Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes (original) (raw)
Abstract
The prokaryotic organisms can be divided into two main groups depending upon whether their cell envelopes contain one membrane (monoderms) or two membranes (diderms). It is important to understand how these and other variations that are observed in the cell envelopes of prokaryotic organisms have originated. In 2009, James Lake proposed that cells with two membranes (primarily Gram-negative bacteria) originated from an ancient endosymbiotic event involving an Actinobacteria and a Clostridia (Lake 2009). However, this Perspective argues that this proposal is based on a number of incorrect assumptions and the data presented in support of this model are also of questionable nature. Thus, there is no reliable evidence to support the endosymbiotic origin of double membrane bacteria. In contrast, many observations suggest that antibiotic selection pressure was an important selective force in prokaryotic evolution and that it likely played a central role in the evolution of diderm (Gram-negative) bacteria. Some bacterial phyla, such as _Deinococcus_-Thermus, which lack lipopolysaccharide (LPS) and yet contain some characteristics of the diderm bacteria, are postulated as evolutionary intermediates (simple diderms) in the transition between the monoderm bacterial taxa and the bacterial groups that have the archetypal LPS-containing outer cell membrane found in Gram-negative bacteria. It is possible to distinguish the two stages in the evolution of diderm-LPS cells (viz. monoderm bacteria → simple diderms lacking LPS → LPS containing archetypal diderm bacteria) by means of conserved inserts in the Hsp70 and Hsp60 proteins. The insert in the Hsp60 protein also distinguishes the traditional Gram-negative diderm bacterial phyla from atypical taxa of diderm bacteria (viz. Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia). The Gram-negative bacterial phyla with an LPS-diderm cell envelope, as defined by the presence of the Hsp60 insert, are indicated to form a monophyletic clade and no loss of the outer membrane from any species from this group seems to have occurred. This argues against the origin of monoderm prokaryotes from diderm bacteria by loss of outer membrane.
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-011-9616-8) contains supplementary material, which is available to authorized users.
Keywords: Bacterial cell envelopes, Origin of the outer cell membrane, Gram-negative bacteria, Lipopolysaccharides, Monoderm and diderm bacteria, Endosymbiotic hypothesis, Antibiotic selection pressure, Conserved inserts in Hsp60 and Hsp70 proteins, Simple monoderms
Introduction—complexity of the bacterial cell envelopes
The prokaryotic organisms are traditionally divided into two main groups i.e. Gram-positive and Gram-negative, based on their Gram-stain retention characteristics (Gram 1884; Stanier et al. 1976). Although the Gram-staining has not proven to be a reliable criterion for the higher-level division or classification of prokaryotic organisms, a more important structural characteristic that generally distinguishes these two types of organisms is the nature of their cell envelopes (Stanier et al_._ 1976; Murray 1986). Most Gram-positive bacteria are bounded by a single cell membrane and they generally contain a relatively thick peptidoglycan layer that is responsible for retaining the Gram-stain. In contrast most “true” Gram-negative bacteria are surrounded by two different cell membranes and they contain only a thin peptidoglycan layer in the periplasmic compartment that is bounded by the inner and outer membranes (Stanier et al_._ 1976; Murray 1986; Truper and Schleifer 1992; Gupta 1998b; Sutcliffe 2010). Although these differences in the cell envelope characteristics of Gram-positive and Gram-negative bacteria have long been known, due to the variability of Gram-staining response and polyphyletic branching of these two groups of bacteria in the 16S rRNA gene and other phylogenetic trees (Olsen and Woese 1993; Ludwig and Klenk 2005), the possibility that the cells with two membranes (diderm bacteria) might be phylogenetically distinct from monoderm prokaryotes was not recognized until 1998. This recognition came from the identification of a 21–23 aa long conserved insert in the Hsp70 family of protein that was uniquely shared by different phyla of diderm bacteria but absent in all other prokaryotes including Archaea (Gupta and Singh 1994; Gupta 1998b, c). The absence of this indel in bacterial lineages such as Mycoplasma that stained Gram-negative (as they lack the peptidoglycan layer) but contained a single membrane, provided evidence that the presence or absence of the outer cell membrane, rather than the Gram-staining response constituted a useful phylogenetic characteristic (Gupta 1998b, 2000; Cavalier-Smith, 2002).
The division of prokaryotic organisms into two distinct groups viz. “monoderms” and “diderms” based upon the presence or absence of the outer membrane and the large insert in the Hsp70 protein (Gupta 1998a, b, c) led to the question regarding which of these two lineages was ancestral and which was derived. Insights into this question was provided by our observation that Hsp70 and another protein, MreB, which is also present in different prokaryotic lineages, have evolved from an ancient gene duplication in the common ancestor of prokaryotes (Gupta and Golding 1993; Gupta 1998b). Thus, the presence or absence of the insert in the MreB protein could be used to determine whether the indel in the Hsp70 protein is an insert or a deletion. Because the MreB protein from different lineages, similar to the Hsp70 from monoderm prokaryotes, did not contain this indel, the absence of the indel was inferred to be the ancestral character state of the Hsp70 protein (Gupta and Golding 1993; Gupta 1998b). Thus, the large indel in the Hsp70 protein was an insert that occurred in an ancestor of the diderm bacteria (Gupta 1998b, c). This observation and a number of other observations (reviewed in Gupta 1998b) indicated that the cells with one membrane are ancestral and that the cells with two membranes originated from them (Gupta and Golding 1993; Gupta 1998b; Koch 2003). A subsequent study by Lake also supported this inference (Lake et al_._ 2007). Some authors have suggested that cells with two membranes evolved prior to those with one membrane (Cavalier-Smith 2006; Griffiths 2007; Valas and Bourne 2009). However, Valas and Bourne place the root of the prokaryotic tree in the Chloroflexi, which are now indicated to have a monoderm rather than a diderm cell envelope (Valas and Bourne 2009; Sutcliffe 2010, 2011). It is also difficult to conceive of any simple model where a cell with both an inner and outer membrane can directly evolve without the initial development of a cell with only a single membrane.
