Exploiting polypharmacology for drug target deconvolution (original) (raw)
Significance
Protein kinase inhibitors represent a major class of anticancer drugs, which are notoriously unspecific. Efforts to exploit the polypharmacology of inhibitors for target deconvolution have met with little success. Our significant contribution is to apply regularized regression to kinase expression and kinase profiling on a large set of inhibitors. By selecting a set of optimally designed kinase inhibitors that span a broad range of kinase specificities, we identified relevant kinases from our model in six cell lines; we then empirically validated a set of specific kinases that regulate cancer cell migration. Using this model, we predict a cell type-specific response to previously untested inhibitors. Broadly, these approaches should prove useful in identifying novel targets and in rational cancer therapy.
Keywords: systems pharmacology, regularized regression, perturbation biology, predictive modeling, cancer cell migration
Abstract
Polypharmacology (action of drugs against multiple targets) represents a tempting avenue for new drug development; unfortunately, methods capable of exploiting the known polypharmacology of drugs for target deconvolution are lacking. Here, we present an ensemble approach using elastic net regularization combined with mRNA expression profiling and previously characterized data on a large set of kinase inhibitors to identify kinases that are important for epithelial and mesenchymal cell migration. By profiling a selected optimal set of 32 kinase inhibitors in a panel against six cell lines, we identified cell type-specific kinases that regulate cell migration. Our discovery of several informative kinases with a previously uncharacterized role in cell migration (such as Mst and Taok family of MAPK kinases in mesenchymal cells) may represent novel targets that warrant further investigation. Target deconvolution using our ensemble approach has the potential to aid in the rational design of more potent but less toxic drug combinations.
For most diseases, the development of specific “one target, one drug” or euphemistically, “magic bullet” therapy, has been difficult to achieve (1). It is even rather difficult to chemically achieve single target specificity. Furthermore, it is now evident that many of the most effective drugs in therapeutic areas as diverse as oncology (such as Gleevec), psychiatry (such as serotonin reuptake inhibitors), and inflammation (such as aspirin) act on multiple rather than single targets—a phenomenon known as polypharmacology (2, 3). Although the pharmaceutical industry and the US Food and Drug Administration (FDA) has for years focused on single targets, it may turn out to be true that hitting multiple targets is preferable, an emerging idea referred to as network pharmacology.
Designing drugs with a specific multitarget profile or designing a rational combination of such drugs is both complex and difficult, but could serve to improve the balance between efficacy and safety compared with single targets agents. Therefore, despite the complexity of designing such drugs, there is an incentive to develop new systems-based methods capable of exploiting the known polypharmacology of drugs to identify the molecular targets of active hits, also called “target deconvolution.” Such methods are not only important for elucidating mechanisms of action but also for identifying effective pathways involved in disease as a preliminary step in rational design of drugs for new targets. Furthermore, if target-specific toxicity and off-target effects could be addressed early in the drug discovery pipeline, the high attrition rate in drug development might be reduced (4).
Recent advances in high-throughput “omics” technologies have led to the development of methods to efficiently and reliably profile drug target selectivities both in vitro and in the cellular environment. One such well-characterized set of drugs is the class of small molecule kinase inhibitors that are widely used to identify cellular signaling pathways and are promising therapeutic agents. To date, several groups have profiled hundreds of known kinase inhibitors against sizable fractions of the 518 human protein kinases (5, 6). The resulting kinase-inhibitor interaction maps have revealed an unexpected number of interactions with off-target kinases, even for well-characterized kinase inhibitors that were thought to be specific (5). This complexity has made it difficult to be certain whether a given pathway is in fact involved in a specific process based on the assumption that an inhibitor is specific for a target kinase. Although this presents a serious obstacle, it still may be possible to use that information in conjunction with advanced computational methods to identify the relevant kinase target or targets. Furthermore, such an approach might also be reversed to achieve improved specificity by designing mixtures of kinase inhibitors.
Our initial goal was to identify the important kinases involved in a specific cellular phenotype from the spectrum of action of unspecific inhibitors on that phenotype. As a first step toward that goal, we demonstrate that, for some classes of drugs and target molecules, analysis of unspecific drug action can be broken down into three steps: first, an in vitro characterization of a complete “drug to target” specificity map for a set of unspecific inhibitors; second, an in vivo phenotypic assay of a subset of these drugs; and third, construction of an optimized model that will identify the primary kinase targets and should also have the bonus of predicting the effects of untested drugs on that phenotype (Figs. 1 and 2_A_). To exemplify our approach, we examined the effect of known kinase inhibitors on cell migration in several cancer cell lines. We apply a well-established variable selection method, called “elastic net regularization” combined with mRNA expression-based profiling of kinases previously executed on a large set of kinase inhibitors to identify kinases (deconvolve) that are important for epithelial and mesenchymal cell migration. Our approach is based on the property that kinase inhibitors have broad specificity, but the spectrum of targets is different for each one. Profiling a set of 32 optimally designed kinase inhibitors against six cell lines, we identified cell type-specific kinases that regulate cell migration. Using gene depletion, we validated a role for a subset of cell type-specific kinases in mesenchymal cancer cell migration. Further, using the same regularized regression model (dubbed “kinome regularization”), we showed that we can also accurately predict a cell type-specific response to previously untested kinase inhibitors. Factoring the analysis this way enables the tradeoff between resources put toward the core drug characterization vs. resources that could be expanded to additional assaying cell lines with new phenotypes.
Fig. 1.

Step by step method: exploiting polypharmacology for drug target deconvolution.
