Gsn-capped actin filaments
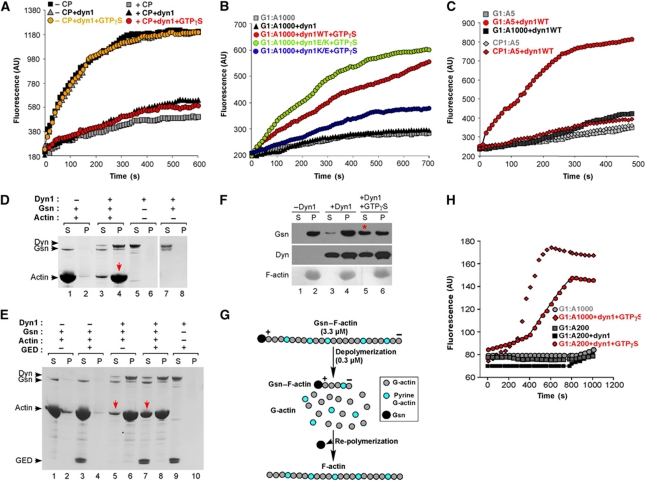
The results presented so far show that direct binding of dynamin to actin promotes actin stress fibre formation in cells. Moreover, we found that short actin filaments in turn promote dynamin assembly. To understand how assembled dynamin might modulate actin dynamics, we first tested whether dynamin can promote actin polymerization. A solution-based actin polymerization assay using pyrene-labelled actin was performed (Fujiwara et al, 2009). In this assay, pyrene fluorescence increases when monomeric actin is assembled into filaments. We tested whether addition of dynamin stimulates actin polymerization from actin seeds by reducing the lag phase, as shown for other actin regulatory proteins such are formins (Moseley et al, 2006). As shown in Figure 5A, addition of dynamin with or without GTPγS had no effect on the rate of actin polymerization. Given the ability of short actin filaments to promote dynamin oligomerization, we next tested whether dynamin rings can induce elongation of actin filaments that are capped by CP. CP is a ubiquitously expressed heterodimeric actin-binding protein that is essential for normal actin dynamics in cells. As filaments in this experiment are relatively long, dynamin oliogmerization is promoted by addition of GTPγS. Addition of dynamin in the presence or absence of GTPγS failed to induce actin polymerization from the CP-capped filaments (Figure 5A). In striking contrast, dynamin induced rapid actin polymerization of filaments that were capped by Gsn (Figure 5B). Interestingly, the actin polymerization was achieved only in the presence of GTPγS, suggesting that only assembled dynamin can promote actin elongation. Importantly, ‘loss-of-function' dyn1K/A mutant was significantly impaired in stimulating actin polymerization (blue circles), whereas ‘gain-of-function' dyn1E/K mutant stimulated actin polymerization better than dyn1WT (green circles). Together, these data suggest that dynamin oligomerization driven by direct dynamin–actin interactions is essential for observed actin polymerization. If this conclusion is correct, it is predicted that Gsn:actin ratios that stimulate dynamin oligomerization (based on the GTPase assays presented in Figure 4B) should promote dynamin oligomerization in the absence of GTPγS. Indeed, dynamin stimulated actin polymerization at Gsn–actin ratios of 1:5 (Figure 5C, red circles), but not those capped by CP (red triangle). Together, these data show that dynamin rings can promote elongation of actin filaments capped by Gsn, but not those capped by CP.
Figure 5.
Dynamin rings dissociate gelsolin from barbed ends and promote actin elongation. (A) Solution-based actin polymerization using actin seeds. Actin seeds were generated by vortexing actin filaments for 20 s immediately before use; 0.2 μM dyn1 was added to 0.8 μM seeds or seeds capped with 5 nM CP protein. At time zero, 2 μM pyrene-labelled monomeric actin was added, and pyrene fluorescence was monitored. (B) Effects of dynamin on actin elongation; 5 μM Gsn–F-actin (G1:A1000) was incubated with 0.2 μM dyn1WT, dyn1E/K and dyn1K/A with or without 200 μM GTPγS. At time zero, 0.5 μM pyrene-labelled monomeric actin was added, and pyrene fluorescence was monitored. (C) Experiment performed as in (B) except that capped actin filaments were generated either by addition of gelsolin or CP at a 1:5 ratio to supply short actin filaments that can promote dynamin oligomerization in the absence of GTPγS. (D, E) Dynamin promotes elongation of short actin filaments capped by gelsolin. Elongation of actin filaments was measured by their ability to pellet under high-speed centrifugation. Actin was polymerized in the presence of gelsolin (G1:A10) for 20 min to generate short actin filaments that stay in the supernatant during high-speed centrifugation; 16.5 μM Gsn–F-actin complexes were incubated with 1 μM dyn1 for 30 min. Subsequently, samples were centrifuged at 150 000 g for 30 min at 22°C. Experiments were performed with dyn1WT (D) and in the presence of 5 μM recombinant GED, as indicated (E). (F) Dynamin rings displace gelsolin from the barbed ends. Effect of dynamin with or without 200 μM GTPγS on the partitioning of Gsn; 20 μM Gsn–F-actin (G1:A300) was incubated with 0.2 μM dyn1 for 30 min at RT. Samples were centrifuged at 150 000 g for 30 min. Gsn was detected using anti-Gsn antibody, dyn1 using anti-dynamin antibody and F-actin using Coomassie