AIM2 inflammasome in infection, cancer and autoimmunity: role in DNA sensing, inflammation and innate immunity (original) (raw)
. Author manuscript; available in PMC: 2017 Feb 1.
Published in final edited form as: Eur J Immunol. 2015 Dec 28;46(2):269–280. doi: 10.1002/eji.201545839
Abstract
Recognition of DNA by the cell is an important immunological signature that marks the initiation of an innate immune response. AIM2 is a cytoplasmic sensor that recognizes dsDNA of microbial or host origin. Upon binding to DNA, AIM2 assembles a multi-protein complex called the inflammasome, which drives pyroptosis and proteolytic cleavage of the pro-inflammatory cytokines pro-IL-1β and pro-IL-18. Release of microbial DNA into the cytoplasm during infection by Francisella, Listeria, Mycobacterium, mouse cytomegalovirus, vaccinia virus, Aspergillus and Plasmodium species leads to activation of the AIM2 inflammasome. In contrast, inappropriate recognition of cytoplasmic self-DNA by AIM2 contributes to the development of psoriasis, dermatitis, arthritis and other autoimmune and inflammatory diseases. Inflammasome-independent functions of AIM2 have also been described, including the regulation of the intestinal stem cell proliferation and the gut microbiota ecology in the control of colorectal cancer. In this review we provide an overview of the latest research on AIM2 inflammasome and its role in infection, cancer and autoimmunity.
Keywords: AIM2 inflammasome, autoimmunity, cancer, DNA sensing, bacterial/viral infection, gut microbiota
Introduction
DNA recognition by innate immune receptors triggers a myriad of immunological responses that are both beneficial and detrimental to the host. The discovery of Toll-like receptor 9 (TLR9) as a membrane-associated sensor of bacterial CpG DNA provides evidence for the existence of host receptors that specifically mediate immune responses to DNA [1]. Translocation of microbial or mammalian DNA into the cytoplasm of host cells further induces transcription of genes encoding type I interferon (IFN) molecules and inflammation independently of TLR9 [2, 3]. Recent advances in the field have identified multiple cytoplasmic DNA sensors which are responsible for transcriptional activity, including cyclic-GMP-AMP synthase (cGAS), STING, DDX41, Ku70, LRRFIP1, DNA-dependent activator of IRFs (DAI, also known as ZBP1) and IFI16 (reviewed elsewhere [4–6]). Of these DNA sensors, cGAS binds to double-stranded DNA (dsDNA), resulting in a conformational change in cGAS that allows it to convert ATP and GTP to a cyclic dinucleotide cyclic-GMP-AMP (cGAMP) [7]. cGAMP then binds and activates STING to induce transcription of genes encoding type I IFN and pro-inflammatory cytokines via the transcription factors IRF3 and NF-κB, respectively (reviewed in [7]). The molecular basis underlying recognition of DNA by the other aforementioned cytoplasmic DNA sensors is less understood.
DNA introduced into the cytoplasm also induces IL-1β secretion and pyroptosis [8, 9], and these responses are dependent on the activity of a cytoplasmic caspase-1-containing complex known as the inflammasome [10]. In 2009, four groups independently identified AIM2 as the sensor that triggers inflammasome activation, pyroptosis and release of IL-1β and IL-18 in response to intracellularly-delivered dsDNA [11–14].
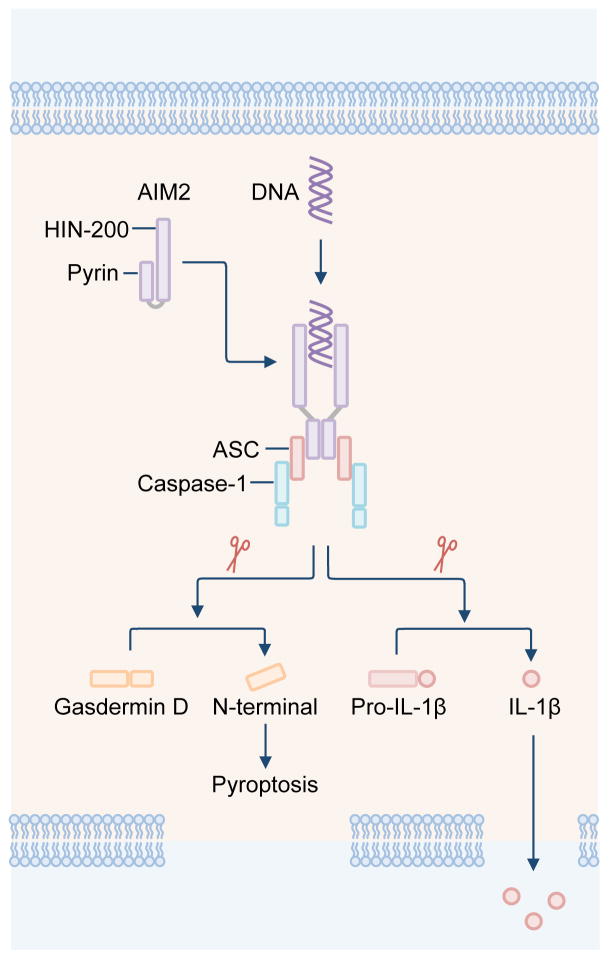
AIM2 consists of a C-terminal HIN-200 domain, which binds directly to dsDNA, and an N-terminal pyrin domain (PYD), which interacts with the PYD of the bipartite PYD-CARD-containing inflammasome adaptor protein ASC (apoptosis-associated speck-like protein containing a carboxy-terminal CARD) [10]. The CARD of ASC binds the CARD of pro-caspase-1, forming a macromolecular complex fulfilling the basic structural elements of an inflammasome (Figure 1) [10]. AIM2, IFI16 and other pyrin and HIN domain-containing (PYHIN) proteins form the AIM2-like receptor family [15–17].
Figure 1. The molecular basis for the activation of the AIM2 inflammasome.
The DNA sensor AIM2 is composed of an N-terminal pyrin domain and a C-terminal HIN-200 domain. The pyrin and HIN-200 domain of AIM2 form an intramolecular complex and are maintained in an autoinhibitory state. Cytoplasmic dsDNA induces activation of AIM2. The HIN-200 domain interacts with dsDNA in a sequence-independent manner, by binding to the sugar-phosphate backbone of dsDNA. The pyrin domain of AIM2 binds to the pyrin domain of ASC. CARD of ASC binds the CARD of pro-caspase-1, forming a macromolecular complex known as the AIM2 inflammasome. Activated caspase-1 drives cleavage of pro-IL-1β and pro-IL-18. Caspase-1 also cleaves the substrate gasdermin D. The N-terminal fragment of gasdermin D induces pyroptosis, allowing mature IL-1β and IL-18 to be released from the cell.
AIM2 recognizes dsDNA in a sequence-independent manner; however, the DNA sequence must be at least 80 base pairs in length [18]. Elucidation of the crystal structure of AIM2 provided insights into the activation mechanism of this DNA-sensing inflammasome. The PYD and HIN-200 domain of AIM2 form an intramolecular complex and are maintained in an autoinhibitory state during homeostasis (Figure 1) [18, 19]. Binding of dsDNA to the HIN-200 domain displaces PYD from the intramolecular complex, liberating PYD for interaction with ASC [18]. The sugar-phosphate backbone of dsDNA interacts with the positively-charged HIN-200 domain via electrostatic attraction, allowing sequence-independent recognition of DNA by AIM2. The PYD of AIM2 can also self-oligomerize to induce the activation of the AIM2 inflammasome [20, 21]. Activation of AIM2 or a related inflammasome sensor NLRP3 initiates polymerization of the PYD of ASC [22, 23], ultimately forming a single and visually distinct inflammasome speck that is readily observed in primary macrophages and DCs [24–29]. Recent work has even suggested that the role of PYD of AIM2 is not to maintain autoinhibition, but to oligomerize and drive filament formation [30].