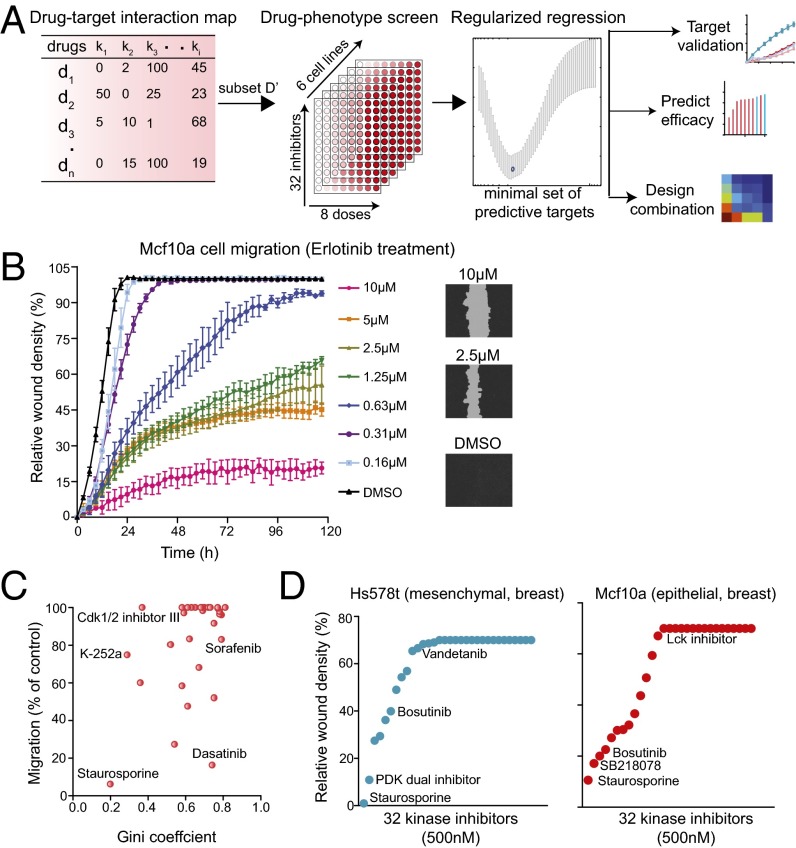
Fig. 2.
An ensemble approach of exploiting polypharmacology for kinase drug target deconvolution. (A) A schematic showing our approach of using a combination of drug-target interaction map, drug phenotypic screen, and regularized regression to exploit the polypharmacology of drugs to identify drug targets, predict efficacy, and rationally design combination therapy. (B) A real-time quantitative phenotypic method to measure cell migration using the scratch wound assay. A plot of relative wound density of Mcf10a cells treated with varying doses of Erlotinib. (Right) Representative images of cells with wound area are also shown. (C) A plot of Gini score of all 32 kinase inhibitors and their effect on relative migration in FOCUS cells. (D) Plots showing the effect of 32 kinase inhibitors on cell migration in Hs578t (mesenchymal) and Mcf10a (epithelial) breast cell lines.
Results
Real-Time Quantitative Measurement of Cell Migration for Phenotypic Screening.
An example of a complex phenotype, for which we might want to identify the underlying molecular pathways, is cell migration. Cell migration is a complicated process involving signaling molecules both in the membrane and in the cytosol and cytoskeletal filaments regulated directly and indirectly by these signaling molecules. It is a hallmark of embryonic development, wound repair, and pathological conditions such as cancer invasion and metastasis (7). Several kinase-driven signaling pathways are known to affect cell migration in a cell type-specific manner, including PI3K, MAPK, focal adhesion kinase, and mammalian target of rapamycin pathways (7). Methods such as Boyden chamber and wound healing assays are well recognized to assess migration of cells in a tissue culture system (8, 9). We optimized a label-free, real-time, high-throughput method of monitoring wound closure using a time lapse imaging system. In this assay, wound closure was monitored in cells plated on 96-well plates treated with varying doses of kinase inhibitors (Fig. 2_B_). An advantage of this approach is that the real-time monitoring allows identification of an optimal time point for dose–response curves and accurate determination of drug-specific EC50 (50% reduction in cell migration compared with DMSO control). For instance, a linear range of wound closure of untreated nontransformed mammary epithelial cells, Mcf10a, had to be measured at a point less than 20 h after wounding the monolayer (Fig. 2_B_). Thus, a 12-h time point (which showed 70% of maximum wound closure) was chosen for dose–response curves to accurately determine the EC50 for various kinase inhibitors. Using this criterion, the EC50 of an epidermal growth factor receptor (EGFR) inhibitor (erlotinib) on migration of Mcf10a cells at 12 h was determined to be 250 nM (a sixfold difference compared with the EC50 measured at the traditional 72-h end point, 1.6 µM, which is distorted by the compression of data at a low dose).
Selection of an Optimal Kinase Inhibitor Set.