staining. (G) Schematic diagram of experiments performed under (H). Pyrene-labelled G-actin (3.3 μM) was polymerized for 1 h in the presence of gelsolin at the indicated ratios (G1:A200 or G1:A1000). Under these conditions, gelsolin capped >99% of the barbed ends. The Gsn–F-actin was then diluted to 0.33 μM in the presence or absence of dynamin. As G- and F-actin coexist in equilibrium, the concentration of G-actin is determined by the critical concentration (_K_d), which is defined by the on and off rates at the filament ends: 0.1 μM at the barbed (+) end, and 0.6 μM or greater at the pointed (−) end. Thus, after dilution to 0.33 μM, which lies between the critical concentrations at the two ends, Gsn–F-actin depolymerizes from the pointed ends, generating a new pool of G-actin. Depolymerization generates ∼0.23 μM pyrene G-actin that can re-polymerize in this assay, but only if the barbed ends become available. (H) Representative time courses of the re-polymerization of actin when 0.33 μM Gsn–actin complexes (G1:A200 or G1:A1000) are incubated in the presence or absence of 0.1 μM dyn1 and with or without 100 μM GTPγS. Of note, 1 μM pyrene actin represents 300–400 fluorescence units. Therefore, 75–100 units represents ∼0.25 μM F-actin, which re-polymerized at the barbed ends.
As an alternative means to measure dynamin-dependent actin polymerization, we performed sedimentation assays. At low salt conditions, dynamin is induced to oligomerize into single rings, which pellet during high-speed centrifugation (Warnock et al, 1996), but at physiological salt concentration, dyn1 stays in the supernatant (Figure 5D, lane 7). Short actin filaments generated at a Gsn:actin ratio of 1:10 did not pellet in this assay (Figure 5D, lane 1). However, when dyn1 was mixed with Gsn–F-actin filaments, the majority of both actin and dynamin were recovered in the pellet (Figure 5D, lane 4). In contrast, dyn1 did not bring short actin filaments capped by CP into the pellet (Supplementary Figure S6D), in agreement with its inability to promote actin elongation of CP-capped filaments (Figure 5C). Together, these data argue that sedimentation of actin filaments in this assay was due to their elongation, which was promoted by actin-dependent dynamin oligomerization.
The sedimentation assay allowed us to further explore the role of dynamin rings in inducing elongation of capped actin filaments. Thus, we performed co-sedimentation assays under conditions that impaired the oligomerization of dyn1. Dynamin's GED is directly involved in dynamin oligomerization through GED–GED contacts (Sever et al, 1999; Zhang and Hinshaw, 2001). Hence, recombinant GED competes for these GED–GED interactions and impairs dynamin oligomerization (Sever et al, 1999). As shown in Figure 5E, GED counteracted the ability of dyn1 to draw Gsn–F-actin complexes into the pellet, leaving more actin and dynamin in the supernatant (compare lanes 5 and 7). Importantly, GED had no significant effect on the fractionation of Gsn–F-actin in the absence of dyn1 (Figure 5E, compare lanes 1, 2 and 3, 4). Thus, pelleting of Gsn–F-actin in this assay was dependent on dynamin oligomerization and because of elongation of actin filaments. Together, these data show that dynamin oligomers can induce actin elongation from filaments capped by Gsn.
Dynamin rings displace Gsn from the barbed ends of actin filaments
Next, we attempted to elucidate the mechanism by which dynamin induces elongation of Gsn-capped actin filaments. Thus, we first investigated whether dynamin itself binds barbed ends by using actin annealing assays (Andrianantoandro et al, 2001). Short actin filaments generated by shearing F-actin can re-associate through end-to-end annealing, which is impaired by saturating concentrations of CPs (Andrianantoandro et al, 2001). G-actin was allowed to polymerize for 1 h, resulting in actin filaments that were on average ∼7.4 μm long (Supplementary Figure S6A, panel 1). Shearing of F-actin by passage through a 27-gauge needle generated short actin filaments that were ∼0.75 μm long (panel 2). After annealing for 2 h, the sheared filaments grew to a length of ∼3.1 μm (panel 3). Addition of dyn1 with or without GTPγS resulted in actin filaments that were ∼3.9 μm long and that appeared crosslinked into bundles (panels 4 and 5). Thus, addition of dynamin did not impair annealing of short actin filaments, suggesting that dynamin, even when oligomerized into rings, did not bind naked barbed ends.
