Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems (original) (raw)
. Author manuscript; available in PMC: 2016 Apr 26.
Published in final edited form as: Biochim Biophys Acta. 2004 Nov 11;1694(1-3):219–234. doi: 10.1016/j.bbamcr.2004.02.013
Abstract
The translocation of DNA across biological membranes is an essential process for many living organisms. In bacteria, type IV secretion systems (T4SS) are used to deliver DNA as well as protein substrates from donor to target cells. The T4SS are structurally complex machines assembled from a dozen or more membrane proteins in response to environmental signals. In Gram-negative bacteria, the conjugation machines are composed of a cell envelope-spanning secretion channel and an extracellular pilus. These dynamic structures (i) direct formation of stable contacts—the mating junction—between donor and recipient cell membranes, (ii) transmit single-stranded DNA as a nucleoprotein particle, as well as protein substrates, across donor and recipient cell membranes, and (iii) mediate disassembly of the mating junction following substrate transfer. This review summarizes recent progress in our understanding of the mechanistic details of DNA trafficking with a focus on the paradigmatic Agrobacterium tumefaciens VirB/D4 T4SS and related conjugation systems.
Keywords: Conjugation, Type IV secretion, DNA transfer, Pathogenesis, Protein translocation, Pilus, Coupling protein
1. Introduction
The type IV secretion systems (T4SS) translocate DNA and protein substrates across the bacterial cell envelope. These systems are classified on the basis of an ancestral relatedness to bacterial conjugation machines [1]. Consequently, members of this family include all conjugation systems of Gram-negative and -positive bacteria and related systems mediating DNA or protein translocation [2,3]. In general, T4SS deliver their substrates to recipient cells via direct cell-to-cell contact, although there are examples of contact-independent protein export and DNA release to and uptake from the extracellular milieu [1,4,5]. The nature of type IV secretion substrates [6,7], the general architectural features of various T4SS [2,3,8], and the cellular consequences of macromolecular trafficking during pathogenesis [1,7,9] have been the subjects of recent reviews.
Here, I will focus on the type IV machine itself, emphasizing the recent work on the Agrobacterium tumefaciens VirB/D4 T4SS and related conjugation machines. In summarizing the recent progress, I hope it is evident that detailed structure–function studies are advancing our state of knowledge of conjugation and related systems extremely rapidly. Within the next few years, a mechanistic understanding of these systems can be expected to approach that of the general secretory pathway (GSP) and other systems reviewed in this compendium.
2. The VirB/D4 T4SS operon structures and regulatory controls
The A. tumefaciens VirB/D4 T4SS is encoded by the virB and virD operons [10]. The virB operon encodes 11 genes, virB1 through virB11. The virB gene products, termed the mating pair formation (Mpf) proteins, elaborate a cell envelope-spanning structure required for substrate transfer, as well as an extracellular filament termed the T pilus that mediates attachment to recipient cells [11]. The virD operon encodes five genes, virD1 through virD5. virD1 and virD2 code for proteins that process the DNA substrate (T-DNA) for transfer; these are termed the DNA transfer and replication (Dtr) proteins [12]. virD3 and virD5 code for proteins that are nonessential for processing or transfer, and virD4 codes for the coupling protein (CP) [13,14]. The VirD4 CP is not involved in T-DNA processing or biogenesis of the T pilus but instead acts together with the Mpf structure to deliver substrates across the cell envelope [10,11].