Although the monoderm or diderm cell structures as exemplified by the model organisms Bacillus subtilis and Escherichia coli are the most common types of cell envelopes present within the bacterial domain, several bacterial taxa are now known that contain atypical outer cell envelopes (or layers) that do not correspond to these model organisms (Sutcliffe 2010, 2011). Sutcliffe (2010) has recently reviewed the work on this subject and it illustrates that the distinction between the monoderm and diderm cell structures is not clear-cut and that the observed differences are important in terms of understanding the origin of the outer cell membrane. For example, the bacteria belonging to the order Corynebacterineae (phylum Actinobacteria), although widely considered as monoderms, have an outer lipid layer composed of mycolic acid molecules which are arranged in a highly ordered form resembling an outer membrane (Brennan and Nikaido 1995; Sutcliffe 2010). Similarly, the bacteria belonging to the phylum Thermotogae contain an outer toga (envelope) consisting primarily of proteins rather than lipids (Reysenbach 2001). Although some characteristics of genes/proteins found in the Thermotogae genomes indicate that they are capable of synthesizing lipids and transporting them to the outer envelope (Sutcliffe 2010), their outer envelope is clearly distinct from all other monoderm and diderm bacteria. Several other phyla of bacteria that are considered as diderms (viz. _Chloroflexi, Deinococcus_-Thermus) lack LPS (Sutcliffe 2010), which is considered to be a defining characteristic of the archetypal outer cell membrane. Sutcliffe has also presented strong arguments that Chloroflexi, which are widely believed to have a diderm cell envelope, are monoderm (Sutcliffe 2011). They lack LPS (as well the genes for various key proteins involved in the synthesis of LPS) and proteins characteristic of outer membranes, such BamA family proteins and outer membrane components of secretion systems. Further, the cell envelopes of some of these species are indicated to be multi-layered with no evidence of lipids in the outer cell layer (Hanada and Pierson 2006). Hence, the outer layer in some Chloroflexi could be composed of polysaccharides or proteins (Sutcliffe 2011). These observations point to the complexity of the bacterial outer membrane structure and indicate that the distinction between monoderm and diderm cell envelopes is not quite simple or straightforward, at least by biochemical means. These observations are important in evaluating any model or hypothesis for the origin of the outer cell membrane. Moreover, it should be taken into account that the mycolic acid based outer membranes of the order Corynebacterineae (phylum Actinobacteria) provide evidence that lipid outer membranes have evolved more than once.
Have diderm bacteria originated via endosymbiosis?—critical evaluation of Lake’s hypothesis and data
Lake has recently proposed that cells with two membranes are the result of an ancient endosymbiotic event involving two monoderm bacteria belonging to the phyla/taxa Actinobacteria and Clostridia (Lake 2009). He reached this inference based upon the presence or absence of a given dataset of proteins in different groups of bacteria. For his analyses, Lake made an important assumption that all prokaryotic organisms belong to one of the five natural and phylogenetically well separated groups viz. i.e. Diderm bacteria (D), Actinobacteria (A), Archaea (R), Bacillus and relatives (B) and Clostridia and relatives (C). Of these five groups, the distinctness of Archaea, and more recently Actinobacteria, is established based upon large numbers of molecular characteristics, including many signature proteins and conserved indels that are uniquely found in all species from these taxa (Olsen and Woese 1997; Gao and Gupta 2005; Walsh and Doolittle 2005; Gao et al_._ 2006; Gao and Gupta 2007; Gupta and Shami 2011). The clade D corresponding to diderm bacteria as defined by Lake contains different bacterial phyla including Thermotogae, Fusobacteria, Deinococcus_-Thermus and Chloroflexi, whose outer cell envelopes are atypical and differ in important respects from archetypical bacterial cell with two membranes (Sutcliffe 2010). Significantly, there is no evidence from any source that different bacterial phyla that Lake places in Clade D form a monophyletic lineage. The presence of the large insert in the Hsp70 protein is indicated to be a marker that can distinguish monoderm and diderm prokaryotes (Gupta 1998b); however, this insert is not found in Thermotogae and Fusobacteria (Gupta 1998b; Singh and Gupta 2009) but is found in the monoderm Chloroflexi (see above). Phylogenetic studies on Fusobacteria indicate that they are more closely related to Clostridia than to any of the diderm bacteria (Griffiths and Gupta 2004; Mira et al._ 2004; Karpathy et al_._ 2007). Therefore, the clade D as defined by Lake does not constitute a monophyletic group based upon either morphological or phylogenetic considerations, which is an essential requirement for analysis of this nature. Further, this clade also includes majority of the known bacterial phyla (including Chloroflexi which are now indicated to be monoderms; Sutcliffe 2011) and the representation of this heterogeneous group by a single entity, as Lake has done, can lead to misleading results.