The differential effects of dynamin rings on filaments capped by Gsn and CP, both of which bind to barbed ends, suggested that dynamin might increase actin polymerization by removing Gsn from the barbed ends. To test this, actin was assembled in the presence of Gsn at a ratio of G1:A300. Under these conditions, >90% of the Gsn pellets with F-actin after high-speed centrifugation (Figure 5F, lane 2). Addition of dyn1 to Gsn–F-actin promoted release of Gsn from F-actin in a GTPγS-dependent manner (Figure 5F, lane 5), whereas F-actin stayed in the pellet (lane 6). These data show that, when oligomerized into rings, dynamin can efficiently displace Gsn from the barbed ends of F-actin.
As an alternative means to examine actin elongation that is dependent on displacement of Gsn from the barbed ends, we performed an actin re-polymerization assay (see model in Figure 5G; Barkalow et al, 1996; Pruyne et al, 2002). As shown in Figure 5H, dyn1 induced actin re-polymerization, but only in the presence of GTPγS (compare black diamonds and red circles with red squares). These data show that dynamin did not inhibit actin depolymerization from the pointed end, and that only dynamin rings can promote actin re-polymerization at the barbed ends capped by Gsn. To our knowledge, dynamin is the first protein that has been shown to induce actin elongation from Gsn-capped actin filaments by displacing Gsn from the barbed ends.
Dynamin–actin interactions promote formation of free barbed ends in cells
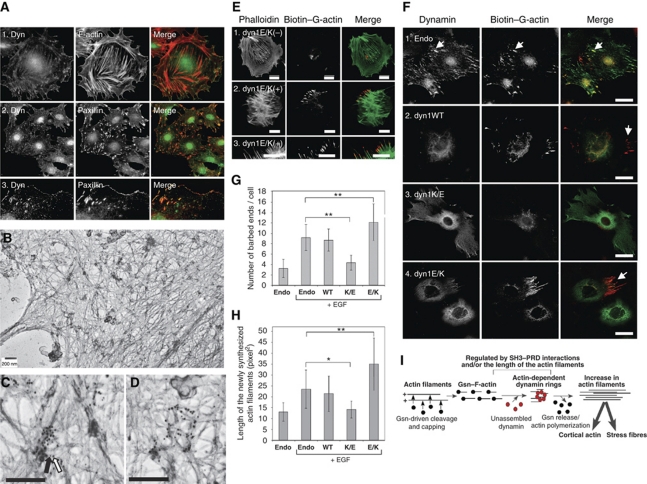
Given the role of dynamin–actin interactions in formation of FAs and stress fibres in cells (Figure 2), together with the biochemical analysis, our data suggested that dynamin rings might regulate actin polymerization at FAs by generating free barbed ends. Thus, we next examined localization of endogenous dynamin in podocytes. As shown before (Kruchten and McNiven, 2006), endogenous dynamin could be detected at FAs, marked by paxillin staining (Figure 6A). To examine whether endogenous dynamin associates with actin filaments, we performed electron microscopy on detergent permeabilized podocytes (Figure 6B; Schliwa et al., 1981). This method visualizes the cortex of cells from which the cytoplasm has been released. Dynamin antigenic sites were detected using monoclonal anti-dynamin antibody followed by 10-nm gold-conjugated secondary antibody (white arrow in Figure 6C), whereas paxilin antigenic sites were detected using anti-paxillin polyclonal antibody followed by 5 nm gold-conjugated secondary antibody (black arrow in Figure 6C). Dynamin antigenic sites were concentrated along actin filaments at distinct locations. Groups of large gold particles co-localized with small gold particles (Figure 6B–D), suggesting that in some instances, dynamin clustered around FAs.
Figure 6.
Dynamin–actin interactions at focal adhesions drive actin polymerization by generating free barbed ends. (A) Dynamin localizes to focal adhesions in podocytes. Podocytes were stained using anti-dynamin antibody, rhodamin phalloidin for F-actin or anti-paxillin antibody. Dynamin co-localizes with paxillin at focal adhesions and focal complexes along the membrane. (B–D) Electron micrographs of the podocyte cytoskeleton. Podocytes contain a thick actin network. Dynamin antigenic sites were visualized in the electron microscope by treatment with monoclonal anti-dynamin antibody followed by secondary antibody labelled with 10 nm gold particles (white arrow). Paxillin antigenic sites were visualized by polyclonal anti-paxillin antibody followed by secondary antibody labelled with 5 nm gold particles (black arrow). Scale bars, 200 nm. (E, F) Dynamin–actin interactions promote formation of free barbed ends in podocytes. Podocytes expressing the indicated dynamin mutants were stimulated with 5 nM EGF to induce de novo actin polymerization. After 5 min, cells were permeabilized in the presence of 0.45 μM biotin-labelled G-actin for 45 s, fixed and stained using rhodamin-conjugated anti-biotin antibody (red). In (E), total F-actin was labelled using FITC phalloidin (green). In (F), cells were stained using anti-dynamin antibody (green). Scale bars, 20 μm. (G, H) Quantification of barbed ends per cell (G) and length of the newly synthesized actin filaments (H). Data represent measurements of >20 cells (except for dyn1K/E where only 10 cells were examined) and are plotted as ±s.d. *P<0.05, **P<0.01. (I) Working model for the role of dynamin oligomerization in regulation of actin cytoskeleton. Short actin filaments are generated by gelsolin-driven cleavage of the actin filaments. Their high local concentration promotes dynamin oligomerization into rings, which in turn displace gelsolin from the barbed end and allow filament extension.