The problem of identifying the optimal set of kinase inhibitors can be viewed as a dimensionality reduction problem. We would like to identify a smaller optimal subset of inhibitors that will be almost as informative about the kinases as the complete set. The intrinsic dimensionality of the kinase-to-inhibitor map can be measured by the number of principal components in the principal component analysis (PCA). For the interaction map, the cumulative percent of variance explained by the first few principal components turns out to be relatively low, which suggests that kinase data are rich and cannot be explained by any set of a small number of dimensions (drugs). In particular, we observed that 26 principal components capture 80% of the variance in the data (Fig. S1 and Table S1). I.e., we can replace 178 dimensions (drugs) with 26 at a relatively small (20%) loss of information about the kinase targets. Once the number of informative variables m is decided on, we selected the m principal variables, chosen so that they preserve most of the variation in the complete dataset. To do this, we used the well-established forward selection procedure termed B4 (10), which associates and retains variables with the highest absolute value in the top m principal components. Table S1 shows the list of top 26 inhibitors selected by the B4 principle variable procedure; 16 of these inhibitors (labeled in bold font) were used in our experiments. An additional 16 reasonably selective inhibitors [Gini coefficient (11) > 0.5 that scores relative selectivity from 0 (nonselective) to 1 (highly selective)] were also chosen, representing what we consider to be a sound set of 32 kinase inhibitors for phenotypic profiling.
Optimally Designed Kinase Inhibitor Screen That Measures Cell Migration as an Aggregate Phenotype.
We treated a panel of six cell lines spanning three different cancer types with a set of 32 optimally designed small molecule kinase inhibitors that collectively target a wide variety of protein kinases (Table S2). Each drug was examined at several different concentrations, and its effect on cell migration was then scored using a quantitative real-time wound closure assay. We used a previously characterized kinase inhibitor-activity interaction matrix to assess the in vitro activity of kinase inhibitors that profiled 300 kinases, including those targeting serine, threonine, and tyrosine (5). This collection of kinase inhibitors spanned kinases with profiles exhibiting very broad selectivity (e.g., staurosporine, which inhibited 82% of all kinases tested at 500 nM) to profiles indicating high selectivity (e.g., lapatinib, which showed measurable inhibition of only 1% of all kinases tested; Fig. S1).
In an ideal world of pharmacology, there would be one completely specific inhibitor for each kinase, and in addition, there might be broader-based inhibitors whose targets represented proper subsets of proteins related by sequence or some other property. The real world is far from that. Most kinase inhibitors affect multiple targets often from diverse subfamilies; often a single drug will hit kinases in very different structural subclasses, making it necessary to deconvolve inhibition data empirically by the polypharmacology of the compounds. However, polypharmacology can be measured directly in vitro by probing recombinant kinases with a drug at a range of concentrations to generate a kinome profile (5) and a Gini coefficient. The Gini coefficient of inhibitors in our screen varied from 0.2 (staurosporine) to 0.81 (masitinib) (Fig. 2_C_). The correlation between the Gini score and inhibition of cell migration in cell lines was low [_R_2 ∼ 0.20 in Friendship of China and US (FOCUS) cells derived from a patient with primary hepatocellular carcinoma], suggesting that only one or a small number of kinases was involved (Fig. 2_C_). Overall, our set of 32 kinase inhibitors, when assayed at a concentration range of 1 nM to 10 µM, which was at least 20 times above the IC50 in vitro, was spread across a wide range of effectiveness in cell migration in all epithelial or mesenchymal cell lines tested (Fig. 2_D_). Several inhibitors had either low or no effect on cell migration (migration efficiencies of >90% of control), whereas some inhibited cell migration very potently (<20% of control) (Fig. 2_D_). Several inhibitors had a moderate to weak effect on cell migration (30–70% of control) (Fig. 2_D_). It was certainly not the case that the most potent inhibitors were most specific, as was clear from Fig. 2_C_, and this therefore made it more difficult to infer which kinases were the most responsible for inhibiting cell migration.
Deconvolution of the Polypharmacology.
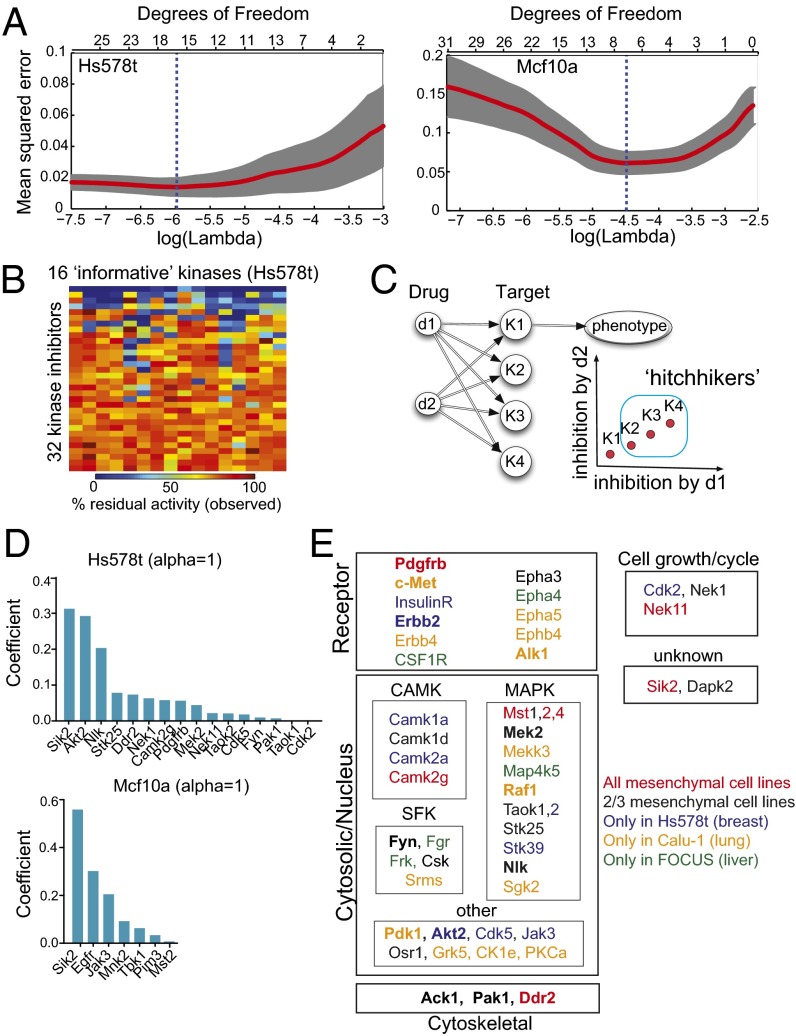
