Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations (original) (raw)
. Author manuscript; available in PMC: 2017 Jan 1.
Published in final edited form as: Cancer. 2015 Nov 5;122(1):20–33. doi: 10.1002/cncr.29706
Abstract
Neuroblastoma is an embryonal malignancy that commonly affects young children and is remarkably heterogenous in its malignant potential. Recently, the genetic basis of neuroblastoma has come into focus, which has catalyzed not only a more comprehensive understanding of neuroblastoma tumorigenesis, but has also revealed novel oncogenic vulnerabilities that are being leveraged therapeutically. Neuroblastoma is a model pediatric solid tumor in its use of recurrent genomic alterations, such as high-level MYCN amplification, for risk stratification. Given the relative paucity of recurrent activating somatic point mutations or gene fusions in primary neuroblastoma tumors studied at initial diagnosis, innovative treatment approaches beyond small molecules targeting mutated or dysregulated kinases will be required moving forward to achieve noticeable improvements in overall patient survival. However, the clonally acquired, oncogenic aberrations in relapsed neuroblastomas are currently being defined and may offer an opportunity to improve patient outcomes with molecularly targeted therapy directed towards aberrantly regulated pathways in relapsed disease. This review will summarize the current state of knowledge of neuroblastoma genetics and genomics, highlighting the improved prognostication and potential therapeutic opportunities that have arisen from recent advances in understanding germline predisposition, recurrent segmental chromosomal alterations, somatic point mutations and translocations, and clonal evolution in relapsed neuroblastoma.
Keywords: Pediatric, Neuroblastoma, GWAS, MYCN, ALK, clonal evolution
Introduction
Neuroblastoma is an embryonal tumor that arises from the aberrant growth of neural crest progenitor cells of the developing sympathetic nervous system. Tumors typically arise in the adrenal medullary tissue or paraspinal sympathetic ganglia and present as masses in the abdomen, chest, or neck.1 Neuroblastoma occurs in very young children with the median age of diagnosis being 17 months.2 It is the most frequent malignancy during infancy, accounts for over 20% of cancers diagnosed during the first year of life, and can be detected in utero..2-3 Although neuroblastoma constitutes only approximately 5% of all pediatric cancer diagnoses, it disproportionately causes up to 10% of childhood cancer mortality.4
A hallmark of neuroblastoma is its heterogeneity in clinical presentation, course and overall prognosis, ranging from infants with tumors that can spontaneously regress, to children with localized tumors with favorable genomic characteristics that have excellent overall survival with limited cytotoxic chemotherapy, to critically ill older children, adolescents or young adults with widely disseminated disease that can grow relentlessly despite intensive multimodal chemoradiotherapy.3, 5-6 While remarkable improvements over the last three decades have been made for children with lower stage disease, children with high-risk neuroblastoma continue to often have incurable disease with survival rates of less than 40% despite dramatic escalations in the intensity of the cytotoxic chemoradiotherapy.1, 5, 7-9
Neuroblastoma serves as a model cancer for the clinical utility of comprehensive characterization of tumor genetic and biological features at the time of diagnosis. The discovery of frequent high-level amplification of the MYCN oncogene on chromosome 2p24 in neuroblastoma and its association with aggressive clinical disease in the 1980's ushered in the era of using prognostic genomic biomarkers in neuroblastoma, and arguably in cancer in general.10-11 Risk stratification in neuroblastoma has evolved over time to encompass both easily measured clinical variables, such as patient age and tumor stage, and precise genomic aberrations that have been found to be predictive of clinical outcome, such as DNA ploidy and specific recurrent segmental chromosomal aberrations.12-16 A multinational, collaborative effort was initiated in 2004 to develop a standardized International Neuroblastoma Risk Group (INRG) classification system.17-18 A pooled cohort of 8800 patients treated over 12 years was analyzed considering 13 potential prognostic factors.17 Four broad prognostic clinical categories were identified (very low-risk, low-risk, intermediate-risk, and high-risk) from analyses of the 7 most significant prognostic factors at the time of tumor diagnosis: age at diagnosis, INRG tumor stage, tumor histological category, grade of tumor differentiation, DNA ploidy, and copy-number status at MYCN and at chromosome 11q. These clinical categories of very low-risk, low-risk, intermediate-risk, and high-risk correlated with the 5-year event-free survival rates of >85%, ≥75% to ≤ 85%, ≥50% to ≤75%, or ≤ 50%, respectively.17 Prospective implementation of this classification system in current pediatric neuroblastoma clinical trials is ongoing and will allow for improved comparison of trial outcomes across different countries.
Neuroblastoma genetic predisposition
Neuroblastoma, like other pediatric embryonal malignancies, has long been thought to arise from the malignant progression of misappropriated fetal tissue during development. However, it was not until this past decade that we have gained a comprehensive understanding of the underlying genetic events driving neuroblastoma tumorigenesis.19-34 Many of the initiating genetic events for both familial and sporadic neuroblastoma have recently been defined and have led not only to an improved understanding of the transforming biological aberrations that contribute to neuroblastoma tumorigenesis, but these investigations have also identified potentially druggable, activated pathways that have been translated to advances in the clinic.19-38 In part, the persistence of the effect of these oncogenes in established tumors may explain the paucity of recurrent, somatic oncogenic driver mutations found in recent whole genome sequencing efforts.39-42
Familial neuroblastoma
Similar to other human malignancies, a small subset of neuroblastoma is inherited in an autosomal dominant manner, and in these families the disease generally occurs at a younger age and is more likely to present with multifocal primary tumors than sporadic tumors, as predicted by Knudson's two-hit hypothesis.43-44 Familial neuroblastoma is rare, only accounting for 1-2% of neuroblastoma cases, and recently gain of function mutations in the anaplastic lymphoma kinase (ALK) gene were discovered as the major cause of highly penetrant, hereditable disease.22, 24 ALK is an orphan (ligand not known) receptor tyrosine kinase that acts as a major oncogenic driver in several human malignancies, including anaplastic large cell lymphoma (ALCL) and non-small cell lung carcinoma (NSLC), but unlike in neuroblastoma, in these malignancies ALK is typically activated via translocation events creating aberrantly expressed fusions transcripts that contribute to cellular transformation and progression.38 In familial neuroblastoma, constitutive ALK activation is achieved through kinase domain mutations and often identical activating mutations have also been found in sporadic neuroblastoma tumors.22, 24, 36-42, 45 However, although the most common ALK germline mutation is also the most common somatic ALK alteration (R1275Q), the most activating somatic ALK variants typically do not appear to be tolerated in the germline.45-46 In the rare cases that the more potent ALK mutations (F1245V and F1174V) have appeared in the germline they result in not only congenital neuroblastoma, but also global developmental and neurocognitive defects suggesting an important role for ALK regulation in the normal development of the CNS.45-46 Furthermore, the penetrance of ALK activating mutations in the germline appears to directly be correlated with the level of aberrant kinase activation.45 Based on these preclinical findings, small molecule inhibitors of ALK's kinase activity have been rapidly, albeit with extensive and thorough preclinical study, translated to neuroblastoma patient care in early phase clinical trials.47
Finally, a small subset of familial and sporadic neuroblastomas that typically occur along with other anomalies of neural-crest derived tissues is caused by loss of function mutations in the paired-like homeobox 2B (PHOX2B) gene on chromosome 4p12 that encodes a transcription factor integral to neural crest development, loss of which disrupts terminal differentiation of neuroblastoma cells.48 PHOX2B mutations are found in a majority of children with congenital central hypoventilation syndrome and some children with Hirschsprung disease, but account for only perhaps 10% of familial neuroblastomas.48-49 Genetic testing for germline ALK or PHOX2B mutations has become the standard of care for children with a family history of neuroblastoma and should also be considered for children with bilateral adrenal tumors. Typically asymptomatic children who are found to harbor an ALK or PHOX2B mutation are generally screened with serial abdominal ultrasounds and urine catecholamine levels approximately every three months until age 5. However, there are currently no data or consensus guidelines to support these clinical practices, and there is an urgent need to address this prospectively. Ongoing efforts are focused on discovering the cause of the remaining 10-20% of familial neuroblastoma that do not harbor obvious ALK of PHOX2B mutations.