Next, we examined whether dynamin–actin interactions underlie the formation of free barbed ends in cells. Formation of barbed ends in cells expressing different dynamin mutants was examined upon EGF stimulation (Figure 6E and F; Symons and Mitchison, 1991; Bailly et al, 1999). In this assay, permeablized cells were allowed to incorporate biotin-labelled G-actin at barbed ends in situ for 45 s (red in Figure 6E and F). Thus, newly polymerized actin incorporated at barbed ends will be labelled red (compare biotin–G-actin and phalloidin staining in Figure 6E). As podocytes are terminally differentiated cells, only ∼5% of all cells exhibited EGF-dependent induction of free barbed ends (Supplementary Figure S6E). To identify cells expressing dynamin mutants, dynamin was subsequently stained using anti-dynamin antibody (green in Figure 6F). Compared with the endogenous control, expression of dyn1WT did not alter the number of cells expressing barbed ends (Supplementary Figure S6E), the number of free barbed ends per cell (Figure 6G) or the size of newly polymerized actin filaments (Figure 6H). Thus, endogenous dynamin is not rate limiting for the formation of free barbed ends. Notably, endogenous dynamin localized at the newly generated free barbed ends (Figure 6F, panel 1, white arrows). In the presence of ‘loss-of-function' dyn1K/E, much fewer cells exhibited barbed ends (Supplementary Figure S6E), and in these, the number and length of newly labelled actin filaments were reduced (Figure 6F–H), consistent with a loss of barbed ends. In contrast, cells expressing the ‘gain-of-function' dyn1E/K exhibited a statistically significant increase in the number and length of newly polymerized actin filaments (Figure 6F–H). Together, our data indicate that direct dynamin–actin interactions promote the formation of free barbed ends, which drive actin polymerization at FAs in podocytes.
Discussion
The data presented in this paper suggest a novel, dynamin-dependent mechanism that promotes actin polymerization (Figure 6I). In this model, proteins such as Gsn first fragment and cap F-actin filaments; the resulting short filaments bind and promote dynamin assembly into rings; dynamin rings in turn displace CPs; finally, the uncapped barbed actin ends undergo polymerization. The model is based on four key observations. First, we discovered a direct, functional interaction between dynamin and actin. Mutations in a previously unrecognized ABD of dynamin reduced actin stress fibre formation and altered the cortical actin cytoskeleton in cultured podocytes and rat hepatocytes. Conversely, a dynamin mutant with increased actin-binding affinity stimulated formation of stress fibres in cells. Therefore, direct binding of dynamin to actin is crucial for maintenance of the actin cytoskeleton. Second, actin directly regulates dynamin self-assembly. Thus, using GTPase assays and electron microscopy, we found that the interaction of short actin filaments with dynamin promotes dynamin self-assembly. Third, assembled dynamin dissociates the actin-CP Gsn from barbed ends in vitro, thereby allowing filament elongation. Fourth, direct dynamin–actin interaction promotes formation of free barbed ends in podocytes. Together, the data establish a feedback loop in which short actin filaments promote formation of dynamin rings, which in turn displace barbed end CPs, thereby allowing actin filament elongation.
A role for dynamin rings in regulating Gsn-dependent actin polymerization
Many cellular processes in which dynamin has been implicated are driven by actin polymerization (e.g. cell motility, endocytosis, cytokinesis). They all depend on rapid bursts of actin filament assembly at specific subcellular locations. Actin polymerization can be achieved either by de novo filament formation, or by severing of pre-existing filaments and elongation of the resulting barbed ends. The extent of filament elongation in vivo is limited by the presence of high-affinity barbed end CPs (for review see Chesarone and Goode, 2009). Our study identifies a novel role for dynamin rings in regulating actin polymerization by displacing barbed end CPs such as Gsn. As dynamin rings displaced Gsn, but not CP, our study suggests specificity in dynamin's interaction with different CPs.
Dynamin's novel role in regulating actin polymerization through Gsn is consistent with the previous studies, which implicate Gsn in the same cellular processes that are regulated by dynamin. Thus, Gsn has been implicated in podosomal assembly (Chellaiah et al, 2000), reorganization of cortical actin filament networks that underlie protruding lamellae (Safiejko-Mroczka and Bell, 2001) and the length of Listeria actin tails (Laine et al, 1998). Indeed, immunofluorescence micrographs revealed that both dynamin and Gsn concentrate just behind motile bacteria, at the junction between the actin filament rocket tail and the bacterium (Laine et al, 1998; Lee and De Camilli, 2002; Orth et al, 2002). Finally, it has been shown that Gsn also has a function in stress-fibre-dependent cell contraction (Arora et al, 1999). While Gsn has been implicated in actin polymerization, the mechanism by which Gsn, once associated with the barbed ends, can be displaced has not been identified (Kuhn and Pollard, 2007). Our study suggests that dynamin rings regulate actin polymerization at the FAs by displacing the CP Gsn. We are currently exploring in detail how dynamin rings promote the de novo formation of FAs.