Activation of the AIM2 inflammasome and other canonical inflammasomes results in a type of inflammatory cell death called pyroptosis [10], which is mediated, in part, by the inflammatory caspase substrate gasdermin D [31, 32] (Figure 1). Activation of the AIM2 inflammasome is tightly regulated by the cell and requires phosphorylation and linear ubiquitination of ASC [33, 34]. Autophagy can mediate degradation of the AIM2 inflammasome to terminate inflammatory responses [35]. Furthermore, several proteins produced in the cell have the ability to inhibit activation of the AIM2 inflammasome [14, 36–43]. These include the PYD-containing proteins POP1 [39] and POP3 [36] in human cells and the bipartite protein containing two HIN domains, p202, in mouse cells [14, 40–43].
Here, we summarize the latest research on the regulation of AIM2 inflammasome, and its role in pathogen recognition after infection, cancer and autoimmunity.
AIM2 in the response to infection
Bacterial infection
During infection of a host cell, microbial DNA and other microbe-associated molecular patterns (MAMPs) are released into the cytoplasm, where they are recognized by cytoplasmic DNA sensors, i.e. cGAS, STING or AIM2. AIM2 resides in the cytoplasm and has been shown to provide immunosurveillance to the pathogenic bacteria Francisella tularensis Live Vaccine Strain (LVS), F. tularensis subspecies novicida (F. novicida), Listeria monocytogenes, Streptococcus pneumonia, Mycobacterium species, Porphyromonas gingivalis, Staphylococcus aureus, Brucella abortus and Chlamydia muridarum (Table 1) [26, 44–61]. F. novicida and F. tularensis LVS are the only bacterial pathogens known to exclusively activate the AIM2 inflammasome in mouse macrophages and DCs, whereas other bacteria have been shown to activate more than one inflammasome receptor in mouse and human cells [44–46]. In the human THP-1 macrophage-like cell line, both AIM2 and NLRP3 contribute to the activation of the inflammasome in response to F. novicida and F. tularensis LVS [48]. Interestingly, caspase-8 is recruited to the AIM2 inflammasome to drive apoptosis in _Francisella_-infected cells in the absence of caspase-1 [47, 62], suggesting a complex interplay between members of the caspase family.
Table 1.
Bacteria recognized by the AIM2 inflammasome
| Bacteria | Cells | Mice |
|---|---|---|
| Francisella tularensis LVS or F. tularensis subspecies novicida (F. novicida) | Reduced caspase-1 activation, IL-1β and IL-18 and pyroptosis in _Aim2_−/− mouse BMDMs or BMDCs [26, 44–47] Reduced IL-1β in shRNA- silenced Aim2 THP1 human monocytic cell line [48] | Reduced serum IL-18 levels 1d p.i. and increased bacterial burden 3d p.i. [44, 45] |
| Listeria monocytogenes | Reduced caspase-1 cleavage, IL-1β release and pyroptosis in _Aim2_-silenced mouse BMDMs [49–52] | N/A |
| Streptococcus pneumoniae | Reduced caspase-1 cleavage, IL-1β and IL-18 release and pyroptosis in _Aim2_-silenced peritoneal mouse macrophages [53] | N/A |
| Mycobacterium tuberculosis | Reduced IL-1β and IL-18 release in _Aim2_−/− and _Aim2_- silenced THP-1 human monocytic cell line [54, 55] | Increased overall, susceptibility, increased bacterial burden in the lungs and liver 4 weeks p.i. [56] Reduced caspase-1 cleavage and increased infiltration of inflammatory cells in lungs. [56] Reduced IL-1β in BALF and IL-18 in serum at 3 weeks p.i. [56] Reduced IFN-γ production by CD4+ T cells [56] |
| Mycobacterium bovis | Reduced caspase-1 cleavage, IL-1β release and pyroptosis in _Aim2_-silenced mouse macrophage J774A.1 [57] | N/A |
| Porphyromonas gingivalis | Reduced caspase-1 cleavage, IL-1β and pyroptosis in siRNA-silenced Aim2 THP-1 human monocytic cell line [58] | N/A |
| Legionella pneumophila Δ_SdhA_ | Reduced caspase-1 cleavage and IL-1β release in _Aim2_−/− mouse BMDMs [78] | N/A |
| Staphylococcus aureus | N/A | Increased susceptibility upon intracranial infection [59] Reduced IL-1β, IL-6, CCL2 and CXCL10 in wound abscess [59] |
| Brucella abortus | Reduced caspase-1 cleavage, IL-1β and cell death in _Aim2_−/− mouse BMDMs [60] | Increased bacterial burden 4 weeks p.i [60] |
| Chlamydia muridarum | Reduced IL-1β and IL-18 in _Aim2_−/− mouse BMDMs [61] | N/A |
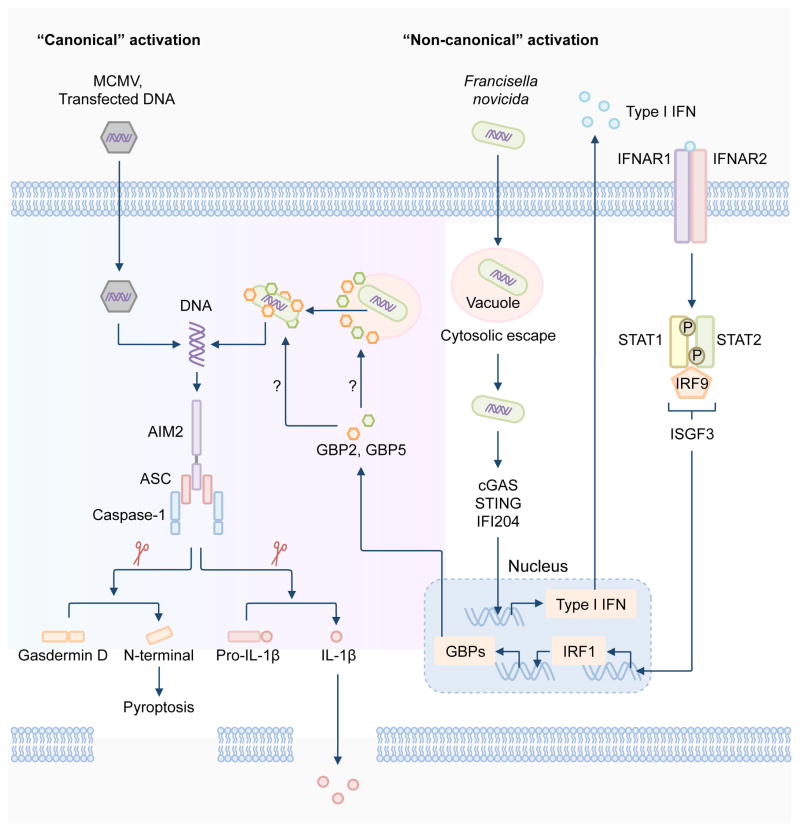
Activation of the AIM2 inflammasome by F. novicida and F. tularensis LVS requires the ability of the bacteria to escape the vacuole into the host cytoplasm, a process mediated by a range of bacterial virulence factors, including the transcriptional regulator MglA and proteins encoded by the _Francisella_-pathogenicity island [26–28, 63, 64]. The _Francisella_-pathogenicity island is a genomic region which contains a cluster of 16 to 19 genes encoding virulence factors of the bacterium [65]. Similarly, L. monocytogenes must escape the vacuole and undergo bacteriolysis in order to induce the activation of the AIM2 inflammasome [49–52, 66]. Type I IFN potentiates the activity of the AIM2 inflammasome during bacterial infection [26–28, 67, 68]. In response to F. novicida infection, the DNA sensors cGAS, IFI204 and STING cooperate to detect small amounts of DNA released by the bacteria to drive production of type I IFN in mouse macrophages [27, 46, 69]. Type I IFN is then released to the outside of the cell, where it binds to the type I IFN receptor (IFNR) in an autocrine manner, activating the IFN-stimulated gene factor 3 (ISGF3) [70]. ISGF3 is comprised of the transcription factors STAT1, STAT2 and IRF9, which drives the transcription of IFN-stimulated genes (ISGs) [70]. A study has now demonstrated that, during infection with F. novicida, signaling via type I IFN induces the expression of the transcription factor IRF1, where IRF1 further drives expression of IFN-inducible GTPases called guanylate-binding proteins (GBPs) [27]. Induction of both IRF1 and GBPs are necessary to fully engage the AIM2 inflammasome by F. novicida infection (Figure 2) [27, 28].