Sporadic neuroblastoma predisposition
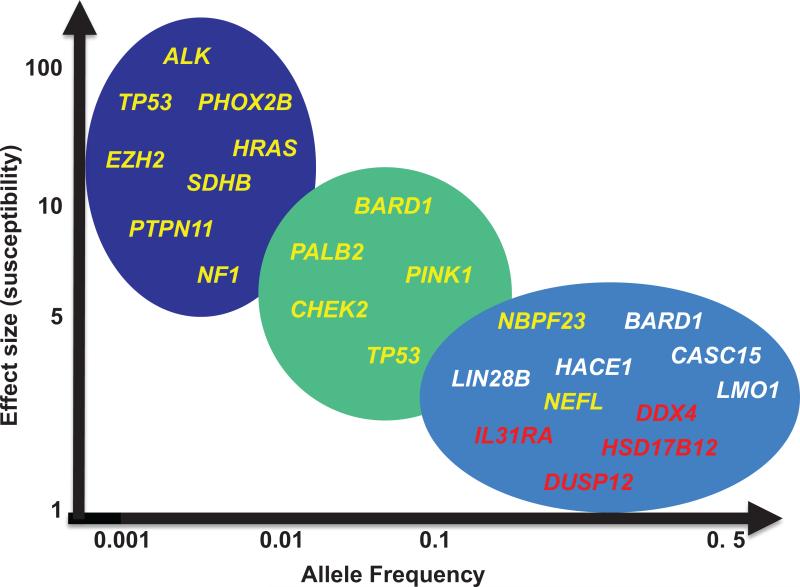
We now know that sporadic (nonfamilial) neuroblastoma is a complex genetic disease in which common polymorphic alleles contribute substantively to malignant transformation. A large genome-wide association study (GWAS) using samples collected by the cooperative efforts of the Children's Oncology Group (COG) utilizing high-density SNP-based microarray technology has begun to define the genetic landscape of sporadic neuroblastoma predisposition and have identified common DNA alleles in CASC15, BARD1, LMO1, DUSP12, DDX4, IL31RA, HSD17B12, HACE1, LIN28B, and NEFL to be significantly associated with neuroblastoma development (Figure 1, Table 1).19-21, 23, 25-34 Although the motivation for this study was to discover neuroblastoma initiating events, it has become clear that many of the neuroblastoma GWAS loci harbor driver oncogenes that are critical in maintaining tumorgenicity in established tumors. These discoveries have collectively illustrated how robust GWAS signals offer genomic landmarks that point toward identification of molecular mechanisms involved in both tumor initiation and malignant progression and, in some instances, offer insights into druggable biological pathways that may be leveraged clinically (Figure 1, Table 1).
Figure 1. Rare and common genomic variants predispose to neuroblastoma.
In addition to ALK and PHOX2B associated familial neuroblastoma, neuroblastoma can also arise in the setting of genetic syndromes with underlying RAS-MAPK pathway germline mutations such as NF1 in Neurofibromatosis type 1,99 PTPN11 in Noonan syndrome,97-98 and HRAS in Costello syndrome (left).97 TP53 mutations associated with Li Fraumeni syndrome,119 EZH2 mutations associated with Weaver syndrome120 and SDHB mutations in familial paraganglioma/pheochromocytoma (PGL/PCC)121 are also rarely associated with neuroblastoma genesis (left). Low frequency alleles in multiple DNA damage response genes (BARD1, CHEK2, PALB2, and TP53) with an intermediate effect size also contribute to neuroblastoma predisposition (middle) and more common alleles with a modest effect size discovered via a GWAS approach also collectively contribute to neuroblastoma genesis (right), and at times specifically to a high-risk (white) or low-risk (red) neuroblastoma phenotype.
Table 1.
Summary of genomic loci significantly associated with neuroblastoma predisposition.
| Genomic Locus | Candidate Gene | Phenotype association | Top SNP | _P_-Value (combined) | MAF cases | Odds Ratio (OR) | Proposed Mechanism | Level of Evidence# |
|---|---|---|---|---|---|---|---|---|
| 6p22 | CASC15/NBAT-1 | High-risk | rs6939340 | 9.33 × 10−15 | 56% | 1.37 | Loss of function | 223, 31, 33 |
| 2q35 | BARD1 | High-risk | rs6435862 | 8.65 × 10−18 | 40% | 1.68 | Gain of function | 221, 32 |
| 11p15 | LMO1 | High-risk | rs110419 | 5.20 ×10−16 | 55% | 1.34 | Gain of function | 120, 34 |
| 6q16 | LIN28B | High-risk | rs17065417 | 1.2 × 10−8 | 8% | 1.38 | Gain of function | 119, 51 |
| 6q16 | HACE1 | High-risk | rs4336470 | 2.7 × 10−7 | 30% | 1.26 | Loss of function | 319 |
| 1q23 | DUSP12 | Low-risk | rs1027702 | 2.07 × 10−6 | 31% | 2.01 | unknown | 429 |
| 5q11 | DDX4 | Low-risk | rs2619046 | 2.94 ×10−6 | 32% | 1.47 | unknown | 429 |
| 5q11 | IL31RA | Low-risk | rs10055201 | 6.54 × 10−7 | 29% | 1.49 | unknown | 429 |
| 11p11 | HSD17B12 | Low-risk | rs11037575 | 4.20 × 10−7 | 39% | 1.67 | unknown | 429 |
| 8p21 | NEFL | - | rs11994014 | 0.005 | 20% | 0.88 | Loss of function | 225 |
| 17p13 | TP53 | - | rs35850753 | 3.43 × 10−14 | 3.6% | 2.70 | Loss of function | 126, 52 |
| 1q21 | NBPF23 | - | CNV | 2.97 × 10−17 | 15% | 2.49 | unknown | 430 |
One of the most significant and robustly replicated association signals from this GWAS enriched in the high-risk subset of neuroblastoma resides in the BRCA1-associated RING domain-1 (BARD1) locus on chromosome 2q35.21 BARD1 has been classically thought of as a tumor suppressor because its protein dimerizes with BRCA1 through their respective RING domains. However, it was found in neuroblastoma that disease associated variations correlate with the expression of an oncogenetically activated isoform, BARD1β, that has growth promoting effects in neuroblastoma models potentially through cooperation with the Aurora family of kinases.32 Importantly, this study not only identified BARD1 as an oncogenic driver of high-risk neuroblastoma tumorigenesis, but also re-identified a potential druggable pathway in neuroblastoma as Aurora A kinase inhibitors have been independently found to be an effective therapy in neuroblastoma models.50 While the common polymorphisms at the BARD1 locus associated with neuroblastoma individually have a modest impact on disease initiation, it is now also evident that rare germline mutations in this gene also contribute to tumorigenesis, likely with a much higher effect size (Figure 1).40 However, whether or not these patients are also susceptible to breast and/or ovarian cancer, and what the functional consequence and penetrance of these mutations are requires further study.