We can envision two different mechanisms by which dynamin rings displace gelosolin from the barbed ends. One possibility is that dynamin binds and displaces Gsn. However, Gsn did not affect the GTPase activity of dynamin, suggesting that these two proteins might not form direct protein–protein interactions. Nevertheless, oligomerization-dependent interaction of dynamin with Gsn on actin filaments cannot be ruled out. A second possibility is that dynamin affects Gsn indirectly. We show that dynamin rings bind and align F-actin filaments into bundles with defined filament–filament spacing (Figure 3E). Thus, it is possible that dynamin rings alter filament geometry/twist and thereby affect the binding properties of other actin-binding/regulatory proteins including Gsn (Stokes and DeRosier, 1987). We favour the second model for the following reasons. Gsn binds to filament ends and changes the twist of actin filaments, which in turn displaces phalloidin (Cooper, 1987). This observation implies that locking F-actin into an unfavourable conformation could disfavour Gsn binding. In contrast, CP is not known to alter actin filament twist, and CP was not displaced by dynamin rings. These data are consistent with the idea that dynamin rings stabilize terminal actin subunits in a conformation that disfavours Gsn binding. By analogy with cofilin, another protein that binds and changes actin geometry (McGough et al, 1997), we speculate that dynamin rings promote the formation of a distinct population of conformationally unique actin filaments, which binds or releases specific sets of actin-binding/regulatory proteins.
Role of dynamin–actin interactions in regulation of dynamin oligomerization
Our study identifies unexpected regulation of dynamin's oligomerization status by actin filaments. Interestingly, while long actin filaments had no effect on dynamin oligomerization, shorter actin filaments (50–700 nm) promoted dynamin oligomerization into single rings. In addition, we have developed a novel, cell-based assay that measures dynamin oligomerization into higher order structures using fluorescence lifetime imaging microscopy. This assay confirms that dynamin oligomerization is promoted by short actin filaments in live cells (Gu et al, manuscript in preparation). As dynamin oligomerization is highly cooperative with respect to dynamin concentration (Warnock et al, 1996), our data suggest that local concentration of short actin filaments promotes actin-dependent dynamin oligomerization at distinct cellular locations. Supporting our hypothesis, we found that dynamin antigenic sites are concentrated at distinct locations on actin filaments, some of which are most likely FAs (Figure 6C and D).
The mechanism by which short actin filaments promote dynamin oligomerization is not readily apparent and is topic of future studies. It is plausible that short filaments exhibit lower steric hindrance, thus allowing more efficient oligomerization of bound dynamin. Regardless of the mechanism, the preference for short filaments is expected to provide temporal and spatial regulation of dynamin assembly.
Importantly, a number of SH3-domain containing proteins that bind/regulate actin such as SNX9 and cortactin have been shown to regulate dynamin oligomerization (Yarar et al, 2007; Mooren et al, 2009). Thus, it seems reasonable to conclude that dynamin oligomerization is regulated not only by the length of actin filaments, but also by PRD–SH3 interactions. In summary, our study provides a direct link between the oligomerization cycle of dynamin and the regulation of the actin cytoskeleton. It further suggests that dynamin's effect on the actin cytoskeleton will ultimately depend on actin binding, its intrinsic oligomerization cycle and other actin regulators that modulate dynamin's assembly state.
Materials and methods
Cells, antibodies, reagents and standard techniques
Mouse podocyte cell lines were grown as described previously (Mundel et al, 1997). Antibodies: anti-dyn1/2 hudy 1 (Upstate Technology, Lake Placid, NY); mouse anti-dyn1 VAM-SV041 (StressGene, Victoria, Canada); anti-dyn2 (Santa Cruz Biotechnology, Delaware Avenue, CA); rabbit polyclonal anti-paxillin (StressGene, Victoria, Canada); mouse monoclonal anti-Gsn (Sigma-Aldrich, Saint Louis, MO); anti-cortactin (Millipore, Billerica, MA). Subcellular fractionation was performed as described (Damke et al, 1994). GTPase assays were performed as described (Leonard et al, 2005). PC:PIP2 vesicles and actin bundling experiments were performed as described (Schafer et al, 2002). Adenoviral infections of cultured podocytes were preformed as described (Sever et al, 2007). Internalization of rhodamine transferrin was performed using HeLa cells, 18 h post-infection with 20 μg ml–1 of rhodmaine-Tfn in PBS containing 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose and 0.2% BSA for 10 min at 37°C. The wound-healing assays were performed as described (Asanuma et al, 2006). Rho type GTPases activation assays were performed using commercial Rho activation kit (Cell Biolabs, San Diego, CA).