The other two proposed main taxa, B and C, are presently part of the phylum Firmicutes (Ludwig and Klenk 2005). This phylum is poorly characterized phylogenetically and no biochemical or molecular marker is known that is uniquely shared by all Firmicutes species. The division of this phylum into the two main prokaryotic taxa, B and C, which according to Lake are naturally and phylogenetically clearly separated, is not accurate and no evidence is presented to support that they form monophyletic lineages. Within the Firmicutes, the Clostridia species (taxa C) in particular are a very heterogeneous assemblage and it has proven difficult to circumscribe this clade by phylogenetic or any other means (Wiegel et al_._ 2006). Recently, several bacterial species that were previously part of the Class Clostridia have been placed in a separate phylum, the Synergistetes (Jumas-Bilak et al_._ 2009; Hugenholtz et al_._ 2009). The species from this phylum, similar to Fusobacteria, contain two membranes and also genes for the key LPS biosynthetic enzymes (Baena et al_._ 1998; Jumas-Bilak et al_._ 2009; Sutcliffe 2010). Moreover, the situation is further complicated by the recent delineation of the Class Negativicutes within the phylum Firmicutes (Marchandin et al_._ 2010), as many representatives of this apparently have outer membranes containing LPS (Sutcliffe 2010). Thus, if the Clade D is defined on the basis of presence of two cell membranes then these taxa should have been part of Clade D rather than Clade C. Therefore, the division of the prokaryotes into the 5 main groups as defined by Lake (2009), on which his entire analysis was based, was based on completely arbitrary considerations and it has no valid phylogenetic, taxonomic or morphological/biochemical basis.
Another serious problem with Lake’s hypothesis (Lake 2009) relates to the quality and accuracy of the data on which his hypothesis was based. Lake examined the presence or absence of proteins from different families into the five proposed taxa and based on these results reached the conclusion that a tree like topology was not supported by the character states of many proteins and that their distribution can only be explained by a ring-like structure involving origin of taxon D by merger of taxa A and C. However, Lake provided no information how widely these proteins were distributed in different groups. To obtain information in this regard, I carried out Blast searches on 24 proteins corresponding to the first, third and fourth row in Table 1 of Lake’s paper (Lake 2009). The results of these analyses, along with those reported by Lake for the same proteins, are presented in Table 1. The character states for these proteins as reported by Lake are also shown in the Table 1. Very surprisingly, the species distribution patterns (or character states) for most of these proteins were very different from those reported by Lake. For example, of the first three proteins in Table 1, which according to Lake supported the pattern [R (+), A (+), B (+), C (−) and D (−)], the first two were present in large numbers of Clostridia (C) as well as several diderm bacteria (D). The third protein (PTH2) was found to be largely specific for Actinobacteria (A) and only 4 hits for Archaea (R) and 1 hit for Bacillus (B), which are barely significant, were observed. Similar major discrepancies were noted for the 15 proteins that were reported to exhibit the pattern [R (−), A (+), B (+), C (+) and D (−)]. For two of these proteins MecA_N and RsbU_N, large numbers of hits from A, B, C and D were observed; For the proteins Cas_Csm6 and DUF624, all significant hits were from the Bacillus group (B); For two other proteins (Omega-Repress and SASP), only 1–3 hits from A were observed, but a similar number of hits were also seen for D (Table 1). For Lactococcin 972, no significant hit for C was observed. Of the 15 proteins in this category, only 5 proteins (DUF1048, DUF939, Etx_Mtx2, L.biotic_A and Phage_holin), at best, indicated the pattern noted by Lake (Lake, 2009). However, for three of these proteins, the total numbers of significant hits from all groups were in the range of 15–17 (including many hits for the same species) and for all 5 of these proteins very few hits were observed from the clade A and C species indicating that their species distribution was extremely limited and they do not provide reliable characteristics. Additionally, for 4 of the 15 proteins in this category, many significant hits were from bacteriophages, indicating that lateral gene transfer for these proteins should be common (Gogarten and Townsend 2005) and their species distribution patterns would not be reliable. Similar discrepancies between the observed and reported patterns were seen for 8 other proteins, which according to Lake supported the pattern [R (+), A (−), B (+), C (+) and D (−)]. Although, comprehensive analyses have not been conducted on all of the proteins that were analyzed by Lake, the results for the 26 proteins presented in Table 1, which correspond to three of the important character states, raise serious concerns regarding the quality and accuracy of the data that was used to infer that the cells with two membranes (clade D) evolved by a merger of taxa A and C.
Table 1.
Distribution patterns of various protein families in the indicated taxa
| Protein name | Accession no. | Species distribution or character state pattern | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reported by Lake | Observed distribution pattern | ||||||||||
| R | A | B | C | D | R | A | B | C | D | ||
| DUF567 | PF04525 | + | + | + | – | – | +3 | +>20 | +>50 | +>20 | +3 |