Type IV translocation systems whose subunits are related to the virB and virD4 gene products are classified as the type IVA subfamily [15]. The virB genes of the known T4SS often comprise a single operon, whereas virD4 is usually genetically linked to the genes involved in T-DNA processing. In the systems studied to date, the T4SS genes are activated in response to perception of environmental signals. For example, phenolic and monosaccharide sugar molecules released from wounded plant cells activate a VirA/VirG two-component regulatory system which in turn induces transcription of the A. tumefaciens virulence (vir) regulon [13]. Additionally, low (1–5 mM) phosphate and an acidic pH in the range of 5.0–5.5 are required for induction. A PhoB-dependent regulatory system is thought to mediate the response to phosphate starvation, whereas acid pH activates transcription from an acid-inducible promoter upstream of the virG response regulator gene and might also protonate phenolic compounds leading to increased membrane permeability and VirA sensory perception [13,16]. Several intracellular pathogens of mammals such as Brucella spp. and Legionella pneumophila also induce expression of their T4SS gene clusters in response to stress, e.g., to starvation and acidic pH [17,18]. It is thus reasonable to predict that plant and mammalian pathogens express T4SS genes and elaborate these translocation systems in response to a combination of specific inducing molecules, e.g., plant phenolics, and more general environmental signals, e.g., acidic pH and low available nutrients, typically present at sites of infection.
Focusing on the virB operon of A. tumefaciens, the signals described above control transcription from the virB promoter. There is also increasing evidence for the existence of multiple transcriptional and posttranscriptional regulatory controls that govern assembly of the VirB/D4 T4SS. At least one internal promoter is thought to transcribe a subset of virB genes in the absence of plant phenolic signals (L. Banta, personal communication). Moreover, it was recently shown that virB6 translation, but not production of a functional VirB6 protein per se, is essential for efficient synthesis of downstream virB gene products. The data suggest virB6 translation might contribute to stabilization of the distal portion of this polycistronic message [19]. Additionally, several closely juxtaposed or overlapping virB genes, e.g., virB1/virB2/virB3/virB4 and virB7/virB8/virB9/virB10, are probably translationally coupled, as supported by results of genetic complementation studies [20]. Several of these virB gene products mutually stabilize each other, suggesting that co-translation serves to coordinate the timing and probably also the stoichiometric synthesis of interacting subunits during machine biogenesis [19–23]. These and other regulatory controls to be discovered probably have evolved to maximize the chance and frequency of gene transfer at an overall minimal energetic cost to the cell.
3. Type IV secretion substrates: processing and recognition
3.1. Formation of the T-strand-relaxase intermediate
To understand the mechanistic details of type IV secretion, it is first necessary to describe the secretion substrates and associated processing reactions (Fig. 1). Early studies showed that a complex of proteins assembled at the origin-of-transfer (oriT) sequence catalyzes a conversion of supercoiled plasmid DNA to the open, relaxed form that is detectable upon exposure to denaturing detergents [24]. This complex of processing proteins at oriT is termed the relaxosome, and the subunit responsible for nicking at oriT the relaxase. The relaxase is a transesterase that preserves energy from cleavage of the phosphodiester backbone of the strand destined for transfer (T-strand) as a stable phosphotyrosyl intermediate [25]. Additionally, conjugal DNA transfer proceeds in a 5′–3′ direction [12], prompting a proposal that the relaxase covalently bound to the 5′ end of the T-strand supplies substrate recognition signals and possibly also a piloting function to direct DNA passage through the secretion channel [26,27]. Such a model has now received very strong experimental support (see below).
Fig. 1.