Plasmid constructs
Plasmid construction for adenovirus production and in vitro transcription and translation was accomplished by Gateway Technology System with Clonase II (Invitrogen). Donor vector was generated by cloning full-length human dyn1 or dyn2 cDNA into the pDONR201 vector using site-specific recombination reaction of BP clonase II enzyme. Actin-binding defective dyn mutant constructs were generated using appropriate site-directed mutagenesis. Resulting fragments were cloned using _Bst_XI and _Cla_I into the pDONR201-containing gene for dyn. IVT expression vector pCITE4 (Invitrogen) was generated using pDONR201 vector and the site-specific recombination reaction of LR clonase II. Adenoviral vectors were generated by recombining pDONR201 with pAd/CMV/V5-DEST. C-terminally His-tagged dynamin was generated by subcloning dyn1 cDNA into the pGE-60 vector (Qiagen) using _Nco_I and _Bam_HI enzymes. All subsequent mutants were generated by subcloning _Bst_XI and _Cla_I fragments from pDONR201 into pQE-60 (Qiagen). Vectors expressing ECFP and EYFP N-terminally tagged dynamin were generated using pECFP and pEYFP vectors (BD Biosciences Clontech). To generate the dyn1 ΔPRD mutant, first, 970-bp PCR products corresponding to nt 1414–2250 of human dynamin1 coding DNA sequence were amplified using specific primers, incorporating three stop codons and _Nar_I restriction site at COOH terminus of PCR products. Subsequently, _Cla_I/_Nar_I digested PCR fragments were subcloned into the corresponding region of full-length dynamin1 cDNA in pDONR201 vector via T/A cloning using pGEM-T-easy vector (Promega). To construct double mutants, dyn1 ΔPRD /EK and −KE, dyn1 EK and –KE mutant constructs were restricted by _Bst_XI and _Cla_I enzymes and then the resulting DNA fragments were subcloned into the corresponding regions of dyn1 ΔPRD cDNA in pDONR201 vector. Adenoviral vectors were generated by recombining pDONR201 with pAD/CMV/V5-DEST, using LR clonase II and production of adenoviruses was accomplished followed by manufacture's manual (Invitrogen). All constructs were verified by DNA sequencing.
Dynamin expression and purification
Non-tagged human dyn1 (isoform 1) was expressed using baculovirus expression system in Tn5 insect cells, and was purified as described (Damke et al, 2001). Baculoviruses expressing C-terminally His-tagged rat dyn2 were generous gift from Dorothy Schaffer (University of Virginia, Charlottesville, VA). Baculoviruses expressing N-terminally His-tagged ΔPRD were generated in this study. C-terminally His-tagged dyn1WT, dyn1K/E, dyn1K/A and dyn1E/K were either expressed in Tn5 insect cells (baculoviral expression), or in Escherichia coli (bacterial expression), and purified on a nickel column as described (Ingerman and Nunnari, 2005). Alternatively, untagged dyn1WT, dyn1K/E, dyn1K/A and dyn1E/K were expressed in HeLa cells and purified using GST-SH3 domain of amphiphysin II as described (Quan and Robinson, 2005). Different purification strategies for dynamin were used to minimize possibility to co-purify putative actin-binding proteins. Activity of recombinant dynamin was measured using GTPase assays (Leonard et al, 2005). Given different strategies to purify recombinant proteins, only recombinant proteins that exhibited catalytic constant _k_cat between 0.5 and 1.5 min−1 (wild-type values) were used in this study.
Actin staining and quantification
Cells expressing different dynamin constructs were stained with anti-dynamin antibody (mouse monoclonal VAM-SV041), anti-paxillin antibody and/or phalloidin (Invitrogen). Images were captured with a Zeiss LSM 5 PASCAL laser scanning microscope and a × 40 objective. For quantification of actin, cells were imaged with above microscope using a fixed exposure of 700 ms for phalloidin. The intensity of the actin staining was measured from whole cells by Image J (v1.4) software. Total fluorescence from uninfected cells and infected was analysed separately, and the staining intensity were normalized to the uninfected cells. The experiment was repeated at least three times. Number of FAs was determined by integrated morphometry analysis performed using Image J (v1.4) on thresholded images to select classified objects of a size range of >1 pixels as FAs, based on anti-paxillin staining. The analysed particles command was used to measure number of FAs. The number of barbed ends in podocytes was examined as described in Chan et al (1998) and is included in Supplementary data. When indicated, data were further analysed using GraphPad Prism (v 4.03) for Windows (GraphPad Software, San Diego, CA) to perform statistical analysis using two-tailed unpaired _t_-tests. Based on this analysis, *P<0.05, **_P_<0.01 and _P_>0.05 is considered not significant.