LIM domain only 1 (LMO1) is another neuroblastoma oncogene that was discovered via this GWAS, and again this association was found to be enriched in the high-risk phenotype.20 In other words, individuals with “risk” alleles in the BARD1 and LMO1 loci are not only more likely to develop neuroblastoma, but if the disease does develop, they are much more likely to develop metastatic disease and have worse clinical outcome probabilities. This concept, that the ultimate clinical phenotype when patients present with neuroblastoma is largely dictated by underlying germline genotypes is yet another unexpected conclusion from this GWAS. LMO1 is a transcription regulator and is the first example in neuroblastoma where a definitive disease causal SNP has been identified. Briefly, a single G>T transversion in the first intron of LMO1 was discovered in a super enhancer element, and the G (risk) allele allows for GATA3 transcription factor binding leading to robust LMO1 expression.20, 34 The T (protective) allele completely abrogates GATA3 binding resulting in dramatically lower LMO1 expression. Many other robust neuroblastoma association signals have been discovered from this GWAS that collectively can be estimated to have a combined odds ratio of close to 20 to develop this disease and the functional follow-up of which have confirmed that many of these genes are also driver genes in established tumors (Figure 1, Table 1).19-21, 23, 25-34, 51
In addition to common SNP variation, neuroblastoma was the first example where germline copy number variation (CNV), another major determinant of human diversity, contributes to cancer susceptibility.30 NBPF23 is a gene on chromosome 1q21.1 that harbors a region that varies in copy number amongst individuals, and hemizygous deletion of this region is highly associated with neuroblastoma. Other yet to be discovered CNVs are also likely to contribute to neuroblastoma tumorigenesis.
Finally, as this GWAS has continued to accrue subjects, it has become statistically feasible to identify rare polymorphisms that contribute to neuroblastoma susceptibility (Figure 1, Table 1). For example, rare germline variants in TP53 have also been found to be integral in neuroblastoma genesis.26 Two SNPs, rs78378222 and rs35850753, that map respectively to the 3' and 5' untranslated regions (UTRs) of TP53, have recently been found to be robustly associated with neuroblastoma predisposition.26 Similar to other malignancies, the disease associated rs7878222 allele is hypomorphic and impairs transcriptional termination and polyadenylation of TP53 transcripts.26, 52 However, the functional implications of the other noncoding SNP, rs35850753, remain unclear but may involve driving TP53 alternative transcript expression as it maps to the 5' UTR of the delta133 TP53 isoform which is transcribed off an alternative, internal promoter. This isoform lacks a TP53 transactivation domain, and has been shown to act in a dominant negative fashion to the tumor suppressive functions of wild-type TP53.53 As somatic TP53 mutations are rare in neuroblastoma, this study suggests alternative mechanisms that neuroblastoma cells may utilize to circumvent the tumor suppressor effects of TP53 in the development of a malignant phenotype.39-42, 54
Somatic genomic aberrations in neuroblastoma: prognostic implications and therapeutic opportunities
The discovery of high-level MYCN amplification on chromosome 2p24 and its association with aggressive clinical disease in the 1980's heralded the beginning of using tumor specific genomic alterations as prognostic biomarkers in neuroblastoma,10-11 and arguably in cancer in general. Since this time, several other recurrent genomic alterations have been robustly validated as having prognostic predictive power in addition to MYCN amplification including DNA ploidy, gain of chromosome 17q and deletions of chromosome arms 1p, 3p, 4p, or 11q.12-17, 55-60 However, while gains or losses of these chromosomal loci are clinically prognostic in neuroblastoma, the candidate oncogenes or tumor suppressor genes at these loci that underlie this difference in tumor biology have yet to be definitively defined.
Chromosomal rearrangements
The ploidy (DNA index) of neuroblastoma primary tumors has become a significant predictor of patient outcome and is a main component of the current INRG classification system.17 Diploid tumors (DNA index = 1) occur in approximately one-third of patients, while two-thirds of neuroblastoma tumors are hyperdiploid (DNA index >1).15 Tumor ploidy is one of the most significant prognostic markers for children less than 18 months old, and infants with hyperdiploid tumors have significantly increased survival.15-17 Since determining tumor DNA index is straightforward it continues to be used as a prognostic marker by most cooperative groups. However, with the advent of next-generation genomic techniques it has become clear that neuroblastoma tumor DNA copy number aberrations can be more precisely segregated into two main global categories that, independent of the specific genomic loci involved, carry clinically prognostic information.55, 61 In general, tumors with gains of whole chromosomes resulting in hyperdiploidy are associated with lower risk disease and favorable clinical outcomes while tumors that have segmental chromosomal alterations are higher risk and are associated with more clinically aggressive disease.55, 61 Additionally, the acquisition of new segmental chromosomal aberrations appears to occur at the time of relapse in both tumors that had or didn't have a segmental genomic profile at diagnosis.62 Undoubtedly, prospective studies are needed to evaluate if the presence of segmental chromosomal aberrations at the time of diagnosis or relapse can be used prognostically. It would be of great clinical utility if the presence of segmental chromosomal aberrations can identify the rare, less favorable subsets within non-high risk neuroblastoma patient groups (i.e. localized disease without MYCN amplification) that is predetermined to relapse or progress after surgery enabling upfront initiation of adjuvant cytotoxic chemotherapy.62-63 One recent clinical trial that has begun to address clinical questions such as these is the European Study, LINES 2009 (Low and Intermediate Risk Neuroblastoma Study; ClinicalTrials.gov identifier NCT01728155).
While segmental chromosomal aberrations are common in neuroblastoma, translocation events that create fusion oncogenes are rarely seen in diagnostic neuroblastomas, but do occur more frequently in relapsed disease after exposure to intensive DNA damaging agents. Structural rearrangements in protein coding genes have been found to occur in primary neuroblastomas, on an average of four per primary tumor39 and up to approximately 20% of predicted fusion transcripts can be detected at the RNA transcript level in neuroblastoma, including fusions transcripts involving the ALK, NBAS, PTPRD, and ODZ4 genes.64 However, although ALK rearrangements leading to a constitutively active ALK fusion gene product are common drivers of the pathogenesis of non-small lung cancer (NSCLC) and pediatric anaplastic large cell lymphoma (ALCL), they are overall rare in neuroblastoma tumors.65-66 Over the coming decade, next generational sequencing technologies coupled with follow-up functional studies should allow for a more comprehensive profiling of the role that genomic structural arrangements and resulting fusion genes play in driving neuroblastoma tumorigenesis. Finally, chromothripsis, or the local shredding of chromosomes, may be common in higher stage neuroblastomas.41 Although while some studies have found this to occur in up to 18% of clinically aggressive tumors41, chromothripsis was not found to be as common in other cohorts of primary neuroblastomas and thus definitively determining how common chromothripsis is in primary neuroblastomas awaits additional next generation sequencing efforts.39-40
Chromosomal allelic gains and oncogenes
The classic genetic aberration of neuroblastoma that is most consistently associated with poor outcome is genomic amplification of the MYCN oncogene on chromosome 2p24.10-11 MYCN amplification occurs in approximately 20% of all primary neuroblastoma tumors and in 50% of high-risk tumors, and correlates strongly with clinically aggressive, advanced stage disease and treatment failure.10-11 Transgenic mice that are engineered to over-express MYCN in their neuroectodermal cells developed neuroblastoma, providing a direct link of this gene to neuroblastoma genesis.67
MYCN encodes a transcription factor that forms heterodimers with MAX and together they collectively activate the transcription of downstream target genes.68 RNAi induced knockdown of MYCN decreases neuroblastoma tumorigenesis by inducing apoptosis and terminal differentiation suggesting that the direct targeting of this protein may be an effective therapeutic strategy.69-70 Targeting MYCN with small molecule inhibitors has proven to be challenging though, due in part to the difficulties of targeting ubiquitously expressed pleotrophic transcription factors, a hurdle that is further exacerbated by MYCN's limited small molecule binding real estate among its two extended alpha helices.71 Thus, although MYCN amplification has been known to be associated with aggressive neuroblastoma for over 30 years, it wasn't until recently that targeting of this protein became a therapeutic reality in neuroblastoma through the development of novel therapeutic strategies to circumvent the requirement of direct MYCN interaction. These strategies include blocking the MYCN/MAX interaction, the development of small molecules disrupting MYCN transcription or stability of the MYCN protein, and by synthesizing drugs targeting synthetic lethal interactions of MYCN (reviewed in 35, 72).