Figure 2. Regulation of the activation of the AIM2 inflammasome.
The AIM2 inflammasome is activated by a number of microbial pathogens and dsDNA ligands, including the DNA virus mouse cytomegalovirus (MCMV), the cytosolic bacterium Francisella novicida and the dsDNA ligand poly(dA:dT). MCMV infection or transfection of poly(dA:dT) leads to “canonical” activation of the AIM2 inflammasome, which does not require the type I interferon (IFN) pathway. F. novicida infection activates the AIM2 inflammasome via a “non-canonical” pathway owing to its requirement for type I IFN, analogous to the non-canonical NLRP3 inflammasome pathway. Intracellular F. novicida releases DNA into the cytoplasm to activate the DNA sensors cGAS, STING and IFI204, which drive transcription of genes encoding type I IFN molecules. It remains unclear why the released DNA is unable to activate AIM2 at this stage, since AIM2 is constitutively expressed in the cell. Type I IFN provides a feedback loop to induce expression of the transcription factor IRF1, which upregulates expression of the IFN-inducible GTPases, including GBP2 and GBP5. GBP2 and GBP5 are recruited to bacterial structures, however, whether they directly target the bacterial membrane or the membrane of intact _Francisella_-containing vacuole is unclear. Nevertheless, GBPs mediate bacterial killing, resulting in abundant release of bacterial DNA for recognition by AIM2. Assembly of the AIM2 inflammasome induces caspase-1-dependent cleavage of pro-IL-1β and pro-IL-18. Caspase-1 also drives cleavage of the substrate gasdermin D to induce pyroptosis.
GBPs are clustered over two locations in the genome of mice. Genes encoding GBP1_,_ GBP2_,_ GBP3_,_ GBP5 and GBP7 are located on chromosome 3, whereas genes encoding GBP4_,_ GBP6_,_ GBP8_,_ GBP9_,_ GBP10 and GBP11 are found on chromosome 5 [71]. Of these, GBP2 and GBP5 are recruited to cytoplasmic F. novicida bacteria to drive bacterial killing, exposing abundant amounts of bacterial DNA for detection by AIM2 [27, 28]. GBP2 and GBP5 function in a non-redundant manner, and reconstitution of either GBP in type I IFN receptor 1-deficient macrophages cannot rescue inflammasome activation, indicating that type I IFN signaling activates GBPs, possibly via expression other IFN inducible proteins [27, 28]. Overall, mice lacking AIM2, caspase-1, IRF1 or GBPs have been shown to secrete reduced levels of IL-18 in response to F. novicida infection and are all hypersusceptible to F. novicida infection compared with wild type mice [27, 28, 44–46]. In agreement with this observation, antibody-mediated neutralization of IL-1β and IL-18 in wild type mice increases susceptibility to F. novicida infection [64].
The precise mechanism of bacterial killing mediated by GBPs remains unknown. Recent studies have shown that the antimicrobial activity of the gp91 subunit of NADPH oxidase (also known as NOX2), and inducible nitric oxide synthase are not required for mediating activation of the AIM2 inflammasome [28, 72]. However, the pharmacological inhibition of reactive oxygen species (ROS) and mitochondrial ROS partially reduces caspase-1 activation, and therefore, the release of IL-1β driven by F. novicida infection [28, 72]. ROS inhibition impairs the expression of IL-1β and TNF, arguing that further evidence is required to convincingly link ROS-mediated bacterial killing and activation of the AIM2 inflammasome [73]. It also remains a mystery as to why DNA molecules that are released to activate cGAS, STING and IFI204 are unable to activate AIM2 at this stage. One possibility is that the concentration of DNA that is sufficient to activate cGAS, STING and IFI204 is lower than the concentration required to activate AIM2. AIM2 is constitutively expressed in the cell, but its expression is also induced by IFN [11, 37, 38]. However, transfection of dsDNA into the cytoplasm can directly activate AIM2 independently of IFN [27, 28, 73], ruling out a requirement for “priming” in activation of the AIM2 inflammasome.
Bacteria have evolved virulence determinants to prevent release of DNA and other bacterial ligands and avoid cytoplasmic detection and clearance by inflammasomes. F. tularensis subspecies tularensis SchuS4 have been shown to induce low levels of inflammasome activation and IL-1β secretion in primary mouse bone marrow-derived macrophages (BMDMs), possibly due to enhanced resistance to H2O2 to protect itself from bacteriolysis, or other virulence factors that confer evasion of the immune system [72]. The putative lipid II flippase, MviN, and RipA, a protein used for intracellular replication, of F. tularensis LVS are both required to dampen AIM2 inflammasome responses [74, 75]. Further studies have shown that F. tularensis LVS or F. novicida mutants lacking MviN, RipA, and several membrane-associated proteins or proteins involved in O-antigen or LPS biosynthesis are hypersusceptible to intracellular lysis and DNA release in macrophages, providing a rationale for why these mutants induce elevated activation of the AIM2 inflammasome [63]. Genes encoding the 5-formyltetrahydrofolate cycloligase within the folate metabolic pathway and pseudouridine synthase in F. tularensis LVS have also been demonstrated to influence the magnitude of AIM2 inflammasome activation [76]. A more recent study identified a clustered, regularly interspaced, short palindromic repeats-CRISPR associated (CRISPR-Cas) system used by F. novicida to strengthen the integrity of its bacterial membrane, leading to reduced DNA release in the cytoplasm [77]. Another example is found in Legionella pneumophila, which encodes an effector protein SdhA, shown to prevent rupture of the _Legionella_-containing vacuole and thereby minimizing the amount of bacterial DNA released into the cytoplasm [78].
There is limited evidence so far to support the existence of mechanisms used by bacteria to directly inhibit or evade activation of the AIM2 inflammasome. However, F. tularensis LVS and the virulent SchuS4 strain do suppress TLR2-dependent responses to reduce the level of pro-IL-1β available for cleavage by the inflammasome [79]. Overall, the AIM2 inflammasome is an effective antimicrobial machinery against certain bacterial pathogens.