Actin purification and assembly
Actin was purified from rabbit skeletal muscle as published in Pardee and Spudich (1982), with an additional gel filtration step (HiLoad 16/60 Superdex 200 pg). Actin was stored as monomer at −80°C in G-buffer (2 mM Tris–HCl, pH 7.4, 0.5 mM ATP, 0.2 mM CaCl2, 0.5 mM β-mercaptoethanol) and was a generous gift of F Nakamura, Brigham and Women's Hospital, Boston, MA. Before each experiment, G-actin was thawed and diluted to final concentration of 25 μM in G-buffer, incubated at 37°C for 1 h and centrifuged at 250 000 g for 30 min to remove aggregates. The G-actin was polymerized by the addition of 0.1 M KCl and 2 mM MgCl2 for 20 min at RT. Thus, F-actin buffer contains 10 mM Tris–HCl, pH 7.4, 0.5 mM ATP, 0.2 mM CaCl2, 0.5 mM β-mercaptoethanol, 2 mM MgCl2, 0.1 M KCl. Gsn–F-actin complexes were generated by adding recombinant human Gsn at the indicated ratios during the polymerization step. Recombinant human Gsn was purified as described in Wen et al (1996), and was a generous gift of F Nakamura, Brigham and Women's Hospital, Boston, MA. Recombinant mouse CP was purified as described in Fujiwara et al (2009) and was a generous gift of Fujiwara Ikuko and John A Hammer, Laboratory of Cell Biology, NIHBI/NIH, Bethesda, MD. Detail description of different actin assays is included in the Supplementary data.
Supplementary Material
Supplementary Data
Review Process File
Acknowledgments
We are indebted to Fujiwara Ikuko and John A Hammer III for providing recombinant CP as well as pyrine-labelled actin. This work was supported by the National Institute of Health (R01 DK64787 to SS) and the NephCure Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrianantoandro E, Blanchoin L, Sept D, McCammon JA, Pollard TD (2001) Kinetic mechanism of end-to-end annealing of actin filaments. J Mol Biol 312: 721–730 [DOI] [PubMed] [Google Scholar]
- Arora PD, Janmey PA, McCulloch CA (1999) A role for gelsolin in stress fiber-dependent cell contraction. Exp Cell Res 250: 155–167 [DOI] [PubMed] [Google Scholar]
- Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P (2006) Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol 8: 485–491 [DOI] [PubMed] [Google Scholar]
- Bailly M, Macaluso F, Cammer M, Chan A, Segall JE, Condeelis JS (1999) Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J Cell Biol 145: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkalow K, Witke W, Kwiatkowski DJ, Hartwig JH (1996) Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J Cell Biol 134: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzaniti A, Neff L, Sanjay A, Horne WC, De Camilli P, Baron R (2005) Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol Biol Cell 16: 3301–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS (1998) EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci 111(Part 2): 199–211 [DOI] [PubMed] [Google Scholar]
- Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA (2000) Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J Cell Biol 148: 665–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL (2009) Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol 21: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA (1987) Effects of cytochalasin and phalloidin on actin. J Cell Biol 105: 1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL (1994) Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol 127: 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Muhlberg AB, Sever S, Sholly S, Warnock DE, Schmid SL (2001) Expression, purification, and functional assays for self-association of dynamin-1. Methods Enzymol 329: 447–457 [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Remmert K, Hammer JA, 3rd. (2009) Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J Biol Chem 285: 2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL (1995) Dynamin self assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374: 190–192 [DOI] [PubMed] [Google Scholar]
- Ingerman E, Nunnari J (2005) A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol 404: 611–619 [DOI] [PubMed] [Google Scholar]
- Kruchten AE, McNiven MA (2006) Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci 119: 1683–1690 [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD (2007) Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J Biol Chem 282: 28014–28024 [DOI] [PubMed] [Google Scholar]
- Laine RO, Phaneuf KL, Cunningham CC, Kwiatkowski D, Azuma T, Southwick FS (1998) Gelsolin, a protein that caps the barbed ends and severs actin filaments, enhances the actin-based motility of Listeria monocytogenes in host cells. Infect Immun 66: 3775–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, De Camilli P (2002) Dynamin at actin tails. Proc Natl Acad Sci USA 99: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M, Song BD, Ramachandran R, Schmid SL (2005) Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol 404: 490–503 [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A (1997) Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol 138: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW (2000) Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol 151: 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Pucadyil T, Ramachandran R, Schmid SL (2009) Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans 37: 1022–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Kotova TI, Moore AJ, Schafer DA (2009) Dynamin2 GTPase and cortactin remodel actin filaments. J Biol Chem 284: 23995–24005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Maiti S, Goode BL (2006) Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol 406: 215–234 [DOI] [PubMed] [Google Scholar]