One such strategy that holds therapeutic promise is the use of bromodomain and extra terminal (BET) inhibitors to specifically downregulate MYCN expression. The family of BET adaptor proteins (BRD2-4) have bromodomain motifs that enable binding to histone tail acetylated lysine residues at sites of open chromatin, recruiting additional chromatin-modifying proteins and thus modulating target gene transcription.73 Small molecules targeting the bromodomains of these chromatin readers inhibit BET recruitment to open chromatin sites and thus decrease target gene transcription.74 Among the genes targeted by the BET family of chromatin readers are the MYC family genes.75-77 Screening of over 673 well-characterized human cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE) using the prototypical BET inhibitor, JQ1, identified MYCN amplification as a robust biomarker for sensitivity to bromodomain inhibition and, not surprisingly, bromodomain inhibition was cytotoxic in both neuroblastoma cell lines and several neuroblastoma mouse models.76 Similar cell cytotoxicity has been seen with other BET inhibitors via blockade of MYCN expression and BET inhibitors have been found to be efficacious in another pediatric embryonal malignancy, medulloblastoma, a subset of which is driven by MYC.77-78 Clearly, bromodomain inhibition may be a therapeutic option for neuroblastoma patients with MYCN amplification and these findings suggest that MYCN/MYC amplification may be a predictive biomarker for BET inhibitor clinical activity.75-78 However, as with other epigenetic based cancer therapies, a major concern lies in the inherent lack of specificity of small molecules targeting these broadly acting gene expression regulators and clearly the array of on- and off-target toxicities needs to be clearly defined in the preclinical arena. Other than MYCN, focal amplifications are rare in diagnostic neuroblastomas, occurring with any significant frequency only at the ALK locus.45
The most common genomic aberration of neuroblastoma cells is the somatic gain of the distal portion of chromosome 17q which occurs in at least 50% of primary tumors, portends an overall poor prognosis, and is frequently associated with other biomarkers of aggressive disease such as MYCN amplification, older age, and chromosome 1p deletion.13, 56 Remarkably though, despite years of investigation, the mechanism by which 17q gain results in a more aggressive malignancy remains to be identified.
Chromosomal allelic deletions and tumor suppressor genes
Allelic losses at chromosome 1p and 11q have also been found to carry integral prognostic information in neuroblastoma.12, 57-58, 79 Loss of heterozygozity (LOH) at chromosome 1p36 occurs in 23-35% of primary neuroblastomas and is associated with other high-risk clinical and genomic features such as older age, MYCN amplification and metastatic disease.12, 57-58, 79 Loss of chromosome 11q occurs in approximately one-third of neuroblastomas and is associated with a poor prognosis, but is inversely associated with MYCN amplification making it a potential biomarker for aggressive sub-phenotypes of those patients without MYCN amplification.12 However, again despite years of tireless investigation, no gene at 1p or 11q has been found to harbor the recurrent inactivating mutations required to fulfill the classic criteria of a bona fide tumor suppressor gene. It is likely that the very large deletions at these (and other) loci are impacting many genes, with aberrant methylation or other epigenetic alterations further downregulating gene expression.
Somatic mutations
Pediatric malignancies in general have a much lower somatic mutation frequency than most adult carciniomas.80 Accordingly, despite recent extensive efforts using next-generation sequencing techniques, few recurrent somatic mutations have been discovered in neuroblastoma tumors at the time of diagnosis (Table 2).39-42 ALK is the most commonly somatically mutated gene in neuroblastoma, with up to 14% of tumors having genetic missense alterations or gene amplification events.39-42 Putative loss-of-function genetic alterations in the RNA-helicase, ATRX, have been found in up to 10% of neuroblastomas and are enriched in older patients and may help explain the more indolent phenotype seen in this population.40-42 Similar to other tumors, neuroblastomas that are deficient in ATRX appear to undergo the telomerase independent telomere maintenance mechanism termed alternate lengthening of telomeres (ALT), which may be a marker of an indolent course with primary chemotherapy resistance81-83 Other chromatin remodeling proteins are also significantly mutated in neuroblastoma as single nucleotide alterations or deletions of the AT-rich interactive domain 1A/1B (ARID1A/1B) proteins have also been detected in a significant subset of clinically aggressive neuroblastomas.39 Other recurrent somatic mutations have been detected rarely in neuroblastoma tumors (≤5% of primary tumors) including genetic alterations in PTPRD, ODZ3, PTPN11, MYCN (P44L), and NRAS.39-41 Clearly, efforts at improving neuroblastoma outcomes cannot focus exclusively on developing small molecules targeting mutated oncogenes and complementary strategies must be defined.
Table 2.
Summary of potentially actionable genomic alterations in neuroblastoma.
| Clinically Actionable Pathway/Gene | Prevalence at diagnosis | Prevalence at relapse | Biomarker | Therapeutic Strategy | Level of evidence |
|---|---|---|---|---|---|
| ALK# | |||||
| Mutation/Focalamplification | 8-14%39-42, 122 | 26-43%89, 91 | ALK mutation/amplification | ALK inhibition35, 38 | 1 |
| RAS-MAPK Pathway# | |||||
| PTPN11 | *Rare40, 122 | #Rare91 | RAS-MAPK pathwaymutation | MEK inhibition91 | 2 |
| FGFR1 | ” | ” | ” | 2 | |
| BRAF | ” | ” | ” | ” | 2 |
| NRAS | ” | ” | ” | ” | 2 |
| KRAS | ” | ” | ” | ” | 2 |
| HRAS | - | ” | ” | ” | 2 |
| NF1 | Rare40 | 9%91 | ” | ” | 2 |
| Cell Cycle Control | |||||
| CDK4/6 amplification | 4%109 | - | MYCN amplification100-101 | CDK4/6 Inhibition100-101 | 2 |
| Cyclin D1 amplification | 2-15%104, 109 | - | ” | ” | 2 |
| CDKN2A deletion | 0-20%109-110, 123 | 13-22%91, 110 | ” | ” | 2 |
| DNA Damage Pathway | |||||
| TP53 | 1-8%40, 110, 123 | 15%110 | TP53 mutation | Unknown | 3 |
| MDM2 amplification | 13%110 | 13%110 | MDM2 amplification | MDM2 inhibition124-126 | 2 |
| Transcriptional Control | |||||
| MYCN amplification | 16-38%10-11, 17 | - | MYCN amplification/mutation | BET inhibition75-76 | 2 |
| MYCN mutation | Rare40 | - | ” | ” | 2 |
| Chromatin Modification | |||||
| ATRX deletion/mutation | 9-22%40, 42 | 17%91 | ATRX deletion/mutation | Unknown | 3 |
| ARID1A/ARID1B deletion/mutation | 11%39 | Rare91 | ARID1A/B deletion/mutation | ” | 3 |
Clonal evolution: current understanding and druggable opportunities for relapsed patients
While clonal evolution has been studied in neuroblastoma since Peter Nowell first introduced the concept in the 1970's, neither clonal evolution, nor the regional tumor heterogeneity that likely proceeds this, has been comprehensively studied in neuroblastoma with next generation sequencing efforts until very recently.62, 84-87 However, it is clear from early studies that clonal evolution is common in neuroblastoma and that relapsed neuroblastoma tumors acquire many additional segmental chromosomal alterations that may have prognostic implications in the relapsed setting.62 More recent focused studies using genomic deep sequencing efforts have suggested that clonal evolution also results in the acquisition of targetable somatic aberrations in known oncogenic pathways, and early evidence suggest that these converge on the mitogen-activated protein kinase (MAPK) pathway.88-91 Although the MAPK pathway is a key driver of oncogenicity in several human malignancies, until recently this was not thought to be the case in neuroblastoma as MAPK pathway mutations are rare in primary neuroblastoma tumors (despite the fact that NRAS was discovered in neuroblastoma).40, 92 However, in a recent comprehensive deep resequencing of 23 relapsed, refractory neuroblastoma cases, 78% of relapsed specimens were found to have a clonally enriched somatic mutation predicted to activate the MAPK pathway, including not only ALK aberrations, but also NRAS, KRAS, HRAS, BRAF, PTPN11, FGFR1, and NF1 somatic aberrations.91 Furthermore, over 60% of human derived neuroblastoma cell lines, the majority of which are established at disease relapse, similarly carry a MAPK pathway activating genomic alteration.91 These findings highlight a potential therapeutic strategy in the relapsed setting, for example with MEK1/2 inhibitors (Table 2). Interestingly, MAPK pathway inhibition has also recently been found to reverse retinoid resistance in neuroblastoma cells as the tumor suppressor NF1 was identified as a key modulator of neuroblastoma cell retinoid sensitivity, an effect that could be reversed with MEK inhibition.93 Although, it has been known for decades that retinoids can induce terminal differentiation of neuroblastoma cells in vitro and the use of isotretinoin along with GD2 targeted immunotherapy has become the standard of neuroblastoma maintenance therapy, the response to retinoids is heterogeneous and retinoids fail to prevent disease relapse in a substantial proportion of neuroblastoma patients.7-8, 94-96
Finally, considering MAPK pathway germline aberrations, rarely neuroblastoma can arise in the setting of certain genetic syndromes such as Noonan,97-98 Costello,97 and Neurofibromatosis type I99 which are associated with PTPN11, HRAS, and NF1 germline mutations, respectively (Figure 1). Taken together, these data support a strategy targeting the MAPK pathway in at least relapsed, refractory neuroblastoma. Inhibiting this pathway may ultimately have the dual effect of downregulating a clonally acquired oncogenic driver pathway in relapsed neuroblastomas as well as enabling the terminal differentiation of these tumors.