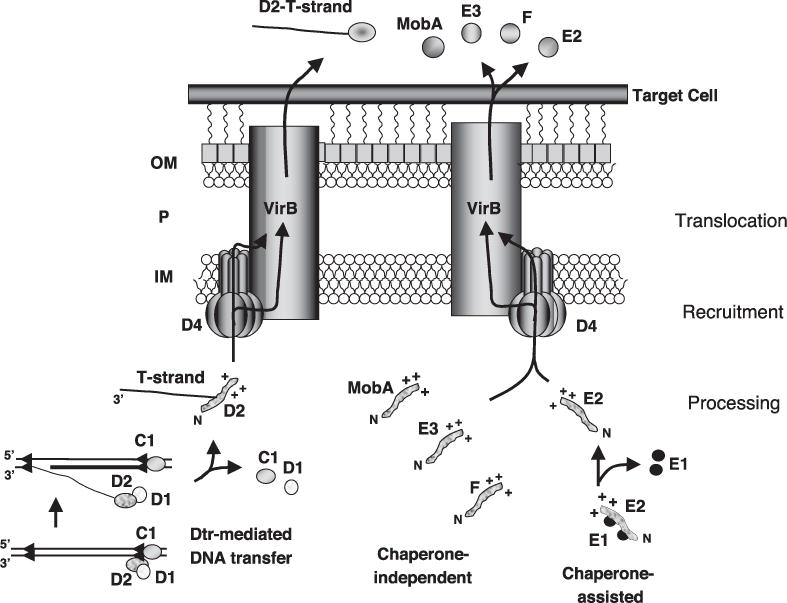
T4SS conjugally transfer DNA and protein substrates to target cells, as illustrated for the A. tumefaciens VirB/D4 T4SS. The transfer DNA (T-DNA) is processed by Dtr proteins (VirD1, VirD2, and VirC1) bound at _oriT_-like border repeat sequences to form the VirD2-T-strand transfer intermediate. The VirB/D4 T4SS recruits the DNA transfer intermediate through binding of the VirD4 coupling protein (CP) to the positively charged C terminus of VirD2, which is unfolded prior to export. The transfer intermediate is translocated across the cell envelope through a secretion channel composed of the VirD4 CP and the VirB mating pair formation (Mpf) proteins; the precise route of translocation is undefined. Translocation of protein substrates similarly proceeds in three steps: (i) processing, which is independent or dependent on chaperone binding to maintain substrate in an unfolded, nonaggregated conformation; (ii) recruitment, mediated by CP binding to positively charged C-termini; and (iii) translocation, via the CP/Mpf secretion channel. IM, inner membrane; P, periplasm; OM, outer membrane.
3.2. Protein substrates
Conjugation systems also export proteins independently of DNA [28–34]. As shown in Fig. 1, these systems translocate protein substrates, most probably in an unfolded conformation, by chaperone-dependent or -independent mechanisms. In this capacity, conjugation systems functionally resemble members of a second T4SS subfamily, the ‘effector translocators’. These systems are used by many pathogens to deliver protein effectors to eukaryotic cells during the course of infection [1]. Several assays have been developed for identifying type IV protein substrates [6], perhaps the most informative being the Cre/loxP recombination system [29,31,32]. As first demonstrated with the A. tumefaciens VirB/D4 T4SS, protein substrates fused to the Cre recombinase mediate transfer of Cre to target cells, as shown by recombination at target loxP sites engineered into target cell genomes [29,31,32]. In each case, transfer requires the VirB/D4 T4SS as well as fusion of the recombinase to the N terminus of the protein substrate [29,31,32].
The question of whether relaxases are bona fide type IV secretion substrates has been a subject of debate. Until very recently, there was fairly convincing—albeit indirect—evidence that the A. tumefaciens VirB/D4 T4SS translocates the VirD2-T-strand and the plasmid RSF1010 MobA-T-strand intermediate to plant cells [26,27]. Now, experiments using the Cre/loxP system have shown unequivocally that both the plasmid RP4 and L. pneumophila Dot/Icm T4SS transfer a Cre∷MobARSF1010 fusion protein to recipient E. coli and L. pneumophila cells, respectively [34]. Of further interest, these T4SS were even capable of translocating Cre∷relaxase independently of a covalent association with the T-strand transfer intermediate. With the Cre/loxP recombination system, the investigators further showed that the L. pneumophila Dot/Icm T4SS delivers Cre fused to two other protein substrates, RalF and LidA, to bacterial recipients, and they also identified a large number of potential Dot/Icm substrates, designated the Sid (Substrates of icm/dot) proteins. The Dot/Icm T4SS was also shown to translocate one of these novel substrates, SidC, to mammalian target cells [34].