| FR47 | PF08445 | + | + | + | – | – | +15* | +>50 | +>50 | +>10 | +>50 |
| PTH2 | PF01981 | + | + | + | – | – | +4* | +>50 | +1* | – | – |
| Cas_Csm6 | YP_82039a | – | + | + | + | – | – | – | +5 | – | – |
| DUF1048 | PF06304 | – | + | + | + | – | – | +7* | +>50 | +9* | – |
| DUF348 | PF03990# | – | + | + | + | – | – | +>50 | +>50 | +>50 | 4 |
| DUF624 | PF04854 | – | + | + | + | – | – | – | +>50 | – | – |
| DUF939 | PF06081 | – | + | + | + | – | – | +7 | +>50 | +11* | – |
| Etx_Mtx2 | PF03318 | – | + | + | + | – | – | +1 | +11* | +7 | – |
| G5 | PF07501# | – | + | + | + | – | – | +>50 | +>50 | +>50 | +4* |
| L.biotic_A | PF04604 | – | + | + | + | – | – | +1 | +12 | +2 | – |
| Lactococcin | PF09683 | – | + | + | + | – | – | +5 | +13* | – | – |
| MecA_N | PF05223 | – | + | + | + | – | – | +>50 | +>50 | +>50 | +>50 |
| Omega Rep | PF07764 | – | + | + | + | – | – | +1 | +28* | +2 | +3 |
| Phage-holin | PF04688# | – | + | + | + | – | – | +3 | +50* | +2 | – |
| Phage_min2 | PF06152# | – | + | + | + | – | – | +8* | +46* | +14* | +1* |
| RsbU_N | PF08673 | – | + | + | + | – | +6 | +>50 | +>50 | +16* | +>50 |
| SASP | PF00269 | – | + | + | + | – | – | +1 | +>50 | +>50 | +2 |
| DUF1002 | PF06207 | + | – | + | + | – | +2* | – | +>50 | +>50 | – |
| DUF1338 | PF07142 | + | – | + | + | – | – | – | +>50 | +17* | – |
| DUF1646 | PF07854 | + | – | + | + | – | +12* | – | +2 | +15* | – |
| DUF964 | PF06133 | + | – | + | + | – | – | – | +>50 | +16* | – |
| DUF988 | PF06177 | + | – | + | + | – | +4 | +7* | +>50 | +>50 | +5 |
| Hth_MGA | PF08280 | + | – | + | + | – | – | – | +>50 | +1* | +1* |
| UPF0154 | PF03672 | + | – | + | + | – | – | – | +>50 | +7* | – |
| YcH | PF07435 | + | – | + | + | – | – | – | +>50 | – | – |
In addition to these important concerns regarding the critical assumptions on which Lake’s analysis was based and the accuracy of his data, the endosymbiotic origin of diderm bacteria by merger of an Actinobacteria and Clostridia is also not supported by several other important observations. First, in all established cases of endosymbiosis (viz. origin of mitochondria from Alphaproteobacteria, or origin of plastids from Cyanobacteria) (Margulis 1993), numerous genes that are distinctive characteristics of the original endosymbiont(s) are commonly retained by all of the derived organisms (Gray 1999). Thus, all plants and photosynthetic eukaryotes contain numerous genes and other characteristics that they uniquely share with cyanobacteria (Gupta et al_._ 2003; Mulkidjanian et al_._ 2006; Gupta and Mathews 2010). Similarly, all eukaryotic organisms, without any exceptions, contain notable fractions of their genes that are derived from either alpha proteobacteria or archaeal ancestors (Gupta 1998b; Rivera and Lake 2004). However, for the two prokaryotic taxa, Actinobacteria and Clostridia, whose merger is postulated to have given rise to the diderm bacteria, no unique molecular or other characteristics have been identified that are commonly shared by all or most species from either taxa A and D or by taxa C and D (Gao et al_._ 2006; Gupta and Gao 2009), which are expected to be very common patterns if the mergers of the taxa A and C gave rise to the taxon D.
Bacterial lineages that might be intermediates in the monoderm-diderm transition
Although the distinction between the monoderm and diderm prokaryotes is very meaningful, it does not represent a major evolutionary transition, such as that seen between prokaryotes and eukaryotes (Szathmary and Smith 1995; Margulis 1996; Mayr 1998; Gupta 1998b). Unlike the latter transition, where no clear intermediates are found, a number of bacterial groups could represent possible intermediates in the transition from monoderm to diderm bacteria. As noted earlier, some bacterial phyla such as Deinococcus_-Thermus and Thermotogae, although they contain some features of the diderm bacteria, they lack LPS which is considered to be a defining characteristic of the archetypical diderm or Gram-negative bacteria (Sutcliffe 2010). In the case of Deinococcus species although they contain an outer membrane, they also possess a thick peptidoglycan layer (~50 nm) and stain Gram-positive similar to various monoderm bacteria (Murray 1992; Gupta 1998b). This observation indicates that in the transition from monoderm to diderm bacteria the outer membrane likely evolved first and this was followed by a reduction in the thickness of the peptidoglycan layer (Gupta 1998b, 2000). The biochemical, structural and phylogenetic characteristics of Deinococcus_-Thermus taxa indicate that the cell envelope in them may represent an intermediate stage in the development of archetypical diderm cell envelope that is characteristic of the traditional Gram-negative phyla. I will refer to this bacterial group lacking LPS and containing some features of the diderm bacteria as “Simple Diderms” in contrast to the LPS-containing archetypical diderm bacteria. The cell envelopes of Thermotogae species may represent an alternate attempt to develop an outer cell membrane. In addition to the above taxa that contain some features of the diderm-prokaryotes, recent work has revealed that a number of bacterial phyla that are either part of the Firmicutes phylum or branch in its proximity (viz. Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia) also contain an outer membrane and the genomes of these species contain genes encoding for LPS biosynthesis (Mira et al. 2004; Karpathy et al_._ 2007; Herlemann et al_._ 2009; Sutcliffe 2010). Because these bacterial phyla are distantly related to the other phyla of traditional Gram-negative bacteria, the relationships of the outer cell envelopes in these two groups is presently unclear (see below).