The cyclin D1/CDK4/CDK6/RB cell cycle regulatory pathway is also a significant contributor to neuroblastoma pathogenesis and activity of this pathway correlates with MYCN amplification, providing a biomarker for potentially susceptible patients.100-109 Although somatic activating mutations in this pathway are rare at diagnosis in neuroblastoma, several genetic aberrations have been found to increase CDK4/CDK6 expression and kinase activity and ultimately drive cell cycle progression including genomic amplification of CCND1 (cyclin D1) and CDK4 or deletion of CDKN2A,40, 102-106, 108-109 and the frequency of these aberrations may be higher in the relapsed neuroblastoma genome (Table 2).91, 110 Overall, approximately one-third of primary diagnostic neuroblastomas may contain genomic aberrations that implicate the G1 cell cycle regulatory genes.102, 109 In a RNAi screen of the protein kinome, CDK4 was identified as one of the key kinases that drives neuroblastoma cell proliferation in neuroblastoma preclinical models.107 Genetic depletion of CDK4 not only decreases neuroblastoma cell proliferation, but also partially restores G-S cell cycle arrest and resensitizes neuroblastoma cells to doxorubicin-mediated cell death.101 Finally, dual pharmacologic inhibition of CDK4/6 with a small molecular inhibitor of CDK4/6 induced cell cycle arrest and cellular senescence in a MYCN amplification dependent manner in neuroblastoma models.100
It is unlikely that a single targeted drug will be curative in the relapsed neuroblastoma setting and acquired resistance to a single agent is a real possibility. Thus, an additional challenge is to identify rational synergistic drug combinations based on the activating genomic parameters present in individual relapsed tumors. Clearly, a NGS-based driven trial is needed in neuroblastoma to define the precise subset of acquired somatic changes in real-time in the relapsed high-risk neuroblastoma genome thereby creating an opportunity to offer these children novel combinatorial therapies targeting the specific pathways that are altered in their tumors (Table 2). Recently, this clinical trial idea has been proposed in the form as the NExt generation PErsonalized Neuroblastoma THErapy (NEPENTHE) trial which is a Phase 1b/2 clinical trial for relapsed/refractory neuroblastoma patients where tumor biopsies and NGS results will be used for novel-novel drug combination therapy assignment that should open in late 2015. Finally, although the current focus of targeted therapy research efforts are directed towards the relapsed high-risk neuroblastoma genome and the activated pathways in this clinical setting, as this landscape becomes more defined it will also be important to consider and study the role of the up-front targeting of these driver pathways in high-risk neuroblastoma treatment.
Disease monitoring and circulating tumor DNA (ctDNA) in neuroblastoma
PCR and immunocytology based methods targeting significantly overexpressed genes in neuroblastoma have been studied and verified as sensitive markers of minimal residual disease.111 However, these methods are reliant on gene expression and are unable to capture genomic alterations either at diagnosis or those that are clonally acquired and may be drivers in the relapsed neuroblastoma tumor. Historically, pediatric oncologists have been reluctant to prescribe an invasive procedure in a relapsed high-risk neuroblastoma patient, especially given the diagnostic sensitivity and specificity of 131I-MIBG scintigraphy. Furthermore, biopsy of relapsed tumors has additional challenges including the fact that they are often in difficult locations (e.g. paraspinal or within bone) and the tumors themselves often demonstrated significant morphologic heterogeneity including large regions of tumor necrosis from prior treatment effect. Additionally, neuroblastoma relapses often occur at multiple anatomic sites and given that regional and metastatic tumor heterogeneity studies of other cancers have shown that distinct driver aberrations can be unique to a metastatic site, a biopsy of one site of neuroblastoma relapse may not comprehensively identify all acquired mutations.112 Consequently, acquired oncogenic vulnerabilities in the relapsed high-risk neuroblastoma tumor may go unrecognized if tumor genomic profiling is reliant on a single biopsy or traditional MRD approaches and a more effective means of comprehensively and noninvasively sampling tumor cells in this setting is needed. Utilizing ctDNA via a “liquid biopsy” may directly address and potentially obviate many of these concerns. Methodologies for detecting ctDNA have evolved such that techniques now exist which can detect rare ctDNA variants in a complex mixture of DNA with high sensitivity, with the ability to detect tumor derived DNA variants (i.e. point mutations) that make up 0.01% or less of the total cell free DNA (cfDNA) in the plasma.113-114 Children with neuroblastoma have been found to have high levels of ctDNA, especially at the time of diagnosis and relapse, and tumor specific genomic alterations have reliably been detected in the serum of these children, including MYCN amplification, gain of chromosome 17q, and most recently ALK mutations using a droplet digital PCR system.114-118 Although potentially useful for monitoring disease burden and response to treatment, the real therapeutic potential of detecting ctDNA might be in relapsed neuroblastoma where the acquisition of actionable somatic aberrations may have been acquired via clonal evolution and may represent a therapeutic opportunity to initiate targeted therapy in these patients as discussed above. Finally, the use of serial ctDNA detection for prognostic purposes has not been studied in neuroblastoma but may also offer an opportunity to capitalize on this technology.