Viral infection
Inflammasome responses play an essential role in the host protection against viral infection [80, 81]. Genetic materials from DNA viruses which enter the cytoplasm can be detected by AIM2, for instance mouse cytomegalovirus (MCMV), vaccinia virus and human papillomaviruses (Table 2) [11, 44, 82]. MCMV and vaccinia viruses robustly induce inflammasome responses in mouse macrophages in an AIM2-dependent manner (Figure 2) [11, 27, 44]. Further, _Aim2_−/− mice infected with MCMV have an impaired ability to secrete IL-18, carry a higher viral titre, and have reduced levels of IFN-γproducing NK cells compared with infected wild type mice [44]. Human papillomaviruses have also been shown to drive IL-1β and IL-18 release in human keratinocytes in an AIM2-dependent manner [82].
Table 2.
Viruses recognized by the AIM2 inflammasome
| Viruses | Cells | Mice |
|---|---|---|
| Mouse cytomegalovirus (DNA virus) | Reduced caspase-1 cleavage and IL-1β release _Aim2_−/− mouse peritoneal macrophages and BMDCs [44] | Increased viral titers 36 h p.i. [44] Reduced serum IL-18 level 36 h p.i. [44] Reduced IFN-γ production [44] |
| Vaccinia virus (DNA virus) | Reduced caspase-1 cleavage and IL-1β release _Aim2_−/− mouse peritoneal macrophages and BMDCs [44] | N/A |
| Human papillomaviruses (DNA virus) | Reduced IL-1β and IL-18 release in _Aim2_-silenced human keratinocytes [82] | N/A |
| Hepatitis B virus (DNA virus) | Reduced gene expression of IL- 1β, IL-18 and caspase-1 in _Aim2_-silenced human glomerular mesangial cell line [83] | N/A |
| Chikungunya virus (RNA virus) | Reduced IL-1β release in _Aim2_- silenced human primary dermal fibroblasts [92] | N/A |
| West Nile virus (RNA virus) | Reduced IL-1β release in _Aim2_- silenced primary human dermal fibroblasts [92] | N/A |
To date, there is no strong evidence in the literature to indicate that DNA viruses other than MCMV and vaccinia virus activate the AIM2 inflammasome. A recent study suggests that in the human glomerular mesangial cell line infected with hepatitis B virus, siRNA-mediated silencing of the gene encoding AIM2 leads to a reduced expression of IL-1β, IL-18 and caspase-1 [83]. Whether AIM2 directly mediates the recognition of viral DNA derived from hepatitis B virus in immune or non-immune cells has not been established. Comparison of the expression of AIM2 in patients with acute and chronic hepatitis B revealed that those at the acute stage expressed higher levels of AIM2 in peripheral blood mononuclear cells [84]. A subsequent study reported that 89.4% of the liver tissues collected from individuals with chronic hepatitis B virus infection were positive for AIM2 expression by immunohistochemistry, compared with only 8.7 % of those with chronic hepatitis C infection [85]. Expression of the gene encoding AIM2 has been reported to be significantly higher in kidney tissues of patients with hepatitis B virus-associated glomerulonephritis compared with patients with chronic glomerulonephritis [83].
In all cases, it is probable that viral DNA binds directly to AIM2 to trigger inflammasome activation, but the precise molecular mechanism that leads to exposure of the viral DNA for sensing by AIM2 is not entirely clear. Unlike F. novicida infection, type I IFN signaling, IRF1 and GBPs are dispensable for the activation of the AIM2 inflammasome by either MCMV infection or transfected dsDNA [27]. However, AIM2 does not respond to all DNA viruses. For example, adenovirus and herpesviruses HSV-1 and MHV-68 activate the NLRP3 or putative IFI16 inflammasome, rather than the AIM2 inflammasome in mouse bone marrow-derived macrophages or mouse thioglycollate-elicited macrophages [8, 44, 80, 81]. During HSV-1 infection of human macrophages, the capsid that encapsulates the viral DNA is degraded by the proteasome, which releases DNA for recognition by IFI16 in the cytoplasm [86], suggesting that the inability of AIM2 to sense viral DNA is probably not due to a lack of viral DNA release in the cytoplasm. It might be possible that certain DNA viruses can strategically inhibit the ability of AIM2 to interact with their DNA. Alternatively, the DNA of certain viruses, such as Hepatitis B virus and other Hepadnaviruses, could be transcribed into RNA templates, which may serve as activators for NLRP3 [87–89]. Indeed, a precedent exists for indirect sensing of DNA by the RNA sensor RIG-I [90, 91]. RNA polymerase III transcribes AT-rich DNA into dsRNA transcripts carrying an uncapped 5′-triphosphate moiety, which has been shown to activate RIG-I [90, 91]. Conversely, a study has reported a role for AIM2 in driving IL-1β secretion in response to the RNA viruses [92]. Silencing of genes encoding AIM2 and caspase-1 reduces proteolytic cleavage and release of IL-1β in human dermal fibroblasts infected with the RNA viruses Chikungunya virus or West Nile virus [92]. How AIM2 might sense RNA viruses is still unclear, and further characterization of the molecular mechanism involved in the activation of the AIM2 inflammasome by viruses is required.
Other pathogens
In addition to bacteria and viruses, AIM2 has been shown to mediate pathogen recognition of, and host defense to, the fungal pathogen Aspergillus fumigatus and the protozoan Plasmodium berghei (Table 3) [93, 94]. AIM2 and NLRP3 function in a redundant fashion to confer inflammasome activation against A. fumigatus infection in mouse bone-marrow derived DCs and mice [93] Furthermore, mice lacking both AIM2 and NLRP3, ASC or caspase-1, and infected with A. fumigatus, are more susceptible than infected wild type mice [93]. The requirement for dual sensing of pathogens by both AIM2 and NLRP3 has also been observed in mouse BMDMs stimulated with _Plasmodium berghei_-infected red blood cells or synthetic and natural hemozoins [94]. AIM2 also directly recognizes A. fumigatus genomic DNA introduced into the cytoplasm by a transfection agent [93] or P. falciparum genomic DNA transported into the cytoplasm by hemozoins [94]. It would be interesting to identify additional pathogens that can activate the AIM2 inflammasome.
Table 3.
Fungi and parasites recognized by the AIM2 inflammasome
| Fungi | Cells | Mice |
|---|---|---|
| Aspergillus fumigatus | Slight reduction in IL-1β and IL-18 in _Aim2_−/− mouse BMDCs owing to redundant roles with NLRP3 [93] | Not susceptible owing to redundant roles with NLRP3 [93] |
| Protozoa | ||
| Plasmodium berghei | Slight reduction in IL-1β and pyroptosis in _Aim2_−/− mouse BMDMs infected with iRBCs, synthetic and natural hemozoin owing to redundant roles with NLRP3 [94] | Decreased neutrophils recruitment in peritoneal cavity owing to redundant roles with NLRP3, after 15 h p.i. [94] |
AIM2 role in cancer biology
AIM2 has also been shown to suppress the development of cancer [95, 96]. The gene encoding AIM2 was originally isolated from human melanoma cells [97]. Reduced expression and frequent frameshift microsatellite instability of the AIM2 have been observed in tumor tissues from patients with colorectal cancer [98–101]. Colorectal cancer patients whose tissues have reduced AIM2 expression have a poorer prognosis compared with those with a higher level of AIM2 expression [98]. Reduced expression of AIM2 has also been reported in prostate cancer [102], whereas increased expression has been detected in nasopharyngeal carcinoma tumors [103, 104], oral squamous cell carcinoma [105], and lung adenocarcinoma [106]. The differential expression of AIM2 in a range of tumor tissues suggests that it may have unique roles in different types of cancer.