In comparing the known protein substrates of conjugation systems and other T4SS, there are no obvious common sequence motifs suggestive of a consensus secretion signal. However, several findings now strongly indicate that such signals reside at the C termini of type IV protein substrates (Fig. 2). Again, the most compelling evidence derives from use of the Cre/loxP system, whereby C-terminal fragments of secretion substrates fused to Cre mediate T4SS-dependent intercellular transfer of the recombinase. For example, the A. tumefaciens VirB/D4 T4SS translocates Cre when fused to C-terminal fragments of the VirE2, VirE3, and VirF secretion substrates (Fig. 1) to plant and yeast cells [29,31,32]. Similarly, the Dot/Icm T4SS translocates Cre when fused to C-terminal fragments of the RalF and LidA substrates to L. pneumophila recipients [34]. Upon close examination, it is noteworthy that the C termini of A. tumefaciens VirB/D4 T4SS substrates carry a conserved Arg-X-Arg motif(s), whereas many T4SS substrates—including relaxases from various conjugation systems—carry an abundance of positively charged residues, mostly Arg, within the last 30–50 residues. Interestingly, VirD3 also possesses an abundance of basic residues at its C terminus and thus might also be exported to plant cells during infection, although this remains to be experimentally tested. By contrast, the VirE1 secretion chaperone—which is not recruited to the VirB/D4 T4SS [32,35]—does not carry such a charge bias at its C terminus (Fig. 2).
Fig. 2.
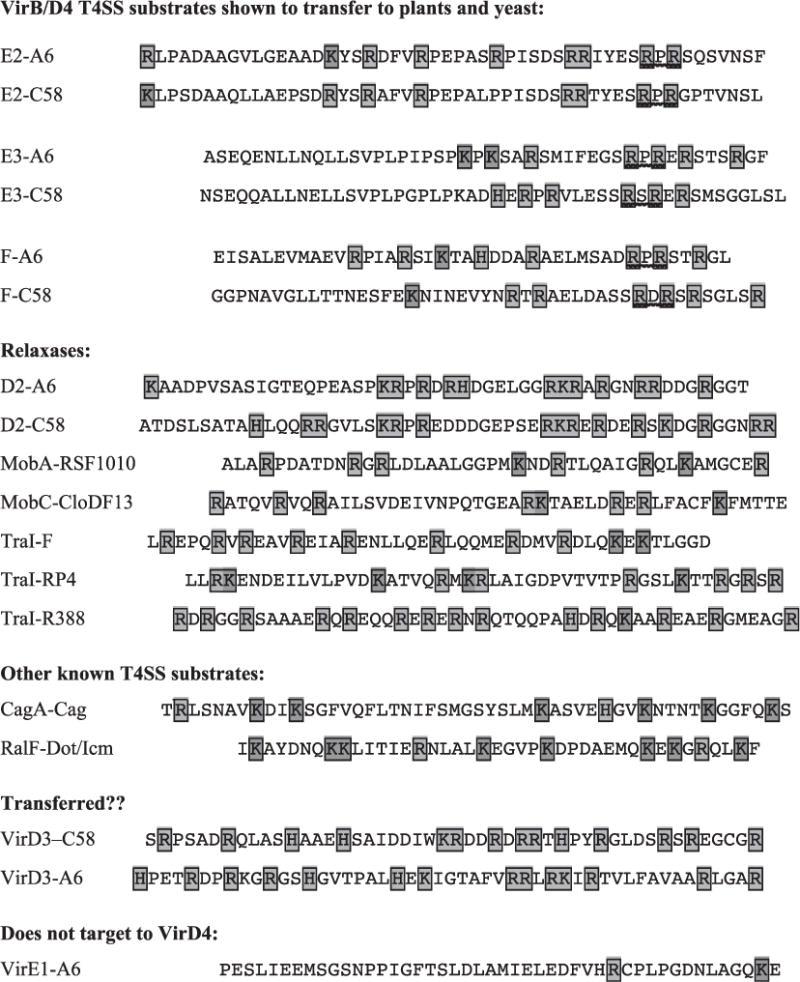
Type IV secretion substrates contain clusters of positively charged residues at their C termini. This charge bias is a postulated feature of the type IV secretion signal recognized by the coupling protein component of the T4SS [31,33,35]. Shown are the C-terminal residues, with Arg (R), Lys (K), and His (H) residues highlighted with box shading and the Arg-X-Arg motifs of VirB/D4 T4SS substrates with wavy underlines. Known or potential substrates include VirB/D4 T4SS substrates encoded by the pTiC58 (C58) and pTiA6NC (A6) plasmids of A. tumefaciens, relaxases encoded by the plasmids listed, CagA exported by the H. pylori Cag T4SS, and RalF exported by the L. pneumophila Dot/Icm T4SS. VirD3 proteins from the C58 and A6 plasmids are candidate VirB/D4 secretion substrates on the basis of a C-terminal charge bias. VirE1 is not predicted to be a substrate on the basis of charge distribution, in agreement with findings that this secretion chaperone is not recruited by the VirD4 CP to the VirB/D4 T4SS [32,35].