To identify a set of kinases that affect cell migration, we used elastic net regularization, which is a technique that regresses a single target variable (quantified cell migration) against a set of predictor variables (the activities of individual kinases) while imposing a penalty on the number of variables that effectively eliminates variables with insignificant contributions. Formally, we modeled the phenotype y as a linear function of kinase activity X, y = _β_0 + Xβ, which is in turn defined by the interaction map M between drugs and kinases. The variable selection step determines which kinases (not which kinase inhibitors) have the greatest explanatory power for the phenotype. We used a standard “leave-one-out cross validation” (LOOCV) to identify a set of informative kinases at the absolute minimum of the least-mean-square error (Fig. 3_A_). The profiles shown in Fig. 3_A_ present two typical optimization scenarios. Degrees of freedom correspond to the number of informative kinases used in regression. As kinases are removed on the left (Hs578t, breast ductal carcinoma), the fitness is roughly flat, which means that extra variables neither helped nor hindered the accuracy of the model, as one would expect from a random variable being factored into a model. Once removing more variables hurts the accuracy, a good list of 16 predictors is found. On the right (Mcf10a), removing variables significantly improves the accuracy at first, indicating that for some kinases the inhibition level works as a proxy identifier for a drug (a variable that leads to overfitting). There is a clearly defined optimal point that gives a set of seven informative kinases. Interestingly, every informative kinase in this set of 16 kinases (in Hs578t) was broadly affected by all 32 inhibitors tested (Fig. 3_B_). Note that our selection procedure eliminates those inhibitors that are collinear in the space of targets. If there were two inhibitors that affected all targets similarly (up to a constant factor), it would be impossible to distinguish which of the affected targets is actually responsible for the phenotype. As illustrated by Fig. 3_C_ where two inhibitors d1 and d2 affect four targets K1–K4 proportionally, if K1 was causally related to the phenotype, it could still appear that the other three kinases would affect the phenotype, because every time K1 is affected, K2–K4 would be affected proportionally. Such false positives would be eliminated by experimental validation. Although our method drastically narrows down the list of candidate kinases from 300 to <30 for each of the six cell lines tested (two are shown in Fig. 3_D_), the reduced set could still contain kinases that are not even expressed in a given cell line, including kinases that are in a collinear group of kinases (Fig. 3_C_). Using mRNA expression profiling of the informative kinase set, we could purge unexpressed kinases and rerun the regression on a pruned set. In this way, we were able to eliminate 12 kinases from consideration in the mesenchymal cell lines (Tables S3 and S4). Overall, we identified a minimal informative set (<10% of all kinases) in each of the epithelial and mesenchymal cell lines and rank ordered these by importance in the cell migration phenotype (Fig. 3_D_ and Table S3).
Fig. 3.
Identification of informative kinases in cell migration using elastic net regularization. (A) Plots show LOOCV error using elastic net regularization fit in HS578t and Mcf10a cell lines. The error bars represent cross-validation error plus 1 SD. The kinases identified at absolute minima (blue dashed line) were termed the most informative kinases. (B) The observed kinase activities of all informative kinases identified using elastic net regularization were affected by the panel of 32 kinase inhibitors used in our screen. A heatmap showing in vitro residual kinase activities of the 16 most informative kinases identified in Hs578t cells against 32 kinase inhibitors tested. (C) An illustration showing if two inhibitors d1 and d2 are affecting four targets K1–K4 proportionally, it would be impossible to distinguish which of the affected targets is actually responsible for the phenotype. (D) Bar graphs showing the nonzero elastic net coefficients associated with the most informative kinases (determined at α = 1) identified in Hs578t and Mcf10a cells are shown. (E) Subcellular and functional annotation of informative kinases identified in all three mesenchymal cancer cells (union of 0.7 ≤ α ≤ 1.0). Kinases with known role in cell migration are listed in bold font.
Kinases Specific to Cell Type.
Having identified a set of specific informative kinases that best predict the phenotype of cell migration, we next asked if these kinases could provide new insight into the functional subclasses or signaling pathways that may play role in cell motility. When we compared the reduced set of informative kinases across the three mesenchymal cell lines, there was appreciable overlap (42%, 21/50) in at least two of the cell lines (Table S3 and Fig. 3_E_). Seven kinases were found in all three mesenchymal cell lines (Camk2g, Ddr2, Mst2, Mst4, Nek11, PDGFRb, and Sik2), and five kinases were found in two epithelial cancer cell lines (Cdk2, Pak1, Mst2, Tbk1, and Sik2). Among the cancer cells lines, three kinases (Ack1, Mst2, and Sik2) were common to both liver cell lines: FOCUS (mesenchymal) and Huh7 (well differentiated hepatocyte derived epithelial cell line), whereas six kinases (Erbb2, Cdk2, Sik2, Jak3, Pak1, and Mst2) were common to both breast cell lines Hs578t (mesenchymal) and Mcf10a (epithelial) (Table S3). Interestingly, five kinases (Camk2g, Ddr2, Pdgfrb, Mst4, and Nek11) were found to be informative in all mesenchymal cell lines and not in any of the two epithelial cell lines. Further, Tbk1 was found to be informative in both epithelial cell lines and not in any of the three mesenchymal cell lines (Table S3 and Fig. 3_E_). Together, these data highlight roles of cell type- specific kinases in cell migration.