Conclusions and future directions
Many significant improvements in neuroblastoma diagnosis, risk stratification, and treatment have been made over the last few decades. However, given that a substantial number of children cannot be cured and those who are often suffer many debilitating long-term side effects, efforts are clearly still needed to better understand the aberrantly expressed genes and activated pathways to identify novel therapeutic targets in neuroblastoma. As next generation genomic tools continue to advance in the coming decade, it is imperative that the integration of these complex genomic, biological and clinical data occurs to best refine risk stratification and treatment strategies. Furthermore, it will be vital to identify at the level of a given tumor, the driver pathways and targetable aberrations so that these can be leveraged clinically if a small molecule or immunotherapeutic approach exists targeting that given alteration. The therapeutically applicable research to generate effective treatments (TARGET) project will continue to play a major role in using a comprehensive molecular profiling approach, including gene expression, methylation, and chromosome copy number analysis, in addition to whole genome and transcriptome sequencing techniques to characterize both primary and relapsed neuroblastomas with a goal of discovering novel therapeutics for high-risk disease (https://ocg.cancer.gov/programs/target/projects/neuroblastoma). Finally, continued work on defining the genetic basis of neuroblastoma and understanding the complex biology at GWAS loci should also identify additional key molecular targets that warrant well designed translational studies.
Relapsed high-risk neuroblastoma patients remain a significant challenge for pediatric oncologists and currently no curative treatment exists. Clonally acquired somatic alterations that occur under the selective pressure of cytotoxic chemoradiotherapy offer clinically tractable targets and this strategy is currently being studied in a prospective trial. Clearly though, efforts at improving neuroblastoma outcomes cannot focus exclusively on developing small molecules targeting mutated or dysregulated oncogenes and complementary strategies must be defined. Recently, the first neuroblastoma immunotherapy strategy utilizing a chimeric monoclonal antibody targeting the disialoganglioside, GD2, on neuroblastoma cells showed improved survival in a randomized phase 3 clinical trial, credentialing immunotherapy for this disease.7 Neuroblastoma is an ideal malignancy to target with immunotherapy as it derives from developing neural crest cells and thus continues to selectively express lineage-specific cell surface markers that may not be widely present on mature, non-embryonic tissues. The focus of current efforts is to determine if more therapeutically ideal, differentially expressed cell surface molecules are present on neuroblastoma cells, and determine if the targeting of these molecules will be efficacious in children with high-risk neuroblastoma, in addition to other aggressive pediatric malignancies. The primary focus of a current Stand Up to Cancer-St. Baldrick's Foundation (SU2C-SBF) co-supported “Pediatric Cancer Dream Team” is to bring the fields of genomics and immunology together to further catalyze the discovery of novel immunotherapeutic targets and deliver novel protein-based and cellular treatments across aggressive pediatric malignancies, including neuroblastoma. Finally, given the complexity of the neuroblastoma genome and common acquisition of mutations at relapse, neoantigen discovery may also serve to be an important aspect of defining targets for immunotherapy. Comprehensive understanding of the genetics and genomics of neuroblastoma has led to major clinical advances over the last few decades and holds great promise for continuing to drive significant improvements in clinical care in the years to come.
Footnotes
The authors have no conflicts of interest to disclose. There are no funders to report for this submission.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [January 2015];Surveillance, Epidemiology and End Results Database. (at http://seer.cancer.gov.)
- 3.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 4.Smith MA, Seibel NL, Altekruse SF, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;28:2625–34. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans AE, Silber JH, Shpilsky A, D'Angio GJ. Successful management of low-stage neuroblastoma without adjuvant therapies: a comparison of two decades, 1972 through 1981 and 1982 through 1992, in a single institution. J Clin Oncol. 1996;14:2504–10. doi: 10.1200/JCO.1996.14.9.2504. [DOI] [PubMed] [Google Scholar]
- 6.Cole WH, Everson TC. Spontaneous regression of cancer: preliminary report. Ann Surg. 1956;144:366–83. doi: 10.1097/00000658-195609000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu AL, Gilman AL, Ozkaynak MF, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363:1324–34. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthay KK, Reynolds CP, Seeger RC, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children's oncology group study. J Clin Oncol. 2009;27:1007–13. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kushner BH, Cheung NK, LaQuaglia MP, et al. Survival from locally invasive or widespread neuroblastoma without cytotoxic therapy. J Clin Oncol. 1996;14:373–81. doi: 10.1200/JCO.1996.14.2.373. [DOI] [PubMed] [Google Scholar]
- 10.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 11.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–6. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 12.Attiyeh EF, London WB, Mosse YP, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–53. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 13.Bown N, Cotterill S, Lastowska M, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–61. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 14.Bourhis J, Benard J, DeVathaire F, et al. Combined analysis of DNA ploidy index and N-myc genomic content in neuroblastoma. Prog Clin Biol Res. 1991;366:107–13. [PubMed] [Google Scholar]
- 15.Look AT, Hayes FA, Shuster JJ, et al. Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol. 1991;9:581–91. doi: 10.1200/JCO.1991.9.4.581. [DOI] [PubMed] [Google Scholar]
- 16.George RE, London WB, Cohn SL, et al. Hyperdiploidy plus nonamplified MYCN confers a favorable prognosis in children 12 to 18 months old with disseminated neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol. 2005;23:6466–73. doi: 10.1200/JCO.2005.05.582. [DOI] [PubMed] [Google Scholar]
- 17.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–97. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monclair T, Brodeur GM, Ambros PF, et al. The International Neuroblastoma Risk Group (INRG) staging system: an INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diskin SJ, Capasso M, Schnepp RW, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–30. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janoueix-Lerosey I, Lequin D, Brugieres L, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 25.Capasso M, Diskin S, Cimmino F, et al. Common Genetic Variants in NEFL Influence Gene Expression and Neuroblastoma Risk. Cancer Res. 2014;74:6913–24. doi: 10.1158/0008-5472.CAN-14-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diskin SJ, Capasso M, Diamond M, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106:dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capasso M, Diskin SJ, Totaro F, et al. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34:605–11. doi: 10.1093/carcin/bgs380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latorre V, Diskin SJ, Diamond MA, et al. Replication of neuroblastoma SNP association at the BARD1 locus in African-Americans. Cancer Epidemiol Biomarkers Prev. 2012;21:658–63. doi: 10.1158/1055-9965.EPI-11-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen le B, Diskin SJ, Capasso M, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–91. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell MR, Penikis A, Oldridge DA, et al. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosse KR, Diskin SJ, Cole KA, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72:2068–78. doi: 10.1158/0008-5472.CAN-11-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey GK, Mitra S, Subhash S, et al. The Risk-Associated Long Noncoding RNA NBAT-1 Controls Neuroblastoma Progression by Regulating Cell Proliferation and Neuronal Differentiation. Cancer Cell. 2014;26:722–37. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Oldridge D, Wood AC, Crimmins I, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 oncogene super-enhancer polymorphism. Nature. 2015 doi: 10.1038/nature15540. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barone G, Anderson J, Pearson AD, Petrie K, Chesler L. New strategies in neuroblastoma: Therapeutic targeting of MYCN and ALK. Clin Cancer Res. 2013;19:5814–21. doi: 10.1158/1078-0432.CCR-13-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George RE, Sanda T, Hanna M, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 38.Carpenter EL, Mosse YP. Targeting ALK in neuroblastoma--preclinical and clinical advancements. Nat Rev Clin Oncol. 2012;9:391–9. doi: 10.1038/nrclinonc.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sausen M, Leary RJ, Jones S, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–7. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–84. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 42.Cheung NK, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–71. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudson AG, Jr., Strong LC. Mutation and cancer: neuroblastoma and pheochromocytoma. Am J Hum Genet. 1972;24:514–32. [PMC free article] [PubMed] [Google Scholar]
- 44.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bresler SC, Weiser DA, Huwe PJ, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26:682–94. doi: 10.1016/j.ccell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Pontual L, Kettaneh D, Gordon CT, et al. Germline gain-of-function mutations of ALK disrupt central nervous system development. Hum Mutat. 2011;32:272–6. doi: 10.1002/humu.21442. [DOI] [PubMed] [Google Scholar]
- 47.Mosse YP, Lim MS, Voss SD, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol. 2013;14:472–80. doi: 10.1016/S1470-2045(13)70095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raabe EH, Laudenslager M, Winter C, et al. Prevalence and functional consequence of PHOX2B mutations in neuroblastoma. Oncogene. 2008;27:469–76. doi: 10.1038/sj.onc.1210659. [DOI] [PubMed] [Google Scholar]
- 49.Amiel J, Laudier B, Attie-Bitach T, et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat Genet. 2003;33:459–61. doi: 10.1038/ng1130. [DOI] [PubMed] [Google Scholar]
- 50.Maris JM, Morton CL, Gorlick R, et al. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP). Pediatr Blood Cancer. 2010;55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molenaar JJ, Domingo-Fernandez R, Ebus ME, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 52.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bourdon JC, Fernandes K, Murray-Zmijewski F, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosoi G, Hara J, Okamura T, et al. Low frequency of the p53 gene mutations in neuroblastoma. Cancer. 1994;73:3087–93. doi: 10.1002/1097-0142(19940615)73:12<3087::aid-cncr2820731230>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 55.Schleiermacher G, Mosseri V, London WB, et al. Segmental chromosomal alterations have prognostic impact in neuroblastoma: a report from the INRG project. Br J Cancer. 2012;107:1418–22. doi: 10.1038/bjc.2012.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bown N, Lastowska M, Cotterill S, et al. 17q gain in neuroblastoma predicts adverse clinical outcome. U.K. Cancer Cytogenetics Group and the U.K. Children's Cancer Study Group. Med Pediatr Oncol. 2001;36:14–9. doi: 10.1002/1096-911X(20010101)36:1<14::AID-MPO1005>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 57.Maris JM, Weiss MJ, Guo C, et al. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: a Children's Cancer Group study. J Clin Oncol. 2000;18:1888–99. doi: 10.1200/JCO.2000.18.9.1888. [DOI] [PubMed] [Google Scholar]
- 58.Caron H, van Sluis P, van Hoeve M, et al. Allelic loss of chromosome 1p36 in neuroblastoma is of preferential maternal origin and correlates with N-myc amplification. Nat Genet. 1993;4:187–90. doi: 10.1038/ng0693-187. [DOI] [PubMed] [Google Scholar]
- 59.Baker DL, Schmidt ML, Cohn SL, et al. Outcome after reduced chemotherapy for intermediate-risk neuroblastoma. N Engl J Med. 2010;363:1313–23. doi: 10.1056/NEJMoa1001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagatell R, Rumcheva P, London WB, et al. Outcomes of children with intermediate-risk neuroblastoma after treatment stratified by MYCN status and tumor cell ploidy. J Clin Oncol. 2005;23:8819–27. doi: 10.1200/JCO.2004.00.2931. [DOI] [PubMed] [Google Scholar]
- 61.Janoueix-Lerosey I, Schleiermacher G, Michels E, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–33. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 62.Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–30. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 63.Morales La Madrid A, Nall MB, Ouyang K, et al. Two cases of localized neuroblastoma with multiple segmental chromosomal alterations and metastatic progression. Pediatr Blood Cancer. 2013;60:332–5. doi: 10.1002/pbc.24311. [DOI] [PubMed] [Google Scholar]
- 64.Boeva V, Jouannet S, Daveau R, et al. Breakpoint features of genomic rearrangements in neuroblastoma with unbalanced translocations and chromothripsis. PLoS One. 2013;8:e72182. doi: 10.1371/journal.pone.0072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fransson S, Hansson M, Ruuth K, et al. Intragenic anaplastic lymphoma kinase (ALK) rearrangements: Translocations as a novel mechanism of ALK activation in neuroblastoma tumors. Genes Chromosomes Cancer. 2015;54:99–109. doi: 10.1002/gcc.22223. [DOI] [PubMed] [Google Scholar]
- 66.Cazes A, Louis-Brennetot C, Mazot P, et al. Characterization of rearrangements involving the ALK gene reveals a novel truncated form associated with tumor aggressiveness in neuroblastoma. Cancer Res. 2013;73:195–204. doi: 10.1158/0008-5472.CAN-12-1242. [DOI] [PubMed] [Google Scholar]
- 67.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cascon A, Robledo M. MAX and MYC: a heritable breakup. Cancer Res. 2012;72:3119–24. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 69.Burkhart CA, Cheng AJ, Madafiglio J, et al. Effects of MYCN antisense oligonucleotide administration on tumorigenesis in a murine model of neuroblastoma. J Natl Cancer Inst. 2003;95:1394–403. doi: 10.1093/jnci/djg045. [DOI] [PubMed] [Google Scholar]
- 70.Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–7. doi: 10.1016/j.bbrc.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luscher B, Larsson LG. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene. 1999;18:2955–66. doi: 10.1038/sj.onc.1202750. [DOI] [PubMed] [Google Scholar]
- 72.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3:a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Josling GA, Selvarajah SA, Petter M, Duffy MF. The role of bromodomain proteins in regulating gene expression. Genes (Basel) 2012;3:320–43. doi: 10.3390/genes3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Puissant A, Frumm SM, Alexe G, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bandopadhayay P, Bergthold G, Nguyen B, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–25. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wyce A, Ganji G, Smitheman KN, et al. BET inhibition silences expression of MYCN and BCL2 and induces cytotoxicity in neuroblastoma tumor models. PLoS One. 2013;8:e72967. doi: 10.1371/journal.pone.0072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.White PS, Thompson PM, Seifried BA, et al. Detailed molecular analysis of 1p36 in neuroblastoma. Med Pediatr Oncol. 2001;36:37–41. doi: 10.1002/1096-911X(20010101)36:1<37::AID-MPO1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 80.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kurihara S, Hiyama E, Onitake Y, Yamaoka E, Hiyama K. Clinical features of ATRX or DAXX mutated neuroblastoma. J Pediatr Surg. 2014;49:1835–8. doi: 10.1016/j.jpedsurg.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 83.Lundberg G, Sehic D, Lansberg JK, et al. Alternative lengthening of telomeres--an enhanced chromosomal instability in aggressive non-MYCN amplified and telomere elongated neuroblastomas. Genes Chromosomes Cancer. 2011;50:250–62. doi: 10.1002/gcc.20850. [DOI] [PubMed] [Google Scholar]