How positive charge clusters contribute to type IV substrate recognition is not known, but recently it was shown that the C terminus mediates an interaction between the VirE2 secretion substrate and the VirD4 CP of the VirB/D4 T4SS. Following up a report that VirD4 localizes at the cell poles of A. tumefaciens [36], cells were engineered to produce a GFP-VirE2 fusion protein and assayed for VirD4-dependent recruitment of the fusion protein to the poles. Indeed, cells displayed polar fluorescence when producing GFP fused to the N terminus, but not to the C terminus, of the substrate (Fig. 3) [35]. Two additional cytological assays, termed cytology two-hybrid (C2H) and bimolecular fluorescence complementation (BiFC), as well as a coupled chemical-cross-linking and immunoprecipitation assay supplied further evidence for the VirD4–VirE2 interaction. By use of BiFC, it was further shown that the C-terminal 100 residues of VirE2 are necessary and sufficient for recruitment of GFP-VirE2 derivatives to VirD4. Although attempts to further delineate the N-terminal boundary of the putative secretion signal were unsuccessful due to problems of protein instability, a small deletion of the extreme C terminus as well as a peptide insertion near the Arg cluster impeded complex formation [35]. These findings, together with the demonstration that a VirE2 C-terminal fragment directs Cre translocation, suggest that the charged C termini of type IV secretion substrates probably contribute to substrate recognition by mediating productive contacts with the CP component of the T4SS (Fig. 1).
Fig. 3.
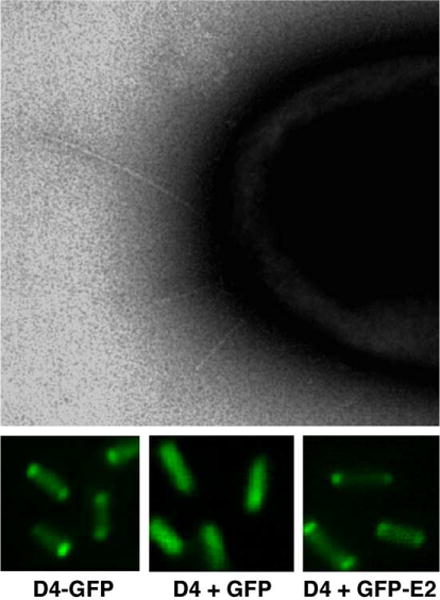
The A. tumefaciens T4SS and a secretion substrate localize at the cell poles. The T pili localize at cell poles as detected by electron microscopy (kindly provided by Alain Bernadac), the VirD4 CP localizes at the cell poles as shown by polar fluorescence of a VirD4-GFP fusion protein, and the VirD4 CP recruits VirE2 to the cell poles as shown by VirD4-dependent polar fluorescence of GFP-VirE2. GFP itself distributes throughout the cytoplasm of A. tumefaciens cells (middle panel) (see Ref. [35]).