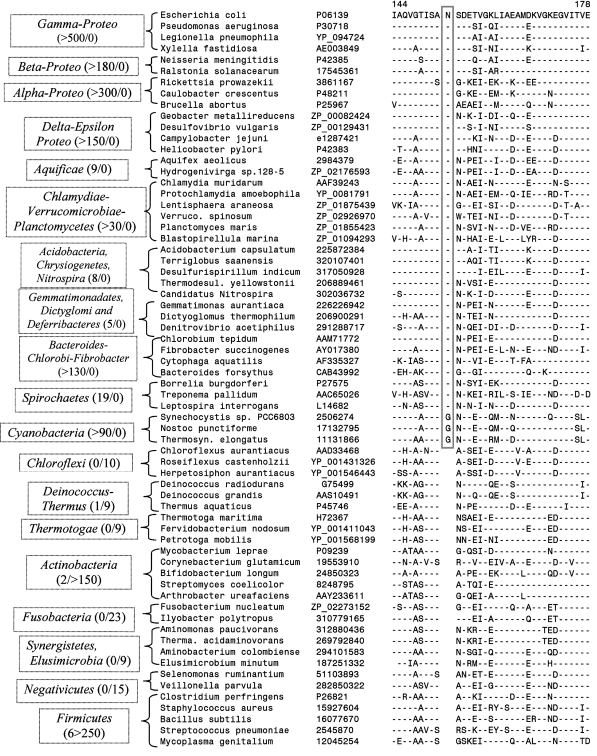
Two conserved inserts that are present in the Hsp70 and Hsp60 proteins provide important insights into the development of outer cell envelopes in bacterial groups. The large insert in the Hsp70 protein that was referred to earlier is a shared characteristic of all bacterial phyla that are traditionally considered to be Gram-negative including the Chloroflexi (likely monoderm, see above) and Deinococcus_-Thermus (Gupta 1998b; Singh and Gupta 2009). However, except for isolated exceptions, this insert is absent from virtually all Actinobacteria, Firmicutes (including Negativicutes), Thermotogae_, Fusobacteria and Synergistetes (as well as Elusimicrobium) (Gupta 1998b; Griffiths and Gupta 2004; Singh and Gupta 2009) (unpublished results). The rare genetic change responsible for this conserved insert was introduced at a very early stage in the evolution of diderm bacteria. This insert provides evidence that the bacterial groups such as Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia that contain an outer cell envelope with LPS are distantly related to the traditional phyla of Gram-negative (diderm) bacteria. Besides the Hsp70 insert, another conserved insert that we have identified in the Hsp60 protein is uniquely present in different phyla of traditional Gram-negative bacteria whose outer cell envelopes contain LPS but it is not found in _Deinococcus_-Thermus, Chloroflexi as well as the above noted phyla of bacteria (Fig. 1). This insert, in addition to further confirming that Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia are distantly related to the traditional Gram-negative bacteria, also provides evidence that the Chloroflexi and _Deinococcus_-Thermus branched prior to all of the phyla of traditional Gram-negative bacteria. Thus, based upon the species distribution patterns of the Hsp70 and Hsp60 inserts, it is possible to infer that the phyla consisting of Chloroflexi and _Deinococcus_-Thermus species branched immediately prior to the clade consisting of different phyla of traditional Gram-negative bacteria (Fig. 2). It should be noted that the conserved insert in the Hsp60 protein is a unique and distinctive property of different species from various phyla of traditional LPS-containing Gram-negative bacteria and this insert provides a reliable molecular marker to identify and circumscribe this clade in molecular terms (Fig. 2). Although the cellular function of this conserved insert is not known, our recent work shows that it is essential for the group of species where it is found as deletion of this insert or any significant changes in it leads to the failure of cell growth (Singh and Gupta 2009).
Fig. 1.
Partial sequence alignment of the Hsp60 protein showing a 1 aa insert (boxed) in a conserved region that is mainly specific for different bacterial phyla corresponding to traditional Gram-negative bacteria that have an outer cell membrane containing lipopolysaccharide. The presence or absence of this insert in all available sequences from different bacterial groups is indicated along with their names. For example, for Gamma-proteobacteria >500 hits corresponding to Hsp60 were observed and all of them contained this insert (i.e. >500 with insert, 0 without insert). Similarly, for the Actinobacteria phylum, >150 hits were observed and of these only 2 contained the insert (2/>150). Only representative sequences from different bacterial phyla are shown here. The absence of this insert in the Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia distinguishes these atypical diderm taxa from all of the phyla of traditional Gram-negative bacteria that contain this insert. The dashes in the alignment indicate that the same amino acid as that found on the top line (i.e. E. coli protein) is present in that position. The accession numbers of sequences are given in the second column. The numbers on the top indicate the position of this sequence in E. coli protein
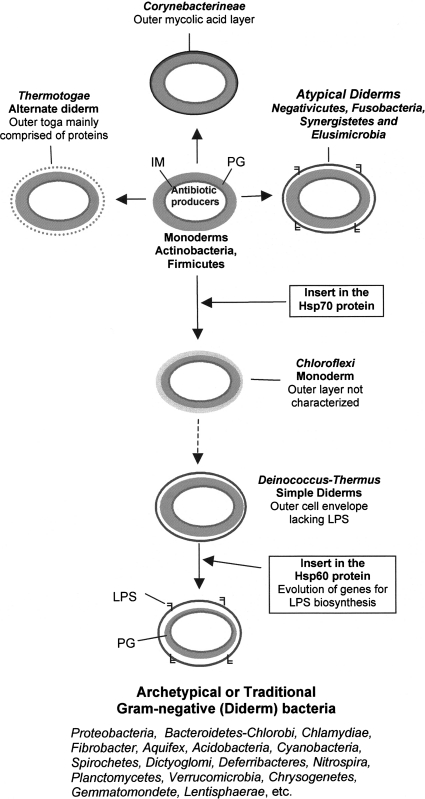
Fig. 2.
A cartoon showing the development of outer cell envelopes in various bacterial lineages in response to antibiotic selection pressure (Gupta 2000). The outer cell envelope in Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia (atypical diderm taxa) is distinguished from traditional diderm Gram-negative bacteria by the absence of the Hsp60 insert. The cell membrane from atypical and traditional Gram-negative bacteria are postulated to show significant differences in their biochemical and functional characteristics. The outer cell envelopes of the archetypical Gram-negative phyla are indicated to have evolved from the Chloroflexi and Deinococcus_-Thermus groups of species. Information regarding species distribution of Hsp70 inserts for most bacterial phyla is provided in earlier work (Griffiths and Gupta 2004; Lake et al._ 2007; Singh and Gupta 2009). Abbreviations: PG peptidoglycan, IM inner membrane, LPS lipopolysaccharides
The bacterial groups consisting of Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia that are also indicated to have an outer membrane with LPS are distinguished from the traditional phyla of Gram-negative bacteria by the absence of the insert in the Hsp60 protein (Fig. 1). It has been reported that Synergistetes species, although they contain an outer membrane, lack the genes for the TolAQR-Pal complex that is required for assembly and maintenance of outer membranes (Hugenholtz et al. 2009). Hence, the nature and the role of the outer membrane in these species could be different from the traditional phyla of Gram-negative bacteria defined by the presence of the Hsp60 insert. Hence, I will refer to these taxa of bacteria as “Atypical diderms” to distinguish them from “Traditional or archetypal” Gram-negative diderm bacteria.