Subcellular and functional annotations of these informative kinases revealed cell type-specific and functionally distinct classes of kinases that are important for mesenchymal cell migration (Fig. 3_E_). Among transmembrane kinases, receptor tyrosine kinases (RTKs) such as PDGFRb, c-Met, Erbb2/4, ephrin, and insulin receptors (IRs) were identified as the most informative. Alk1 was identified only in Calu-1 (non–small-cell lung cancer cell line). Among cytosolic kinases, three major subfamilies of kinases [CAMK, Src family kinases (SFK), and MAPK family) emerged. Finally, two cytoskeletal kinases were also identified (Ack1 and Pak1), along with DDR2, which was previously shown to localize in focal adhesions (12). A set of kinases previously known for important roles in cell growth or cell cycle was also identified to affect cell migration (Cdk2, Nek1, and Nek11) (13–15).
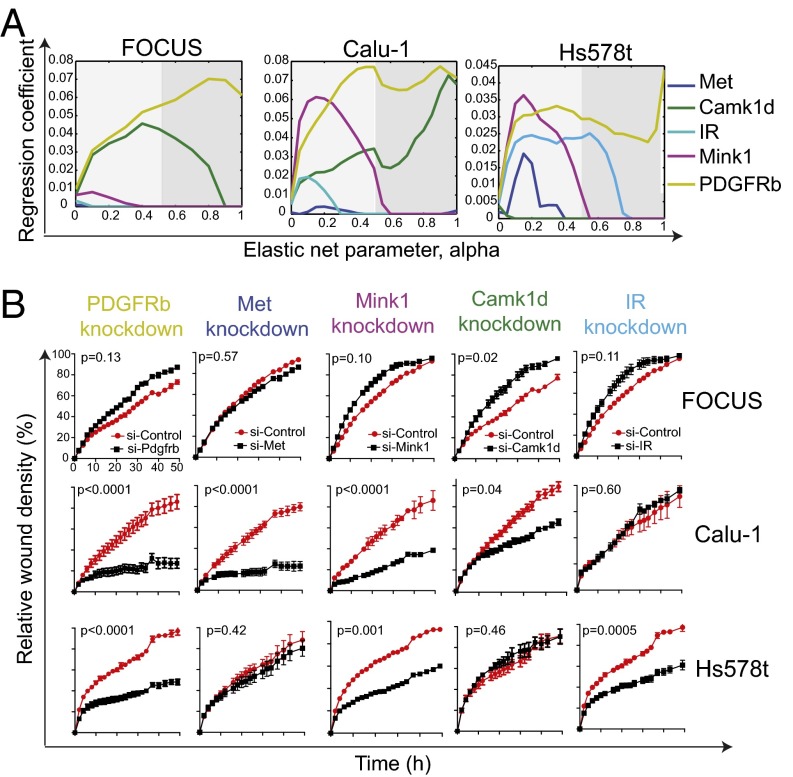
The list of predictors (kinases) identified by the regularized regression is very robust to the parameters of optimization (Fig. 4_A_). In particular, although the elastic net penalty parameter α controls how many predictors get chosen, we observe that the same predictor variables emerge as significant for a wide range of parameter values. We should pay particular attention to the kinases selected at the top of the list (with the highest coefficients). It is well known (16) that the ridge penalty setting (α = 0) shrinks the coefficients of correlated predictors toward each other, whereas the lasso setting (α = 1) tends to pick one of them and discard the others. The elastic net penalty mixes these two; if predictors are correlated in groups, an α ∼ 0.5 tends to select the groups in or out together. Taking this into consideration, we explored all values of α between 0 and 1, focusing on α > 0.5 (Fig. 4_A_). PDGFRb is a great example of a predictor that is selected as an important predictor in all mesenchymal cell lines independently of α, whereas IR is not.
Fig. 4.
Validation of cell type-specific kinases that are determinant of mesenchymal cell migration. (A) Evolution of regression coefficients. Plots showing regression coefficients for respective kinases against value of elastic-net penalty α. Nonzero regression coefficients for kinases picked at α > 0.5 (gray region) are considered significantly informative. (B) RNAi-mediated knockdown of cell type-specific kinases in mesenchymal cancer cells. Plots showing relative wound density of indicated mesenchymal cell lines transfected with either a siRNA targeting the indicated kinase or against a scrambled control. Data are the mean of at least three independent samples and error bars indicate SEM.
To validate the role of the cell type-specific kinases that we predicted to be important in mesenchymal cell migration, we looked at the effects of depleting of these kinases in gene knockdown experiments. Using a pooled set of four siRNA, we knocked down the expression of five kinases in all three mesenchymal cell lines tested and measured their effect on cell migration. As predicted, knocking down the expression of PDGFRb, Met, Mink1, and Camk1d significantly decreased migration, whereas, as a control, knocking down the expression of IR had no effect on cell migration in Calu-1 lung cancer cells (Fig. 3_B_ and Fig. S2). In Hs578t breast cancer cells, PDGFRb, Mink1, and IR were validated to be important for cell migration, whereas knockdown of Met and Camk1d had no observable effect on cell migration (Fig. 3_B_ and Fig. S2), consistent with the elastic net regression predictions. Also, knockdown of only Camk1d in FOCUS cells significantly decreased cell migration, whereas knockdown of PDGFRb, Met, Mink1, and IR had no affect (Fig. 4_B_).