All conjugation systems—at least of Gram-negative bacteria—possess a CP and therefore can be predicted to export substrates via CP-dependent recognition of C-terminal signals. A few members of the effector translocator subfamily do not possess a recognizable CP subunit [1]. Of these, the Bordetella pertussis Ptl T4SS is the only system for which a substrate, pertussis toxin (PT), is known. For secretion of PT subunits across the IM, B. pertussis employs the GSP, and consequently these subunits carry characteristic GSP secretion signals at their N termini. Upon delivery across the IM, the subunits assemble as the PT holotoxin for delivery by the Ptl T4SS across the OM. How PT holotoxin is recognized in the periplasm by the Ptl system is not known [37].
4. Initiation of T-strand transfer: substrate recruitment by the coupling protein
By the mid-1990s it was well-established that a given T4SS, e.g., encoded by plasmids R388 (IncW) or RP4 (IncPα) or the A. tumefaciens VirB/D4 system, translocates a restricted set of substrates that includes the cognate plasmid or oncogenic T-DNA, one or a few mobilizable plasmids such as ColE1 or RSF1010, and one or a few proteins [12]. The mobilizable plasmids carry oriT sequences and the Mob processing proteins acting at oriT, but lack the genes for their own T4SS. To identify machine components involved in substrate recognition, several investigators constructed chimeric systems composed of a CP from one T4SS and an Mpf structure from a second T4SS. Such chimeric T4SS were shown to be functional, and, furthermore, these systems exported substrates characteristically translocated by the T4SS from which the CP was derived. For example, TraG from plasmid RP4 substituted for TrwB of plasmid R388—and vice versa—with respect to RSF1010 mobilization. Yet, the chimeric TraGRP4/MpfR388 system failed to transfer R388, and the TrwBR388/MpfRP4 system failed to transfer RP4. Similarly, both VirD4 and the Ti plasmid TraG CPs of A. tumefaciens substituted for TraGRP4 with respect to RSF1010 mobilization, but not transfer of RP4 [14,38–40]. These findings strongly suggested that the CP links the Dtr processing proteins bound at _oriT_—the relaxosome—to the T4SS, hence the origin of the term ‘coupling protein’.
A couple of CPs were then purified and shown to bind DNA in vitro [41–43]. These DNA-binding studies demonstrated that single-stranded (ss) DNA is preferred over double-stranded (ds) DNA as expected, yet binding was also sequence nonspecific. Thus, it was proposed that the CP recognizes DNA substrates by virtue of interactions with the relaxase and other relaxosomal subunits bound at _oriT_—not the ssDNA intermediate per se. Indeed, in vitro studies have now demonstrated several CP–relaxase interactions. The TraGRP4 CP interacts with TraIRP4 relaxase/helicase [43] and with Mob relaxase of the mobilizable plasmid pBHR1 [44]. The TrwBR388 CP interacts with TrwCR388 relaxase/helicase, and the TraDF CP interacts with TraMF, a Dtr protein that participates in processing by binding at sites near the F plasmid oriT sequence [45]. As noted above, several relaxases possess an abundance of positively charged residues at their C termini (Fig. 2); these residues likely mediate productive contacts with the CP. However, in view of the fact that a given T4SS translocates a restricted set of substrates, it is also likely that other C-terminal signals, e.g., a structural motif presented by folding of the C-terminal domain, probably also contributes to substrate selection.
Upon recruitment, how does the CP mediate the next step(s) of transfer? Recent structural studies have begun to shed light on the answer to this question. Topology studies have shown that CPs are composed of an N-proximal region that includes two transmembrane helices and a small periplasmic domain, and a large C-terminal region that resides in the cytoplasm (Fig. 4) [46,47]. Very interestingly, a crystal structure solved for the soluble domain of TrwBR388 (TrwBΔN70) showed that six equivalent protomers form a spherical particle of overall dimensions 110 Å in diameter and 90 Å in height. This ring-like structure possesses a central channel of 20 Å in diameter, constricted to 8 Å at the entrance facing the cytoplasm. This channel traverses the structure, possibly connecting cytoplasm with periplasm [48,49]. An appendix corresponding to the N-terminal TM domain was discernible by image averaging of electron micrographs [42]. This overall structure bears a striking resemblance to the F1-ATPase α3β3 heterohexamer, whereas the structure of the soluble domain closely resembles DNA ring helicases and other proteins that translocate along ss- or ds-DNA (Fig. 4) [50].