The mechanism for AIM2 in the regulation of tumorigenesis has been described in a mouse model of colitis-associated colorectal cancer [95, 96]. Two groups have recently demonstrated that AIM2 operates independently of the inflammasome to prevent colorectal cancer [95, 96]. Both studies found that _Aim2_−/− mice developed severe colitis, polyps and higher tumor burden upon administration of AOM and DSS [95, 96]. Although differential production of the major inflammatory mediators, including TNF and IL-6, was not observed between wild type and _Aim2_−/− mice, proliferation of enterocytes was more pronounced in _Aim2_−/− mice [95, 96]. Indeed, murine fibroblasts and colon cancer cell lines expressing an AIM2-encoding construct have been shown to have an impaired ability to undergo proliferation [40, 107]. Overexpression of AIM2 in colon cancer cell lines also induces cell cycle arrest, and the transformed cells exhibit a delayed transition from the late S-phase to the G2/M phase [107], suggesting that AIM2 plays a proliferation-inhibitory role in these cancer cell lines.
Furthermore, in another mouse model of intestinal cancer, _Aim2_−/− mice carrying aberrant activating β-catenin mutations failed to prevent the expansion of cancer-associated stem cells in the small and large intestine [95]. Similarly, _Aim2_−/− mice harboring the heterozygous mutation in the adenomatous polyposis coli (Apc) gene developed more tumors than mice carrying only the mutant Apc gene [96]. Intestinal stem cells lacking AIM2 proliferated more than wild type intestinal stem cells in organoid culture, and this proliferation was associated with increased activation of the kinase AKT [95, 96]. Wilson and colleagues identified DNA-PK, a kinase that can phosphorylate and activate AKT [108, 109], as a binding partner of AIM2, whereby AIM2 suppresses the activation of DNA-PK and DNA-PK-dependent phosphorylation of AKT at the serine residue 473 [96]. Indeed, treatment of _Aim2_−/− mice with an AKT inhibitor reduced tumor burden in _Aim2_−/− mice with colitis [96].
Our laboratory performed further analysis and found that _Aim2_−/− mice harbored a microbial ecology different to that of wild type mice [95]. Reciprocal exchange of the microbiota between _Aim2_−/− and wild type mice by means of co-housing substantially reduced tumorigenesis in _Aim2_−/− mice and increased tumorigenesis in wild type mice [95]. Collectively, these studies provide insights into the function of AIM2 in colorectal cancer, and highlight that potential therapies that inhibit the AKT pathway can be further investigated for treatment of cancer associated with AIM2 mutations [40, 95, 96, 107].
AIM2 in inflammatory, autoimmune and other pathological conditions
Given that the host DNA is normally sequestered in the nucleus or mitochondria, the accumulation of host DNA in the cytosol, due to impaired degradation or clearance or excess uptake of extracellular DNA from dying neighboring cells, could induce inflammation. For instance, accumulated DNA can serve as an endogenous danger signal, and has been shown to trigger AIM2-dependent release of IL-1β in skin cells, contributing to the pathogenesis of psoriasis [38]. Scavenging of DNA by the antimicrobial cathelicidin peptide LL-37 produced by the inflamed skin of psoriasis patients prevents overt activation the AIM2 inflammasome and IL-1β release [38]. Increased AIM2 expression has been observed in patients with acute and chronic skin conditions, including psoriasis, atopic dermatitis, venous ulcera, contact dermatitis and experimental wounds in humans [38, 110]. In addition, expression of the gene encoding AIM2 is elevated in immune cells of male patients with systemic lupus erythematosus (SLE) and both increases and decreases in AIM2 expression have been observed in female patients with SLE [111, 112]. Further, DNA methylation of the gene encoding AIM2 is reduced in patients with SLE compared with their healthy siblings [113], suggesting that differential expression or epigenetic changes could be linked to development of the disease.
Increased expression of AIM2 has been reported in patients with inflammatory bowel diseases and liver inflammation [114–116]. For example, elevated expression of AIM2 has been detected in ascitic fluid macrophages collected from cirrhotic patients, compared with PBMCs from the same patients, or with CD14+ macrophages from peripheral blood mononuclear cells of healthy individuals [115]. Increased expression of AIM2 and NLRP3 and elevated activation of caspase-1 and maturation of IL-1β have been found in liver tissues of mice with steatohepatitis [116].
Moreover, there is evidence to suggest that AIM2 is involved in inflammation and cell death of the brain. Cell-free DNA fragments are more frequently detected in the cerebrospinal fluid (CSF) of patients with traumatic brain injury than in CSF from nontrauma patients [117]. When human embryonic cortical neurons, which express AIM2 inflammasome components and have been shown to be capable of IL-1β release and cell death upon AIM2 activation with poly(dA:dT) transfection [117], were exposed to the CSF of traumatic brain injury patients, they exhibited increased AIM2 expression and caspase-1 activation compared with embryonic cortical neurons which had been exposed to the CSF of non-trauma patients [117]. A further study has shown that mice lacking AIM2 are more protected than wild type mice to ischemic brain injury [118]. Upon induction of focal cerebral ischemia, less activation of microglial cells and recruitment of leukocytes were found in _Aim2_−/− mice compared with wild type mice [118].
The inability to degrade self-DNA also contributes to the pathogenesis of autoimmune polyarthritis. Mice lacking the lysosomal endonuclease DNAse II (_Dnase II_−/− mice) are embryonically lethal, owing to an impaired ability to degrade self-DNA by macrophages [119]. Genomic deletion of type I IFN receptor (IFNAR) rescued _Dnase II_−/− mice from embryonic lethality [120]; however, the mice lacking both IFNAR and DNAse II (_Dnase II_−/−_Ifnar_−/− mice) would eventually develop polyarthritis [121]. Intriguingly, genomic deletion of AIM2 was shown to prevent inflammasome activation, inflammatory cytokine production, macrophage infiltration in the joint and the development of arthritis in _Dnase II_−/−_Ifnar_−/− mice [122, 123]. Genomic deletion of STING was also shown to protect _Dnase II_−/−_Ifnar_−/− mice from joint inflammation [122], indicating that multiple DNA sensors might contribute to the inappropriate DNA recognition driving clinical manifestation. Overall, aberrant activation of AIM2 from self-DNA is a key driver of inflammatory and autoimmune diseases.
Conclusions
A wide range of microbial pathogens is sensed by AIM2 in mammalian cells. Recognition of DNA from pathogens by AIM2 leads to protective inflammasome-mediated host responses. Recent studies have demonstrated that AIM2 inflammasome also plays important roles in non-microbial diseases, highlighting the multifaceted nature of AIM2 beyond immunity to infectious diseases. In colorectal cancer, AIM2 orchestrates inflammasome-independent functions by suppressing stem cell proliferation, and contributes to maintenance of a healthy gut microbiota. The role of AIM2 inflammasome in other types of cancer should be further explored. Moreover, inappropriate recognition of self-DNA by AIM2 triggers detrimental inflammatory responses, leading to superficial and systemic inflammation. Inhibiting AIM2 inflammasome activity using synthetic inhibitors, such as suppressive oligodeoxynucleotides [124], or harnessing the power of endogenous AIM2 inhibitors, such as pyrin-containing proteins [36, 39] or antimicrobial cathelicidin peptides [38], could be investigated for their potential to control unwanted inflammation. An additional line of research could focus on understanding the complementary relationship between AIM2 and a plethora of other DNA sensors in the context of different cell types, tissues and organs. A holistic understanding of the biology of AIM2 could lead to improved immunosurveillance in the fight against infectious diseases and cancer, while avoiding debilitating inflammatory and autoimmune diseases.