Evaluating the Predictive Capacity of the Elastic Net Method for New Drugs.
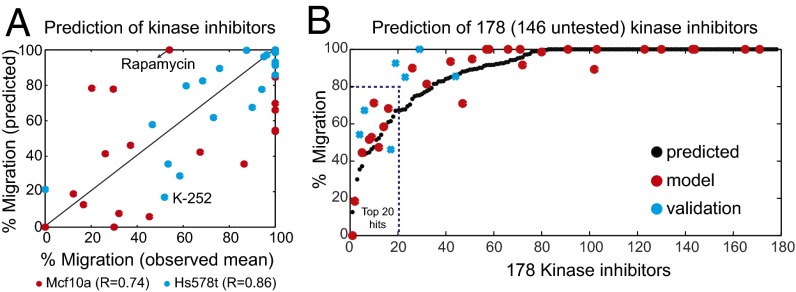
A strong test of any method would be to be able to predict the response of a previously untested perturbation. Therefore, we asked if we can predict a cell type-specific response to an unseen kinase inhibitor using regularized regression. From in vitro target kinase profiling measurements of a test molecule, we should be able to translate this inhibition profile into a prediction of efficacy toward the specific phenotypic marker, in this case cell migration. Because drug action is equated in our approach to a tuple of kinase inhibitions <_X_>, we can extrapolate the phenotype to drugs that were previously characterized in the interaction map M but not measured in the phenotypic assay. We evaluated the prediction power by a LOOCV procedure. A model to predict what phenotype a given drug will induce is constructed using a training set of drugs. LOOCV on a set of k drugs corresponds to k independent experiments, where each time k − 1 drugs are used to build the model and make a prediction for a single held-out drug; then prediction is validated empirically. There is no clear way to define the desired predictive power, because it is unclear how to trade off between precision and recall in different parts of phenotypic value range. Identifying a drug as ineffective is equally important to correctly ordering the efficacy of effective drugs. We present a scatter plot of measured (abscissa) against predicted (ordinate) values (Fig. 5_A_). A strong correlation (Pearson correlation of 0.86 and 0.74, respectively) between observed and predicted migration of both Hs578t (mesenchymal) and Mcf10a (epithelial) cells provides strong evidence for the predictive capacity of our model. A detail discussion on why some drugs are poorly predicted (e.g., rapamycin in Hs578t and K-252 in Mcf10a) is provided in SI Materials and Methods.
Fig. 5.
Predicting the effect of kinase inhibitors on cell migration. (A) Plot showing correlation between observed and predicted migration of HS578t and Mcf10a cells treated with 32 kinase inhibitors. Unpredictable drugs (rapamycin in Hs578t and K-252 in Mcf10a) are also indicated. (B) Informative nonzero variables determined by elastic net regularization were used to predict the efficacy of 178 (146 untested) small molecule kinase inhibitors. The black circles denote predicted values of 178 kinase inhibitors, whereas red circles denote experimental validated 32 kinase inhibitor and blue crosses denote 7 previously unseen inhibitors.
Informative nonzero variables determined by elastic net regularization were next used to predict the efficacy of 178 small molecule kinase inhibitors (146 previously untested; Fig. 5_B_ and Fig. S3). The top 20 kinase inhibitors (hits) predicted to affect cell migration in the FOCUS cell line is highlighted in Fig. 5_B_. Most of the kinase inhibitors had a negligible effect on the phenotype. The analysis predicts that only 20 of 178 drugs would cause as much as a 20% reduction in migration. Of these, the effect of seven previously untested inhibitors was validated experimentally, shown as blue circles in Fig. 5_B_ (Table S5).
Discussion
Polypharmacology is simply the action of drugs against multiple targets. It is both a conundrum for the pharmaceutical industry and a tempting avenue for new approaches to drug development (17). Although industry has usually sought to develop drugs with high affinity and high specificity, there is acknowledgment that these two properties are not a prerequisite for either efficacy or safety. One problem in studying polypharmacology is the need for theoretical ways of using it and predicting it. Large scale unbiased screening of drug combinations is difficult and expensive and therefore must be limited to simple screens. High content screens, such as cell behavior or, worse yet, whole animals, are completely prohibitive. Until these problems can be solved, the challenges are daunting to bring large-scale screening to the stage where proper clinical trials can be designed. An important contribution of this study is first to acknowledge and then to exploit the idea of polypharmacology of drug target deconvolution and to open up some combined experimental and statistical approaches to this area.