Fig. 4.
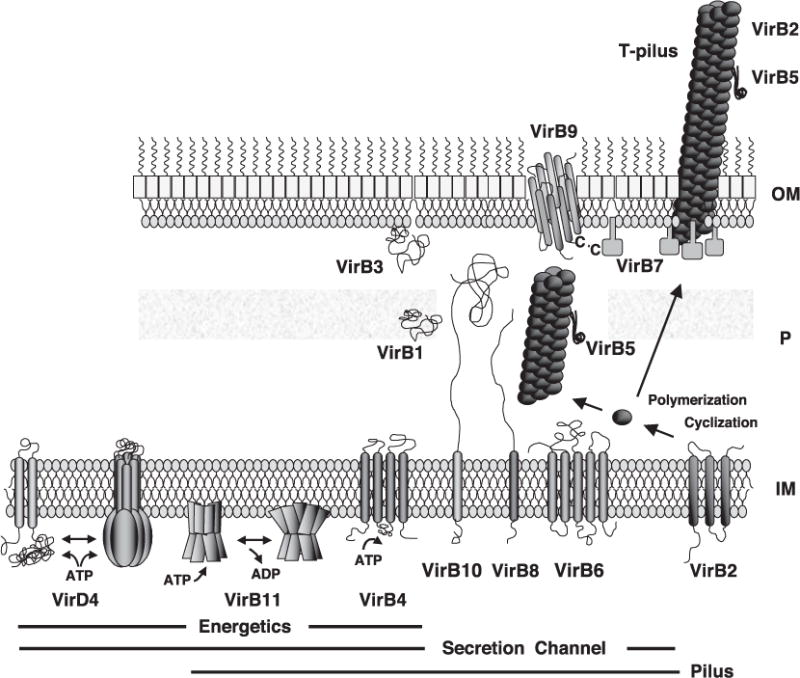
Topologies and oligomeric structures of VirB/D4 T4SS components. The VirD4 CP undergoes monomer–hexamer transitions possibly mediated by ATP binding; the VirB11 ATPase undergoes dramatic conformational changes in its N-terminal domain as a function of ATP binding and ADP release; VirB4 is also postulated to bind and hydrolyze ATP. These components plus the channel subunits shown are postulated to form a membrane-spanning secretion channel. A disulfide-linked dimer of VirB7 lipoprotein and secretin-like VirB9 are postulated to form a pore at the OM for pilus protusion and/or substrate transfer. Independently of the VirD4 CP, the VirB proteins direct polymerization of the T pilus, which may or may not be physically joined to the secretion channel. IM, inner membrane; P, periplasm; OM, outer membrane.
The oligomeric state of CPs in solution was also characterized and results suggest the situation in vivo is probably more complex than was revealed by the X-ray crystal structure. For example, the TrwBΔN70 purified as a monomer, raising the possibility that the hexamer identified by crystallography derived from the precipitation conditions employed during crystallization, e.g., high protein concentration and dehydration. Moreover, the full-length protein containing the N-terminal transmembrane stem purified both as a monomer and as a hexamer [42]. This finding led to a proposal that this CP undergoes monomer–hexamer transitions in vivo, possibly mediated by nucleotide and/or substrate binding (Fig. 4). Such dynamic structural changes might play an important role in substrate translocation. In other studies, purified full-length TraGRP4 and TraDF formed large multimers of >15 subunits; these were probably aggregates as indicated by their broad sedimentation profiles in glycerol gradients [43]. By contrast, HP0524Cag deleted of its N-terminal membrane-spanning domain purified as a multimer of ~4–5 subunits, whereas corresponding TraGRP4 and TrwBR388 truncation derivatives lacking the N-terminal transmembrane domain purified exclusively as monomers [43,51]. Together, these findings point to the importance of the N-terminal transmembrane domain for CP oligomerization in vivo, whereas its presence at least for some CPs seems to induce aggregation in vitro.