Acknowledgments
Research studies from the lab are supported by the US National Institutes of Health (AR056296, CA163507 and AI101935 to T.-D.K.), the American Lebanese Syrian Associated Charities (to T.-D.K.), and the R.G. Menzies Early Career Fellowship from the National Health and Medical Research Council of Australia (to S.M.M.).
Abbreviations
ASC
apoptosis-associated speck-like protein containing a CARD
GBP
guanylate-binding protein
IRF
interferon-regulatory factor
ISGF3
IFN-stimulated gene factor 3
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 2.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 3.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. J Immunol. 2013;190:1911–1918. doi: 10.4049/jimmunol.1203162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat N, Fitzgerald KA. Recognition of cytosolic DNA by cGAS and other STING-dependent sensors. Eur J Immunol. 2014;44:634–640. doi: 10.1002/eji.201344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavlar T, Ablasser A, Hornung V. Induction of type I IFNs by intracellular DNA-sensing pathways. Immunol Cell Biol. 2012;90:474–482. doi: 10.1038/icb.2012.11. [DOI] [PubMed] [Google Scholar]
- 7.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 8.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 9.Stacey KJ, Ross IL, Hume DA. Electroporation and DNA-dependent cell death in murine macrophages. Immunol Cell Biol. 1993;71( Pt 2):75–85. doi: 10.1038/icb.1993.8. [DOI] [PubMed] [Google Scholar]
- 10.Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 14.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 15.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–13235. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol. 2014;426:1420–1427. doi: 10.1016/j.jmb.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou X, Niu X. The NMR solution structure of AIM2 PYD domain from Mus musculus reveals a distinct alpha2-alpha3 helix conformation from its human homologues. Biochem Biophys Res Commun. 2015;461:396–400. doi: 10.1016/j.bbrc.2015.04.046. [DOI] [PubMed] [Google Scholar]
- 22.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Man SM, Hopkins LJ, Nugent E, Cox S, Gluck IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, Bryant CE. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belhocine K, Monack DM. Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells. Cell Microbiol. 2012;14:71–80. doi: 10.1111/j.1462-5822.2011.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, Degrandi D, Pfeffer K, Yamamoto M, Henry T, Broz P. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juruj C, Lelogeais V, Pierini R, Perret M, Py BF, Jamilloux Y, Broz P, Ader F, Faure M, Henry T. Caspase-1 activity affects AIM2 speck formation/stability through a negative feedback loop. Front Cell Infect Microbiol. 2013;3:14. doi: 10.3389/fcimb.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, Sohn J. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun. 2015;6:7827. doi: 10.1038/ncomms8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015 doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 32.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signaling. Nature. 2015 doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 33.Hara H, Tsuchiya K, Kawamura I, Fang R, Hernandez-Cuellar E, Shen Y, Mizuguchi J, Schweighoffer E, Tybulewicz V, Mitsuyama M. Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck-like aggregates and inflammasome activity. Nat Immunol. 2013;14:1247–1255. doi: 10.1038/ni.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodgers MA, Bowman JW, Fujita H, Orazio N, Shi M, Liang Q, Amatya R, Kelly TJ, Iwai K, Ting J, Jung JU. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J Exp Med. 2014;211:1333–1347. doi: 10.1084/jem.20132486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV, Wallin MC, Gangopadhyay A, Forte E, Gottwein E, Perlman H, Reed JC, Greaves DR, Dorfleutner A, Stehlik C. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol. 2014;15:343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, Simanski M, Glaser R, Harder J, Hornung V, Gallo RL, Ruzicka T, Besch R, Schauber J. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Almeida L, Khare S, Misharin AV, Patel R, Ratsimandresy RA, Wallin MC, Perlman H, Greaves DR, Hoffman HM, Dorfleutner A, Stehlik C. The PYRIN Domain-only Protein POP1 Inhibits Inflammasome Assembly and Ameliorates Inflammatory Disease. Immunity. 2015;43:264–276. doi: 10.1016/j.immuni.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choubey D, Walter S, Geng Y, Xin H. Cytoplasmic localization of the interferon-inducible protein that is encoded by the AIM2 (absent in melanoma) gene from the 200-gene family. FEBS Lett. 2000;474:38–42. doi: 10.1016/s0014-5793(00)01571-4. [DOI] [PubMed] [Google Scholar]
- 41.Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, Sagulenko V, Thygesen SJ, Choubey D, Hornung V, Walz T, Stacey KJ, Wu H. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sester DP, Sagulenko V, Thygesen SJ, Cridland JA, Loi YS, Cridland SO, Masters SL, Genske U, Hornung V, Andoniou CE, Sweet MJ, Degli-Esposti MA, Schroder K, Stacey KJ. Deficient NLRP3 and AIM2 Inflammasome Function in Autoimmune NZB Mice. J Immunol. 2015;195:1233–1241. doi: 10.4049/jimmunol.1402859. [DOI] [PubMed] [Google Scholar]
- 44.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, Dixit VM, Monack DM. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierini R, Juruj C, Perret M, Jones CL, Mangeot P, Weiss DS, Henry T. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 2012;19:1709–1721. doi: 10.1038/cdd.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atianand MK, Duffy EB, Shah A, Kar S, Malik M, Harton JA. Francisella tularensis reveals a disparity between human and mouse NLRP3 inflammasome activation. J Biol Chem. 2011;286:39033–39042. doi: 10.1074/jbc.M111.244079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren SE, Armstrong A, Hamilton MK, Mao DP, Leaf IA, Miao EA, Aderem A. Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. J Immunol. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J Clin Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang R, Tsuchiya K, Kawamura I, Shen Y, Hara H, Sakai S, Yamamoto T, Fernandes-Alnemri T, Yang R, Hernandez-Cuellar E, Dewamitta SR, Xu Y, Qu H, Alnemri ES, Mitsuyama M. Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. J Immunol. 2011;187:4890–4899. doi: 10.4049/jimmunol.1100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, Ablasser A. Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host Microbe. 2015;17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Saiga H, Nieuwenhuizen N, Gengenbacher M, Koehler AB, Schuerer S, Moura-Alves P, Wagner I, Mollenkopf HJ, Dorhoi A, Kaufmann SH. The Recombinant BCG DeltaureC::hly Vaccine Targets the AIM2 Inflammasome to Induce Autophagy and Inflammation. J Infect Dis. 2015;211:1831–1841. doi: 10.1093/infdis/jiu675. [DOI] [PubMed] [Google Scholar]