Antibiotic selection pressure as a driving force for the evolution of diderm bacteria
The question can be asked what selective forces were responsible for the evolution of diderm bacteria from monoderm bacteria. Lake speculates that the acquisition of photosynthetic ability from Clostridia may have been important in this regard (Lake 2009). However, photosynthetic ability within the Clostridia (phylum Firmicutes) is only found within a single family Heliobacteriaceae that contains a total of 7 species (Madigan 1992). Of these, the genome of Heliobacterium modesticaldum has been sequenced and the different genes/proteins from it show no specific affiliation to the diderm bacteria (Sattley et al_._ 2008). Additionally, photosynthetic ability within diderm bacteria is found in only 4 of the more than 20 phyla (Blankenship and Hartman 1998; Gupta 2003), which argues against it being the main selective force for the development of outer membrane. In contrast to Lake’s proposal, I have suggested that the outer membrane of diderm bacteria has evolved as a defense mechanism in response to the evolution of antibiotic selection pressures (Gupta 1998b, 2000). The main arguments in support of this view are as follow: (i) The monoderm bacteria, which include Streptomyces, are the main producers of most of the known antibiotics (Davies 1994; Wright 2007); (ii) The production of antibiotics by some organisms gives them tremendous selective advantage over non-producing or antibiotic-sensitive bacteria (Cavalier-Smith 1992; Davies 1994); (iii) Resistance to antibiotics can develop by a variety of mechanisms including: changes in their target genes; inactivation of antibiotics by different enzymes; reducing antibiotic entry into cells by different mechanisms; and expulsion of the antibiotics by drug efflux pumps. (Nikaido 1989; Davies 1994; Spratt 1994; Wright 2007); (iv) Gram-positive bacteria in general display higher sensitivity to antibiotic than Gram-negative bacteria (Nikaido 1989; Spratt 1994).
Based upon these observations, it is easy to conjecture that early in the evolutionary history of microbes when one group of Gram-positive bacteria (viz. Streptomyces) developed mechanisms to produce antibiotics, survival of most of the other bacteria that were sensitive to these antibiotics was at stake. To survive in this strongly selective environment, sensitive bacteria evolved a number of strategies to protect themselves from the cytotoxic effects of these antibiotics (see Fig. 2) (Spratt 1994; Gupta 2000). One of these strategies that was likely employed by Archaea was to mutationally change the target sites of different antibiotics, as various processes that are normally inhibited by antibiotics such as protein synthesis, RNA synthesis and cell wall biosynthesis are resistant to their effects in Archaea (Gupta 1998b, 1998c, 2000). The emergence of Archaea from Gram-positive bacteria in response to antibiotic selection pressure is also supported by a recent detailed study by Valas and Bourne (2011). Another important strategy to escape from the effects of antibiotics was to develop an outer protective layer (membrane) that would retard the entry of antibiotics into the cells (Nikaido 1989; Gupta 2000). In Gram-negative bacteria, many enzymes involved in the inactivation of antibiotics are localized in the periplasmic compartment (or intermembrane space), which further aids in antibiotic resistance (Nikaido 1989, 2003; Davies 1994; Spratt 1994). This strategy was independently employed by a number of bacterial groups leading to development of outer envelopes of differing biochemical properties (Fig. 2). Thus, the layered outer cell envelopes of the Chloroflexi and the diverse diderm cell envelopes of the Corynebacterineae, Thermotogae, _Deinococcus_-Thermus, Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia could represent various attempts of developing an outer protective barrier (Fig. 2). Moreover, the outer cell envelopes in some of these lineages (viz. _Deinococcus_-Thermus and Chloroflexi; Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia) could also be related or derived from each other. The absence of the Hsp70 insert in Thermotogae, Corynebacterineae, Negativicutes, Synergistetes, Fusobacteria and Elusimicrobia indicates that the outer cell membranes or layers in these taxa represent earlier stages (or alternative attempts) to develop a protective barrier in comparison to the outer envelopes of Chloroflexi and _Deinococcus_-Thermus, which have the Hsp70 insert. Of these different evolutionary experiments to develop an outer cell envelope, the structural characteristics of the outer cell envelope in _Deinococcus_-Thermus were apparently most successful and this lineage led to the eventual development of the archetypal diderm membranes that are found in different phyla of traditional Gram-negative bacteria (Fig. 2).
As the _Deinococcus_-Thermus species lack the genes for LPS biosynthesis, it can be hypothesised that the subsequent evolution of these genes in either some species from this group or a closely related bacterium led to the development of an archetypical LPS-containing outer cell envelope characteristic of various traditional Gram-negative phyla. This development and other changes that accompanied the evolution of this new diderm-LPS cell were apparently evolutionarily highly successful as it led to the emergence of much of the microbial diversity (i.e. majority of the bacterial phyla) that is seen today (Ludwig and Klenk 2005; Sutcliffe 2010). It is important to note that the evolution of this archetypical LPS-containing diderm cell envelope, whose presence shows excellent correlation with the presence of the insert in the Hsp60 protein (see Fig. 1 and Table 1 in Sutcliffe 2010), was an important and apparently irreversible evolutionary development, as none of the species from this clade defined by the Hsp60 insert have lost the outer membrane. The fact that the outer cell membrane has not been lost from any of the >1000 of species that are part of the archetypical diderm clade encompassing the majority of bacterial phyla (Fig. 1), also argues strongly against the origin of monoderm prokaryotes from diderm bacteria by the loss of outer membrane and the hypothesis that the cells with two membranes evolved prior to those with one membrane (Cavalier-Smith 2006; Griffiths 2007; Valas and Bourne 2009).