We used the elastic net regularization combined with expression profiling and previously characterized data on a large set of kinase inhibitors to identify kinases that are important for epithelial and mesenchymal cell migration. Profiling a sound set of 32 kinase inhibitors in a panel against six cell lines, we identified cell type-specific kinases that affect cell migration. Many of the informative kinases identified had previously well-established roles in cell migration (e.g., all RTKs and cytoskeletal kinases identified in mesenchymal cells; Fig. 3_E_), validating our approach. Interestingly, our approach uncovered informative kinases that were selective for specific cell types. For example, IR was informative of cell migration in breast cancer cell line, Hs578t, consistent with its established role in breast cancer progression (18). Similarly, Met and Erbb4 were identified as informative in the lung cancer cell line, Calu-1, consistent with previous studies (19, 20). PDGFRb and Epha3 were identified as informative kinases in all mesenchymal cell lines in agreement with their previously known role in mesenchymal cell migration and their contribution to tumor progression and metastasis (21, 22). More importantly, our discovery of cell type-specific multiple informative RTKs may support the idea of extensive redundancy of RTK-transduced signaling in cancer cells (23). RTK-mediated signaling pathways share multiple downstream signaling elements, and inhibiting the dominant RTK often results in the compensatory recruitment of downstream components by secondary RTKs. An example of a dominant RTK includes ErbB2, whereas secondary RTKs such as c-Met, PDGFR, and IGF-1R have been reported (24, 25). These RTK coactivation events converge at a number of downstream signaling pathways such as PI3K/Akt and MAPK/ERK (26). Systems-wide analyses of tumors have also identified RTK coactivation as an important mechanism through which cancer cells attain chemo-resistance (23). Thus, our data showing prevalence of multiple cell type-specific informative RTKs regulating cell migration may in part explain acquired resistance to drugs targeting oncogenic kinases. Targeting multiple redundant informative kinases (such as Erbb2, Pdgfrb, and insulin signaling in Hs578t cells) could offer support for rational and effective strategies to combat drug resistance.
Camk2g, Ddr2, Mst4, and Nek11 were found to be informative in all mesenchymal cell lines but not in any of the two epithelial cell lines, suggesting a preferential role for these kinases in mesenchymal cell migration. Of these, Ddr2 is a critical regulator and marker for epithelial-to-mesenchymal transition (27). Further, Tbk1 was found to be informative in both epithelial cell lines but not in any of the three mesenchymal cell lines. Previous studies have shown Tbk1 is an essential element of innate immunity signaling in most epithelial and stromal cell types (28). Together, these data may support the idea of distinct kinases or pathways that regulate epithelial and mesenchymal cell migration (29). Further, using gene depletion experiments, we validated roles of cell type-specific kinases in regulating mesenchymal cancer cell migration (Fig. 4_B_). Thus, using information gained from the ensemble approach, we cannot only identify cell type-specific targets but can also begin to consider drugs that will only selectively affect one cell type (e.g., tumor cells) while leaving other cell types (e.g., normal cells) alone, thereby minimizing possible adverse effects. Such approaches can be used to distinguish cell types that differ in different tissues, different cell states (analogous to the epithelial and mesenchymal tumors from the same tissue of origin), and cells from different genetic backgrounds. Profiling an informative set of inhibitors against tumor cells from individual tumors could become relatively inexpensive and might in the future even be used to identify appropriate individual therapies. Finally, our discovery of several informative kinases with no previously characterized role in cell migration (such as the Mst and Taok families of MAPK kinases in mesenchymal cells) may correspond to novel targets that could warrant further investigation.
We were frankly surprised by how well this approach worked, as judged by extensive cross-validation and validation of previously untested inhibitors, which begs the question of the intrinsic value of this approach compared with genetic and pseudogenetic approaches like RNAi. In genetics, the gold standard is the null mutation. Whereas there is no question that analysis of such mutations has yielded invaluable data, it is worth pointing out that in physical chemistry, the gold standard is quite different: an infinitesimal perturbation of a system at equilibrium or steady state. Although drug effects have the serious problem of acting on multiple and usually unknown targets, they have the advantage of generating dose–responses, producing small perturbations, which can be closer to the initial state than to the null state and may be closer to the physiological state of a whole organism exposed to a drug. If there were a way to deconvolute the meaningful targets of these generally unspecific drugs, some of the mechanistic value of this quantitative and limited perturbation could be captured. Furthermore, drugs can be added acutely, whereas genetic manipulations and even RNAi usually require days, during which time, adaptations have time to occur. In addition, unlike genetic or RNAi approaches, small molecule inhibitors can easily be adapted for both primary and difficult-to-transfect cell lines, as well as for in vivo models. Finally, using a regularized regression model, cell type-specific response to unseen kinase inhibitors can be predicted (Fig. 5_B_, Fig. S4, and Table S5), a clear advantage over other approaches. The effect of combination of kinase inhibitors may also be predicted with some success (Figs. S4 and S5). Future investigations involving in vitro probing of more kinases at multiple doses and more sophisticated modeling should improve the predictive power of combination treatments. The use of these inexpensive and simple procedures could tip the balance toward the use of more cell lines and new phenotypes over highly expensive and complex assays in only a few cell lines.
Several assumptions underlie the regularized regression approach. We limited our analysis only to those kinases that have been currently profiled. We assumed that different inhibitors accumulate similarly inside the cell. We also assumed that the kinase in vitro behaves similarly to the kinase within the cell, ignoring various types of regulation that may affect total activity and potentially even specificity. No assumptions were made regarding relative abundance, subcellular localization of kinases or the timing of drug-target interactions. Some of these assumptions could be optimized by future studies of both mRNA and protein concentration and the activity of kinases in concentrated extracts from cell lines using advanced sequencing and mass spectrometry-based methods. We were hampered by the fact that the published in vitro kinase measurements were at a single dose (500 nM). With little effort, multiple doses could easily be examined, and this should greatly increase the power of our analysis to capture the polypharmacology of drugs. Nevertheless, our analytical methods are well positioned to take advantage of these improvements and in particular the increase in the number of kinases available for screening (currently ∼400/518 kinases). Finally, future studies could also benefit from more advanced methods of machine learning, including, e.g., adaptive Bayesian lasso regression (30). Broadly, this approach is also generally applicable to other classes of enzyme inhibitors such as deacetylases and methyltransferases, for which informative target profiles can be obtained. The combination rule from the FDA requires a mechanistic rationale to justify use of the investigational drugs in combination at various stages of development. Target deconvolution using our ensemble approach has the potential to aid in the rational design of more potent but less toxic drug combinations, thereby bringing rational polypharmacology to bear on some of the most recalcitrant medical problems.