- 56.Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M, Yamamoto M, Takeda K. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol. 2012;24:637–644. doi: 10.1093/intimm/dxs062. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Zhou X, Kouadir M, Shi F, Ding T, Liu C, Liu J, Wang M, Yang L, Yin X, Zhao D. The AIM2 inflammasome is involved in macrophage activation during infection with virulent Mycobacterium bovis strain. J Infect Dis. 2013;208:1849–1858. doi: 10.1093/infdis/jit347. [DOI] [PubMed] [Google Scholar]
- 58.Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect Immun. 2014;82:112–123. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanamsagar R, Aldrich A, Kielian T. Critical role for the AIM2 inflammasome during acute CNS bacterial infection. J Neurochem. 2014;129:704–711. doi: 10.1111/jnc.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomes MT, Campos PC, Oliveira FS, Corsetti PP, Bortoluci KR, Cunha LD, Zamboni DS, Oliveira SC. Critical role of ASC inflammasomes and bacterial type IV secretion system in caspase-1 activation and host innate resistance to Brucella abortus infection. J Immunol. 2013;190:3629–3638. doi: 10.4049/jimmunol.1202817. [DOI] [PubMed] [Google Scholar]
- 61.Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, Yamamoto M, Nagarajan UM, Miao EA, Coers J. Guanylate Binding Proteins Enable Rapid Activation of Canonical and Noncanonical Inflammasomes in Chlamydia-Infected Macrophages. Infect Immun. 2015;83:4740–4749. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, Roberts TL, Schroder K, Vince JE, Hill JM, Silke J, Stacey KJ. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013;20:1149–1160. doi: 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 66.Tsuchiya K, Hara H, Kawamura I, Nomura T, Yamamoto T, Daim S, Dewamitta SR, Shen Y, Fang R, Mitsuyama M. Involvement of absent in melanoma 2 in inflammasome activation in macrophages infected with Listeria monocytogenes. J Immunol. 2010;185:1186–1195. doi: 10.4049/jimmunol.1001058. [DOI] [PubMed] [Google Scholar]
- 67.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang R, Hara H, Sakai S, Hernandez-Cuellar E, Mitsuyama M, Kawamura I, Tsuchiya K. Type I interferon signaling regulates activation of the absent in melanoma 2 inflammasome during Streptococcus pneumoniae infection. Infect Immun. 2014;82:2310–2317. doi: 10.1128/IAI.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J Immunol. 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, Huang DC, Soldati-Favre D, Horie K, Takeda J, Takeda K. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Crane DD, Bauler TJ, Wehrly TD, Bosio CM. Mitochondrial ROS potentiates indirect activation of the AIM2 inflammasome. Front Microbiol. 2014;5:438. doi: 10.3389/fmicb.2014.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bauernfeind F, Bartok E, Rieger A, Franchi L, Nunez G, Hornung V. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J Immunol. 2011;187:613–617. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang MT, Mortensen BL, Taxman DJ, Craven RR, Taft-Benz S, Kijek TM, Fuller JR, Davis BK, Allen IC, Brickey WJ, Gris D, Wen H, Kawula TH, Ting JP. Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J Immunol. 2010;185:5476–5485. doi: 10.4049/jimmunol.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ulland TK, Buchan BW, Ketterer MR, Fernandes-Alnemri T, Meyerholz DK, Apicella MA, Alnemri ES, Jones BD, Nauseef WM, Sutterwala FS. Cutting edge: mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J Immunol. 2010;185:2670–2674. doi: 10.4049/jimmunol.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulland TK, Janowski AM, Buchan BW, Faron M, Cassel SL, Jones BD, Sutterwala FS. Francisella tularensis live vaccine strain folate metabolism and pseudouridine synthase gene mutants modulate macrophage caspase-1 activation. Infect Immun. 2013;81:201–208. doi: 10.1128/IAI.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin CY, Ratner HK, Llewellyn AC, Jones CL, Laroui H, Merlin D, Zhou P, Endtz HP, Weiss DS. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci U S A. 2014;111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge J, Gong YN, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc Natl Acad Sci U S A. 2012;109:6193–6198. doi: 10.1073/pnas.1117490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dotson RJ, Rabadi SM, Westcott EL, Bradley S, Catlett SV, Banik S, Harton JA, Bakshi CS, Malik M. Repression of inflammasome by Francisella tularensis during early stages of infection. J Biol Chem. 2013;288:23844–23857. doi: 10.1074/jbc.M113.490086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol. 2010;10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lupfer C, Malik A, Kanneganti TD. Inflammasome control of viral infection. Curr Opin Virol. 2015;12:38–46. doi: 10.1016/j.coviro.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reinholz M, Kawakami Y, Salzer S, Kreuter A, Dombrowski Y, Koglin S, Kresse S, Ruzicka T, Schauber J. HPV16 activates the AIM2 inflammasome in keratinocytes. Arch Dermatol Res. 2013;305:723–732. doi: 10.1007/s00403-013-1375-0. [DOI] [PubMed] [Google Scholar]
- 83.Zhen J, Zhang L, Pan J, Ma S, Yu X, Li X, Chen S, Du W. AIM2 mediates inflammation-associated renal damage in hepatitis B virus-associated glomerulonephritis by regulating caspase-1, IL-1beta, and IL-18. Mediators Inflamm. 2014;2014:190860. doi: 10.1155/2014/190860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu DL, Xu GH, Lu SM, Ma BL, Miao NZ, Liu XB, Cheng YP, Feng JH, Liu ZG, Feng D, Na L, Li WQ, Zhao YR. Correlation of AIM2 expression in peripheral blood mononuclear cells from humans with acute and chronic hepatitis B. Hum Immunol. 2013;74:514–521. doi: 10.1016/j.humimm.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 85.Han Y, Chen Z, Hou R, Yan D, Liu C, Chen S, Li X, Du W. Expression of AIM2 is correlated with increased inflammation in chronic hepatitis B patients. Virol J. 2015;12:129. doi: 10.1186/s12985-015-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Horan KA, Hansen K, Jakobsen MR, Holm CK, Soby S, Unterholzner L, Thompson M, West JA, Iversen MB, Rasmussen SB, Ellermann-Eriksen S, Kurt-Jones E, Landolfo S, Damania B, Melchjorsen J, Bowie AG, Fitzgerald KA, Paludan SR. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Nunez G. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 88.Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kailasan Vanaja S, Rathinam VA, Atianand MK, Kalantari P, Skehan B, Fitzgerald KA, Leong JM. Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proc Natl Acad Sci U S A. 2014;111:7765–7770. doi: 10.1073/pnas.1400075111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ekchariyawat P, Hamel R, Bernard E, Wichit S, Surasombatpattana P, Talignani L, Thomas F, Choumet V, Yssel H, Despres P, Briant L, Misse D. Inflammasome signaling pathways exert antiviral effect against Chikungunya virus in human dermal fibroblasts. Infect Genet Evol. 2015;32:401–408. doi: 10.1016/j.meegid.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 93.Karki R, Man SM, Malireddi RK, Gurung P, Vogel P, Lamkanfi M, Kanneganti TD. Concerted Activation of the AIM2 and NLRP3 Inflammasomes Orchestrates Host Protection against Aspergillus Infection. Cell Host Microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kalantari P, DeOliveira RB, Chan J, Corbett Y, Rathinam V, Stutz A, Latz E, Gazzinelli RT, Golenbock DT, Fitzgerald KA. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell Rep. 2014;6:196–210. doi: 10.1016/j.celrep.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, Neale G, Olsen SR, Carter RA, McGoldrick DJ, Wu G, Finkelstein D, Vogel P, Gilbertson RJ, Kanneganti TD. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, Muhlbauer M, Chou WC, Barker BR, Jobin C, Allbritton NL, Ramsden DA, Davis BK, Ting JP. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med. 2015;21:906–913. doi: 10.1038/nm.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS, Trent JM. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 98.Dihlmann S, Tao S, Echterdiek F, Herpel E, Jansen L, Chang-Claude J, Brenner H, Hoffmeister M, Kloor M. Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int J Cancer. 2014;135:2387–2396. doi: 10.1002/ijc.28891. [DOI] [PubMed] [Google Scholar]