The possible relationship of the taxa consisting of atypical diderms (viz. Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia) to the traditional LPS-diderm Gram-negative bacteria is presently unclear. It is quite likely that all of these atypical diderm taxa that show close affiliation to the Firmicutes are related to each other and therefore the diderm-LPS characteristics exhibited by them has a common origin. Although the presence of a diderm-LPS phenotype in these two groups (i.e. atypical diderm and traditional diderms) can be explained by lateral transfer of various genes that are involved in the formation of outer cell membrane as well LPS biosynthesis between these groups, it is also possible that the outer membranes in these two groups have evolved independently and that the cell membrane organization and function in these two groups of prokaryotes may differ from each other in important aspects. Hence, further comparative studies on the biochemical and functional characteristics of the outer membrane characteristics from these two groups of bacteria should be of much interest.
In conclusion, the data presented here represent a significant criticism of the recently proposed ‘prokaryotic endosymbiosis’ hypothesis (Lake 2009). During the preparation of this Perspective, other criticisms of this hypothesis based on other grounds also appeared (Swithers et al. 2011). Alternative mechanisms for the evolution of outer membranes therefore need to be proposed and, as hypothesized here, it is plausible that antibiotic selection pressure was one of the main drivers in this important step in bacterial evolution.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by a research grant from the Natural Science and Engineering Research Council of Canada. I thank Dr. Iain Sutcliffe for many helpful discussions and comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Baena S, Fardeau ML, Labat M, et al. Aminobacterium colombiensegen. nov. sp. nov., an amino acid-degrading anaerobe isolated from anaerobic sludge. Anaerobe. 1998;4:241–250. doi: 10.1006/anae.1998.0170. [DOI] [PubMed] [Google Scholar]
- Blankenship RE, Hartman H. The origin and evolution of oxygenic photosynthesis. Trends Biochem Sci. 1998;23:94–97. doi: 10.1016/S0968-0004(98)01186-4. [DOI] [PubMed] [Google Scholar]
- Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origins of secondary metabolism. Ciba Found Symp. 1992;171:64–80. doi: 10.1002/9780470514344.ch5. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 2002;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Rooting the tree of life by transition analyses. Biol Direct. 2006;1:19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- Gao B, Gupta RS. Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int J Syst Evol Microbiol. 2005;55:2401–2412. doi: 10.1099/ijs.0.63785-0. [DOI] [PubMed] [Google Scholar]
- Gao B, Gupta RS. Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics. 2007;8:86. doi: 10.1186/1471-2164-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Parmanathan R, Gupta RS. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie van Leeuwenhoek. 2006;90:69–91. doi: 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Gram C. Ueber die isolierte farbung der Schizomyceten in Schnitt und Trockenpraparaten. Fortschr Med. 1884;2:185–189. [Google Scholar]
- Gray MW. Evolution of organellar genomes. Curr Opin Genet Dev. 1999;9:678–687. doi: 10.1016/S0959-437X(99)00030-1. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Cell evolution and the problem of membrane topology. Nat Rev Mol Cell Biol. 2007;8:1018–1024. doi: 10.1038/nrm2287. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. Signature sequences in diverse proteins provide evidence for the late divergence of the order Aquificales. Int Microbiol. 2004;7:41–52. [PubMed] [Google Scholar]
- Gupta RS. Life’s third domain (Archaea): an established fact or an endangered paradigm? A new proposal for classification of organisms based on protein sequences and cell structure. Theor Popul Biol. 1998;54:91–104. doi: 10.1006/tpbi.1998.1376. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. What are archaebacteria: Life’s third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Mol Microbiol. 1998;29:695–708. doi: 10.1046/j.1365-2958.1998.00978.x. [DOI] [PubMed] [Google Scholar]
- Gupta RS. The natural evolutionary relationships among prokaryotes. Crit Rev Microbiol. 2000;26:111–131. doi: 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Evolutionary relationships among photosynthetic bacteria. Photosynth Res. 2003;76:173–183. doi: 10.1023/A:1024999314839. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Gao B. Phylogenomic analyses of clostridia and identification of novel protein signatures that are specific to the genus Clostridium sensu stricto (cluster I) Int J Syst Evol Microbiol. 2009;59:285–294. doi: 10.1099/ijs.0.001792-0. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Golding GB. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Mathews DW. Signature proteins for the major clades of cyanobacteria. BMC Evol Biol. 2010;10:24. doi: 10.1186/1471-2148-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Shami A. Molecular signatures for the Crenarchaeota and the Thaumarchaeota. Antonie van Leeuwenhoek. 2011;99:133–157. doi: 10.1007/s10482-010-9488-3. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Singh B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol. 1994;4:1104–1114. doi: 10.1016/S0960-9822(00)00249-9. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Pereira M, Chandrasekera C, Johari V. Molecular signatures in protein sequences that are characteristic of Cyanobacteria and plastid homologues. Int J Syst Evol Microbiol. 2003;53:1833–1842. doi: 10.1099/ijs.0.02720-0. [DOI] [PubMed] [Google Scholar]
- Hanada S, Pierson BK. The family Chloroflexiae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The prokaryotes: a handbook on the biology of bacteria. New York: Springer; 2006. pp. 815–842. [Google Scholar]