Materials and Methods
Kinetic Wound Healing Assay for Cell Migration.
The effect of kinase inhibitors on cell migration was studied using a wound healing assay. Briefly, cells were plated on 96-well plates, and a wound was scratched with a wound scratcher. Inhibitors at different doses were added immediately after wound scratching, and wound confluence was monitored with an Incucyte Live-Cell Imaging System (Essen Instruments). All of the molecules used in this study are known to be bioactive and therefore must penetrate the cells, but comparative pharmacokinetics/bioactivity of this large collection of inhibitors is not known. Inhibitors were dissolved in DMSO and diluted, and then a constant volume of DMSO was added to each well, with a no drug DMSO control. The percentage migration at 500 nM calculated using the full dose–response curves for each of the inhibitors was used as a response variable for regression.
Implementation of Elastic Net.
Regularized regression in this project was done using the “Glmnet” package for MATLAB (www.stanford.edu/∼hastie/glmnet_matlab). Further details can be found in SI Materials and Methods.
Supplementary Material
Supporting Information
Acknowledgments
This study was supported by awards from the National Institutes of Health (R01 HD073104 and R01 GM103785). We also thank V. Savova, A. Klein, and B. Hayete for helpful comments. T.S.G. is a Human Frontier Science Program Fellow.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lee J, Bogyo M. Target deconvolution techniques in modern phenotypic profiling. Curr Opin Chem Biol. 2013;17(1):118–126. doi: 10.1016/j.cbpa.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24(7):805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 4.Terstappen GC, Schlüpen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6(11):891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 5.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 7.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 8.Chen C-Y, Hsiau K-C, Chung C. Measurement of chondrocyte chemotaxis using a Boyden chamber: A model of receptor-mediated cell migration combined with cell sedimentation. Math Med Biol. 2013;30(3):213–239. doi: 10.1093/imammb/dqs022. [DOI] [PubMed] [Google Scholar]
- 9.Yue PY, Leung EP, Mak NK, Wong RN. A simplified method for quantifying cell migration/wound healing in 96-well plates. J Biomol Screen. 2010;15(4):427–433. doi: 10.1177/1087057110361772. [DOI] [PubMed] [Google Scholar]
- 10.Jolliffe I. Principal Component Analysis. New York: Wiley; 2005. [Google Scholar]
- 11.Graczyk PP. Gini coefficient: A new way to express selectivity of kinase inhibitors against a family of kinases. J Med Chem. 2007;50(23):5773–5779. doi: 10.1021/jm070562u. [DOI] [PubMed] [Google Scholar]
- 12.Kuo J-C, Han X, Hsiao C-T, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13(4):383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 14.Pelegrini AL, et al. Nek1 silencing slows down DNA repair and blocks DNA damage-induced cell cycle arrest. Mutagenesis. 2010;25(5):447–454. doi: 10.1093/mutage/geq026. [DOI] [PubMed] [Google Scholar]
- 15.Melixetian M, Klein DK, Sørensen CS, Helin K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol. 2009;11(10):1247–1253. doi: 10.1038/ncb1969. [DOI] [PubMed] [Google Scholar]
- 16.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 17.Peters J-U. Polypharmacology - foe or friend? J Med Chem. 2013;56(22):8955–8971. doi: 10.1021/jm400856t. [DOI] [PubMed] [Google Scholar]
- 18.Law JH, et al. Phosphorylated insulin-like growth factor-i/insulin receptor is present in all breast cancer subtypes and is related to poor survival. Cancer Res. 2008;68(24):10238–10246. doi: 10.1158/0008-5472.CAN-08-2755. [DOI] [PubMed] [Google Scholar]
- 19.Garofalo M, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18(1):74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Starr A, et al. ErbB4 increases the proliferation potential of human lung cancer cells and its blockage can be used as a target for anti-cancer therapy. Int J Cancer. 2006;119(2):269–274. doi: 10.1002/ijc.21818. [DOI] [PubMed] [Google Scholar]
- 21.Jechlinger M, et al. Autocrine PDGFR signaling promotes mammary cancer metastasis. J Clin Invest. 2006;116(6):1561–1570. doi: 10.1172/JCI24652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day BW, et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell. 2013;23(2):238–248. doi: 10.1016/j.ccr.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Wilson TR, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, et al. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-i receptor in breast cancer cells resistant to herceptin. Cancer Res. 2010;70(3):1204–1214. doi: 10.1158/0008-5472.CAN-09-3321. [DOI] [PubMed] [Google Scholar]
- 25.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 26.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70(10):3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh LA, Nawshad A, Medici D. Discoidin domain receptor 2 is a critical regulator of epithelial-mesenchymal transition. Matrix Biol. 2011;30(4):243–247. doi: 10.1016/j.matbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou Y-H, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41(4):458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson S, et al. A systems view of epithelial-mesenchymal transition signaling states. Clin Exp Metastasis. 2011;28(2):137–155. doi: 10.1007/s10585-010-9367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyung M, Gill J, Ghosh M, Casella G. Penalized regression, standard errors, and Bayesian lassos. Bayesian Anal. 2010;5(2):369–411. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information