- 99.Schulmann K, Brasch FE, Kunstmann E, Engel C, Pagenstecher C, Vogelsang H, Kruger S, Vogel T, Knaebel HP, Ruschoff J, Hahn SA, Knebel-Doeberitz MV, Moeslein G, Meltzer SJ, Schackert HK, Tympner C, Mangold E, Schmiegel W. HNPCC-associated small bowel cancer: clinical and molecular characteristics. Gastroenterology. 2005;128:590–599. doi: 10.1053/j.gastro.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 100.Woerner SM, Kloor M, Schwitalle Y, Youmans H, Doeberitz M, Gebert J, Dihlmann S. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes Chromosomes Cancer. 2007;46:1080–1089. doi: 10.1002/gcc.20493. [DOI] [PubMed] [Google Scholar]
- 101.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ponomareva L, Liu H, Duan X, Dickerson E, Shen H, Panchanathan R, Choubey D. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol Cancer Res. 2013;11:1193–1202. doi: 10.1158/1541-7786.MCR-13-0145. [DOI] [PubMed] [Google Scholar]
- 103.Wang LJ, Hsu CW, Chen CC, Liang Y, Chen LC, Ojcius DM, Tsang NM, Hsueh C, Wu CC, Chang YS. Interactome-wide analysis identifies end-binding protein 1 as a crucial component for the speck-like particle formation of activated absence in melanoma 2 (AIM2) inflammasomes. Mol Cell Proteomics. 2012;11:1230–1244. doi: 10.1074/mcp.M112.020594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen LC, Wang LJ, Tsang NM, Ojcius DM, Chen CC, Ouyang CN, Hsueh C, Liang Y, Chang KP, Chen CC, Chang YS. Tumour inflammasome-derived IL-1beta recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol Med. 2012;4:1276–1293. doi: 10.1002/emmm.201201569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kondo Y, Nagai K, Nakahata S, Saito Y, Ichikawa T, Suekane A, Taki T, Iwakawa R, Enari M, Taniwaki M, Yokota J, Sakoda S, Morishita K. Overexpression of the DNA sensor proteins, absent in melanoma 2 and interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation. Cancer Sci. 2012;103:782–790. doi: 10.1111/j.1349-7006.2012.02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kong H, Wang Y, Zeng X, Wang Z, Wang H, Xie W. Differential expression of inflammasomes in lung cancer cell lines and tissues. Tumour Biol. 2015 doi: 10.1007/s13277-015-3473-4. [DOI] [PubMed] [Google Scholar]
- 107.Patsos G, Germann A, Gebert J, Dihlmann S. Restoration of absent in melanoma 2 (AIM2) induces G2/M cell cycle arrest and promotes invasion of colorectal cancer cells. Int J Cancer. 2010;126:1838–1849. doi: 10.1002/ijc.24905. [DOI] [PubMed] [Google Scholar]
- 108.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 109.Lu D, Huang J, Basu A. Protein kinase Cepsilon activates protein kinase B/Akt via DNA-PK to protect against tumor necrosis factor-alpha-induced cell death. J Biol Chem. 2006;281:22799–22807. doi: 10.1074/jbc.M603390200. [DOI] [PubMed] [Google Scholar]
- 110.de Koning HD, Bergboer JG, van den Bogaard EH, van Vlijmen-Willems IM, Rodijk-Olthuis D, Simon A, Zeeuwen PL, Schalkwijk J. Strong induction of AIM2 expression in human epidermis in acute and chronic inflammatory skin conditions. Exp Dermatol. 2012;21:961–964. doi: 10.1111/exd.12037. [DOI] [PubMed] [Google Scholar]
- 111.Yang CA, Huang ST, Chiang BL. Sex-dependent differential activation of NLRP3 and AIM2 inflammasomes in SLE macrophages. Rheumatology (Oxford) 2015;54:324–331. doi: 10.1093/rheumatology/keu318. [DOI] [PubMed] [Google Scholar]
- 112.Kimkong I, Avihingsanon Y, Hirankarn N. Expression profile of HIN200 in leukocytes and renal biopsy of SLE patients by real-time RT-PCR. Lupus. 2009;18:1066–1072. doi: 10.1177/0961203309106699. [DOI] [PubMed] [Google Scholar]
- 113.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O’Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vanhove W, Peeters PM, Staelens D, Schraenen A, Van der Goten J, Cleynen I, De Schepper S, Van Lommel L, Reynaert NL, Schuit F, Van Assche G, Ferrante M, De Hertogh G, Wouters EF, Rutgeerts P, Vermeire S, Nys K, Arijs I. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015 doi: 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 115.Lozano-Ruiz B, Bachiller V, Garcia-Martinez I, Zapater P, Gomez-Hurtado I, Moratalla A, Gimenez P, Bellot P, Frances R, Such J, Gonzalez-Navajas JM. Absent in melanoma 2 triggers a heightened inflammasome response in ascitic fluid macrophages of patients with cirrhosis. J Hepatol. 2015;62:64–71. doi: 10.1016/j.jhep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 116.Csak T, Pillai A, Ganz M, Lippai D, Petrasek J, Park JK, Kodys K, Dolganiuc A, Kurt-Jones EA, Szabo G. Both bone marrow-derived and non-bone marrow-derived cells contribute to AIM2 and NLRP3 inflammasome activation in a MyD88-dependent manner in dietary steatohepatitis. Liver Int. 2014;34:1402–1413. doi: 10.1111/liv.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Adamczak SE, de Rivero Vaccari JP, Dale G, Brand FJ, 3rd, Nonner D, Bullock MR, Dahl GP, Dietrich WD, Keane RW. Pyroptotic neuronal cell death mediated by the AIM2 inflammasome. J Cereb Blood Flow Metab. 2014;34:621–629. doi: 10.1038/jcbfm.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Denes A, Coutts G, Lenart N, Cruickshank SM, Pelegrin P, Skinner J, Rothwell N, Allan SM, Brough D. AIM2 and NLRC4 inflammasomes contribute with ASC to acute brain injury independently of NLRP3. Proc Natl Acad Sci U S A. 2015;112:4050–4055. doi: 10.1073/pnas.1419090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 2001;292:1546–1549. doi: 10.1126/science.292.5521.1546. [DOI] [PubMed] [Google Scholar]
- 120.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 121.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 122.Baum R, Sharma S, Carpenter S, Li QZ, Busto P, Fitzgerald KA, Marshak-Rothstein A, Gravallese EM. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. J Immunol. 2015;194:873–877. doi: 10.4049/jimmunol.1402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jakobs C, Perner S, Hornung V. AIM2 Drives Joint Inflammation in a Self-DNA Triggered Model of Chronic Polyarthritis. PLoS One. 2015;10:e0131702. doi: 10.1371/journal.pone.0131702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kaminski JJ, Schattgen SA, Tzeng TC, Bode C, Klinman DM, Fitzgerald KA. Synthetic oligodeoxynucleotides containing suppressive TTAGGG motifs inhibit AIM2 inflammasome activation. J Immunol. 2013;191:3876–3883. doi: 10.4049/jimmunol.1300530. [DOI] [PMC free article] [PubMed] [Google Scholar]