- Herlemann DP, Geissinger O, Ikeda-Ohtsubo W, et al. Genomic analysis of “Elusimicrobium minutum,” the first cultivated representative of the phylum “Elusimicrobia” (formerly termite group 1) Appl Environ Microbiol. 2009;75:2841–2849. doi: 10.1128/AEM.02698-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Hooper SD, Kyrpides NC. Focus: synergistetes. Environ Microbiol. 2009;11:1327–1329. doi: 10.1111/j.1462-2920.2009.01949.x. [DOI] [PubMed] [Google Scholar]
- Jumas-Bilak E, Roudiere L, Marchandin H. Description of ‘Synergistetes’ phyl. nov. and emended description of the phylum ‘Deferribacteres’ and of the family Syntrophomonadaceae, phylum ‘Firmicutes’. Int J Syst Evol Microbiol. 2009;59:1028–1035. doi: 10.1099/ijs.0.006718-0. [DOI] [PubMed] [Google Scholar]
- Karpathy SE, Qin X, Gioia J, et al. Genome sequence of Fusobacterium nucleatum subspecies polymorphum—a genetically tractable fusobacterium. PLoS One. 2007;2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. Were Gram-positive rods the first bacteria? Trends Microbiol. 2003;11:166–170. doi: 10.1016/S0966-842X(03)00063-5. [DOI] [PubMed] [Google Scholar]
- Lake JA. Evidence for an early prokaryotic endosymbiosis. Nature. 2009;460:967–971. doi: 10.1038/nature08183. [DOI] [PubMed] [Google Scholar]
- Lake JA, Herbold CW, Rivera MC, Servin JA, Skophammer RG. Rooting the tree of life using nonubiquitous genes. Mol Biol Evol. 2007;24:130–136. doi: 10.1093/molbev/msl140. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Klenk H-P. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s manual of systematic bacteriology. Berlin: Springer; 2005. pp. 49–65. [Google Scholar]
- Madigan MT. The family Heliobacteriaceae. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, editors. The prokaryotes. New York: Springer; 1992. pp. 1981–1992. [Google Scholar]
- Marchandin H, Teyssier C, Campos J, et al. Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. 2010;60:1271–1279. doi: 10.1099/ijs.0.013102-0. [DOI] [PubMed] [Google Scholar]
- Margulis L. Symbiosis in cell evolution. New York: W.H. Freeman and Company; 1993. [Google Scholar]
- Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci USA. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Two empires or three? Proc Natl Acad Sci USA. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Pushker R, Legault BA, Moreira D, Rodriguez-Valera F. Evolutionary relationships of Fusobacterium nucleatum based on phylogenetic analysis and comparative genomics. BMC Evol Biol. 2004;4:50. doi: 10.1186/1471-2148-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Koonin EV, Makarova KS, et al. The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci USA. 2006;103:13126–13131. doi: 10.1073/pnas.0605709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RGE. The higher taxa, or, a place for everything? In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Bergey’s manual of systematic bacteriology. Baltimore: The Williams and Wilkins; 1986. pp. 31–34. [Google Scholar]
- Murray RGE. The family Deinococcaceae. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, editors. The prokaryotes. New York: Springer; 1992. pp. 3732–3744. [Google Scholar]
- Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR. Archaeal genomics: an overview. Cell. 1997;89:991–994. doi: 10.1016/S0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- Reysenbach A-L. Phylum BII. Thermotogae phy. nov. In: Boone DR, Castenholz RW, editors. Bergey’s manual of systematic bacteriology. Berlin: Springer; 2001. pp. 369–387. [Google Scholar]
- Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- Sattley WM, Madigan MT, Swingley WD, et al. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J Bacteriol. 2008;190:4687–4696. doi: 10.1128/JB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Gupta RS. Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol Genet Genomics. 2009;281:361–373. doi: 10.1007/s00438-008-0417-3. [DOI] [PubMed] [Google Scholar]
- Spratt BG. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- Stanier RY, Adelberg EA, Ingraham JL. The microbial world. New Jersey: Prentice-Hall Inc.; 1976. [Google Scholar]
- Sutcliffe IC. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010;18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC. Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ Microbiol. 2011;13:279–282. doi: 10.1111/j.1462-2920.2010.02339.x. [DOI] [PubMed] [Google Scholar]
- Swithers KS, Fournier GP, Gogarten JP, Lapierre P (2011) Reassessment of the lineage fusion hypothesis for the origin of double membrane bacteria. Nature precedings. http://precedings.nature.com/documents/5812/version/1 [DOI] [PMC free article] [PubMed]
- Szathmary E, Smith JM. The major evolutionary transitions. Nature. 1995;374:227–232. doi: 10.1038/374227a0. [DOI] [PubMed] [Google Scholar]
- Truper HG, Schleifer KH. Prokaryote characterization and identification. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, editors. The prokaryotes. New York: Springer; 1992. pp. 126–148. [Google Scholar]
- Valas RE, Bourne PE. Structural analysis of polarizing indels: an emerging consensus on the root of the tree of life. Biol Direct. 2009;4:30. doi: 10.1186/1745-6150-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valas RE, Bourne PE. The origin of a derived superkingdom: how a gram-positive bacterium crossed the desert to become an archaeon. Biol Direct. 2011;6:16. doi: 10.1186/1745-6150-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DA, Doolittle WF. The real ‘domains’ of life. Curr Biol. 2005;15:R237–R240. doi: 10.1016/j.cub.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Wiegel J, Tanner R, Rainey FA. An introduction to the family Clostridiaceae. In: Dworkin M, Falkow S, Schleifer KH, Stackebrandt E, editors. The prokaryotes. New York: Springer; 2006. pp. 654–678. [Google Scholar]
- Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.