- 84.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 85.Mora J, Cheung NK, Gerald WL. Genetic heterogeneity and clonal evolution in neuroblastoma. Br J Cancer. 2001;85:182–9. doi: 10.1054/bjoc.2001.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gotoh T, Sugihara H, Matsumura T, Katsura K, Takamatsu T, Sawada T. Human neuroblastoma demonstrating clonal evolution in vivo. Genes Chromosomes Cancer. 1998;22:42–9. doi: 10.1002/(sici)1098-2264(199805)22:1<42::aid-gcc6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 87.Bohm N, Lampert F. [DNA-ploidy and proliferation in metastatic neuroblastoma]. Klin Padiatr. 1982;194:270–4. doi: 10.1055/s-2008-1033816. [DOI] [PubMed] [Google Scholar]
- 88.Wei JS, Johansson P, Chen L, et al. Massively parallel sequencing reveals an accumulation of de novo mutations and an activating mutation of LPAR1 in a patient with metastatic neuroblastoma. PLoS One. 2013;8:e77731. doi: 10.1371/journal.pone.0077731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schleiermacher G, Javanmardi N, Bernard V, et al. Emergence of new ALK mutations at relapse of neuroblastoma. J Clin Oncol. 2014;32:2727–34. doi: 10.1200/JCO.2013.54.0674. [DOI] [PubMed] [Google Scholar]
- 90.Martinsson T, Eriksson T, Abrahamsson J, et al. Appearance of the novel activating F1174S ALK mutation in neuroblastoma correlates with aggressive tumor progression and unresponsiveness to therapy. Cancer Res. 2011;71:98–105. doi: 10.1158/0008-5472.CAN-10-2366. [DOI] [PubMed] [Google Scholar]
- 91.Eleveld TF, Oldridge DA, Bernard V, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015 doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bentires-Alj M, Paez JG, David FS, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–20. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 93.Holzel M, Huang S, Koster J, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–29. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sidell N, Altman A, Haussler MR, Seeger RC. Effects of retinoic acid (RA) on the growth and phenotypic expression of several human neuroblastoma cell lines. Exp Cell Res. 1983;148:21–30. doi: 10.1016/0014-4827(83)90184-2. [DOI] [PubMed] [Google Scholar]
- 95.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children's Cancer Group. N Engl J Med. 1999;341:1165–73. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 96.Helson L, Helson C. Human neuroblastoma cells and 13-cis-retinoic acid. J Neurooncol. 1985;3:39–41. doi: 10.1007/BF00165170. [DOI] [PubMed] [Google Scholar]
- 97.Kratz CP, Rapisuwon S, Reed H, Hasle H, Rosenberg PS. Cancer in Noonan, Costello, cardiofaciocutaneous and LEOPARD syndromes. Am J Med Genet C Semin Med Genet. 2011;157C:83–9. doi: 10.1002/ajmg.c.30300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mutesa L, Pierquin G, Janin N, et al. Germline PTPN11 missense mutation in a case of Noonan syndrome associated with mediastinal and retroperitoneal neuroblastic tumors. Cancer Genet Cytogenet. 2008;182:40–2. doi: 10.1016/j.cancergencyto.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 99.Origone P, Defferrari R, Mazzocco K, Lo Cunsolo C, De Bernardi B, Tonini GP. Homozygous inactivation of NF1 gene in a patient with familial NF1 and disseminated neuroblastoma. Am J Med Genet A. 2003;118A:309–13. doi: 10.1002/ajmg.a.10167. [DOI] [PubMed] [Google Scholar]
- 100.Rader J, Russell MR, Hart LS, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19:6173–82. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gogolin S, Ehemann V, Becker G, et al. CDK4 inhibition restores G(1)-S arrest in MYCN-amplified neuroblastoma cells in the context of doxorubicin-induced DNA damage. Cell Cycle. 2013;12:1091–104. doi: 10.4161/cc.24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molenaar JJ, Ebus ME, Koster J, et al. Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 2008;68:2599–609. doi: 10.1158/0008-5472.CAN-07-5032. [DOI] [PubMed] [Google Scholar]
- 103.Krasnoselsky AL, Whiteford CC, Wei JS, et al. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene. 2005;24:1533–41. doi: 10.1038/sj.onc.1208341. [DOI] [PubMed] [Google Scholar]
- 104.Molenaar JJ, van Sluis P, Boon K, Versteeg R, Caron HN. Rearrangements and increased expression of cyclin D1 (CCND1) in neuroblastoma. Genes Chromosomes Cancer. 2003;36:242–9. doi: 10.1002/gcc.10166. [DOI] [PubMed] [Google Scholar]
- 105.Thompson PM, Maris JM, Hogarty MD, et al. Homozygous deletion of CDKN2A (p16INK4a/p14ARF) but not within 1p36 or at other tumor suppressor loci in neuroblastoma. Cancer Res. 2001;61:679–86. [PubMed] [Google Scholar]
- 106.Easton J, Wei T, Lahti JM, Kidd VJ. Disruption of the cyclin D/cyclin-dependent kinase/INK4/retinoblastoma protein regulatory pathway in human neuroblastoma. Cancer Res. 1998;58:2624–32. [PubMed] [Google Scholar]
- 107.Cole KA, Huggins J, Laquaglia M, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci U S A. 2011;108:3336–41. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caren H, Erichsen J, Olsson L, et al. High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: four cases of homozygous deletions of the CDKN2A gene. BMC Genomics. 2008;9:353. doi: 10.1186/1471-2164-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Molenaar JJ, Koster J, Ebus ME, et al. Copy number defects of G1-cell cycle genes in neuroblastoma are frequent and correlate with high expression of E2F target genes and a poor prognosis. Genes Chromosomes Cancer. 2012;51:10–9. doi: 10.1002/gcc.20926. [DOI] [PubMed] [Google Scholar]
- 110.Carr-Wilkinson J, O'Toole K, Wood KM, et al. High Frequency of p53/MDM2/p14ARF Pathway Abnormalities in Relapsed Neuroblastoma. Clin Cancer Res. 2010;16:1108–18. doi: 10.1158/1078-0432.CCR-09-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beiske K, Burchill SA, Cheung IY, et al. Consensus criteria for sensitive detection of minimal neuroblastoma cells in bone marrow, blood and stem cell preparations by immunocytology and QRT-PCR: recommendations by the International Neuroblastoma Risk Group Task Force. Br J Cancer. 2009;100:1627–37. doi: 10.1038/sj.bjc.6605029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diaz LA, Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kojima M, Hiyama E, Fukuba I, et al. Detection of MYCN amplification using blood plasma: noninvasive therapy evaluation and prediction of prognosis in neuroblastoma. Pediatr Surg Int. 2013;29:1139–45. doi: 10.1007/s00383-013-3374-9. [DOI] [PubMed] [Google Scholar]
- 116.Gotoh T, Hosoi H, Iehara T, et al. Prediction of MYCN amplification in neuroblastoma using serum DNA and real-time quantitative polymerase chain reaction. J Clin Oncol. 2005;23:5205–10. doi: 10.1200/JCO.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 117.Combaret V, Brejon S, Iacono I, et al. Determination of 17q gain in patients with neuroblastoma by analysis of circulating DNA. Pediatr Blood Cancer. 2011;56:757–61. doi: 10.1002/pbc.22816. [DOI] [PubMed] [Google Scholar]
- 118.Combaret V, Iacono I, Bellini A, et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med. 2015;4:540–50. doi: 10.1002/cam4.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Birch JM, Alston RD, McNally RJ, et al. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20:4621–8. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 120.Tatton-Brown K, Murray A, Hanks S, et al. Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am J Med Genet A. 2013;161A:2972–80. doi: 10.1002/ajmg.a.36229. [DOI] [PubMed] [Google Scholar]
- 121.Schimke RN, Collins DL, Stolle CA. Paraganglioma, neuroblastoma, and a SDHB mutation: Resolution of a 30-year-old mystery. Am J Med Genet A. 2010;152A:1531–5. doi: 10.1002/ajmg.a.33384. [DOI] [PubMed] [Google Scholar]
- 122.Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–57. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Omura-Minamisawa M, Diccianni MB, Chang RC, et al. p16/p14(ARF) cell cycle regulatory pathways in primary neuroblastoma: p16 expression is associated with advanced stage disease. Clin Cancer Res. 2001;7:3481–90. [PubMed] [Google Scholar]
- 124.Chen L, Rousseau RF, Middleton SA, et al. Pre-clinical evaluation of the MDM2-p53 antagonist RG7388 alone and in combination with chemotherapy in neuroblastoma. Oncotarget. 2015;6:10207–21. doi: 10.18632/oncotarget.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gamble LD, Kees UR, Tweddle DA, Lunec J. MYCN sensitizes neuroblastoma to the MDM2-p53 antagonists Nutlin-3 and MI-63. Oncogene. 2012;31:752–63. doi: 10.1038/onc.2011.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Van Maerken T, Rihani A, Dreidax D, et al. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Mol Cancer Ther. 2011;10:983–93. doi: 10.1158/1535-7163.MCT-10-1090. [DOI] [PubMed] [Google Scholar]