- Muhlberg AB, Warnock DE, Schmid SL (1997) Domain structure and intramolecular regulation of dynamin GTPase. EMBO J 16: 6676–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundel P, Reiser J, Borja AZ, Pavenstadt H, Davidson GR, Kriz W, Zeller R (1997) Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp Cell Res 236: 248–258 [DOI] [PubMed] [Google Scholar]
- Oh J, Reiser J, Mundel P (2004) Dynamic (re)organization of the podocyte actin cytoskeleton in the nephrotic syndrome. Pediatr Nephrol 19: 130–137 [DOI] [PubMed] [Google Scholar]
- Orth JD, Krueger EW, Cao H, McNiven MA (2002) The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc Natl Acad Sci USA 99: 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, McNiven MA (2003) Dynamin at the actin-membrane interface. Curr Opin Cell Biol 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA (1982) Purification of muscle actin. Methods Enzymol 85(Part B): 164–181 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA (2009) Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C (2002) Role of formins in actin assembly: nucleation and barbed-end association. Science 297: 612–615 [DOI] [PubMed] [Google Scholar]
- Quan A, Robinson PJ (2005) Rapid purification of native dynamin I and colorimetric GTPase assay. Methods Enzymol 404: 556–569 [DOI] [PubMed] [Google Scholar]
- Safiejko-Mroczka B, Bell PB Jr (2001) Reorganization of the actin cytoskeleton in the protruding lamellae of human fibroblasts. Cell Motil Cytoskeleton 50: 13–32 [DOI] [PubMed] [Google Scholar]
- Saleem MA, Zavadil J, Bailly M, McGee K, Witherden IR, Pavenstadt H, Hsu H, Sanday J, Satchell SC, Lennon R, Ni L, Bottinger EP, Mundel P, Mathieson PW (2008) The molecular and functional phenotype of glomerular podocytes reveals key features of contractile smooth muscle cells. Am J Physiol Renal Physiol 295: F959–F970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA (2004) Regulating actin dynamics at membranes: a focus on dynamin. Traffic 5: 463–469 [DOI] [PubMed] [Google Scholar]
- Schafer DA, Weed SA, Binns D, Karginov AV, Parsons JT, Cooper JA (2002) Dynamin2 and cortactin regulate actin assembly and filament organization. Curr Biol 12: 1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, van Blerkom J, Porter K (1981) Stabilization of the cytoplasmic ground substance in detergent-opened cells and a structural and biochemical analysis of its composition. Proc Natl Acad Sci USA 78: 4329–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Altintas M, Nankoe S, Moller C, Ko D, Wei C, Henderson J, del Re E, Hsing L, Erickson A (2007) Proteolytic processing of dynamin by cytoplasmic cathepsin L defines a mechanism for proteinuric kidney disease. J Clin Invest 117: 2095–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Muhlberg AB, Schmid SL (1999) Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature 398: 481–486 [DOI] [PubMed] [Google Scholar]
- Stokes DL, DeRosier DJ (1987) The variable twist of actin and its modulation by actin-binding proteins. J Cell Biol 104: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons MH, Mitchison TJ (1991) Control of actin polymerization in live and permeabilized fibroblasts. J Cell Biol 114: 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum AG, de Graaf JH, Schuuring-Scholtes E, Kluin PM, Fan YX, Zhan X, Moolenaar WH, Schuuring E (2003) Alternative splicing of the actin binding domain of human cortactin affects cell migration. J Biol Chem 278: 45672–45679 [DOI] [PubMed] [Google Scholar]
- Van Troys M, Dewitte D, Goethals M, Carlier MF, Vandekerckhove J, Ampe C (1996) The actin binding site of thymosin beta 4 mapped by mutational analysis. EMBO J 15: 201–210 [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Baba T, Schmid SL (1997) Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol Biol Cell 8: 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Hinshaw JE, Schmid SL (1996) Dynamin self assembly stimulates its GTPase activity. 271: 22310–22314 [DOI] [PubMed]
- Watts RG, Howard TH (1992) Evidence for a gelsolin-rich, labile F-actin pool in human polymorphonuclear leukocytes. Cell Motil Cytoskeleton 21: 25–37 [DOI] [PubMed] [Google Scholar]
- Weins A, Kenlan P, Herbert S, Le TC, Villegas I, Kaplan BS, Appel GB, Pollak MR (2005) Mutational and biological analysis of alpha-actinin-4 in focal segmental glomerulosclerosis. J Am Soc Nephrol 16: 3694–3701 [DOI] [PubMed] [Google Scholar]
- Wen D, Corina K, Chow EP, Miller S, Janmey PA, Pepinsky RB (1996) The plasma and cytoplasmic forms of human gelsolin differ in disulfide structure. Biochemistry 35: 9700–9709 [DOI] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL (2007) SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell 13: 43–56 [DOI] [PubMed] [Google Scholar]
- Yin HL, Hartwig JH, Maruyama K, Stossel TP (1981) Ca2+ control of actin filament length. Effects of macrophage gelsolin on actin polymerization. J Biol Chem 256: 9693–9697 [PubMed] [Google Scholar]
- Zhang P, Hinshaw JE (2001) Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol 3: 922–926 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Review Process File