In addition to a role in oligomerization, the N-terminal region of A. tumefaciens VirD4 directs localization of the CP predominantly to the cell poles [36]. A deletion mutagenesis study established that residues in the periplasmic loop contribute to polar positioning. This VirD4 localization and an observation that the T pilus is polar-localized (Fig. 3) [11] suggest that this T4SS localizes at the cell poles. Interestingly, however, a few years ago it was reported that some VirB proteins, notably VirB8, VirB9, and VirB10, form foci around the cell periphery [52]. Since then, similar types of foci with the TrhC (VirB4 homolog) subunit of the plasmid R27 T4SS were observed [53]. Such foci might correspond to artifacts from protein overproduction, but it is also possible that such complexes represent physiologically relevant intermediate- or even terminal-pathway structures.
Recently, the nature of CP interactions with Mpf subunits has been explored. TraG of plasmid R27 from Salmonella enterica was found to interact with the VirB10-like protein, TrhBR27 [54]. As discussed below, the VirB10-like proteins are probably structural scaffolds required for assembly of the transenvelope T4SS. As expected from their membrane topologies, the TraGR27 CP interacted with the N-terminal region of TrhBR27 bearing a small cytoplasmic domain, a TM, and a portion of the periplasmic region. A second study further showed that the N-terminal domains of both the TrwBR388 CP and the VirB10-like TrwER388 protein mediate their interaction [55]. Consistent with the results obtained with the chimeric T4SS described above, this study also showed that CPs from three different T4SS interacted with the VirB10-like proteins of the cognate system as well as other systems. Additionally, TrwBR388 CP was shown to interact with the TrwCR388 relaxase, establishing a direct link between the relaxosome and the Mpf structure [55].
In summary, the CP recruits DNA substrates to the secretion apparatus through contacts with the DNA-processing proteins (Figs. 1 and 4). Then, through the VirB10 and perhaps other contacts, the CP coordinates passage of the T-strand through the Mpf channel. Precisely how this latter reaction occurs is not known, but the CP structures suggest one possibility—the CP acts as a translocase to deliver the substrate across the inner membrane. In this context, it is interesting to note that CPs share limited sequence similarities with two DNA translocases, Bacillus subtilis SpoIIIE and E. coli FtsK [56,57].
5. Evidence for a transenvelope secretion channel: the Mpf structure
All of the VirB proteins except for VirB1 are required for substrate export [20]. Surprisingly, however, high-resolution electron microscopy studies of mating cells, e.g., RP4- carrying donor and recipient mating pairs, thus far have not identified a supramolecular organelle at cellular junctions. Instead, an electron dense layer was seen between tightly juxtaposed donor and recipient cell surfaces [58,59]. Junctions at the poles were estimated to be ~150–200 nm in length, whereas those along the cell body extended up to 1500 nm in length. No T4SS organelles reminiscent of the basal body of flagella or needle complexes of type III secretion systems (T3SS) were evident. Corresponding fractionation studies of several conjugation systems have shown that most subunits co-partition with both membranes in sucrose gradients and/or with a membrane fraction whose density is intermediate between those of the IM and OM [60–63]. The T4SS might thus assemble at zones of membrane adhesion, which in turn could correspond to the sites of electron density when mating pairs are examined by electron microscopy [59].
Despite a lack of ultrastructural resolution, several findings support the existence of an envelope-spanning secretory apparatus. For example, the presumptive VirB/D4 mating channel or subcomplexes thereof can be isolated by membrane solubilization with nonionic detergents, e.g., dodecyl maltoside, octylglucoside, or Triton X-100. In one study, high-molecular-weight VirB-containing species were detected by blue native gel electrophoresis and size exclusion chromatography. At least two large complexes were identified, one composed of the pilus-associated proteins VirB2, VirB5, and VirB7, and the second composed of several other VirB proteins and VirD4 [64]. Independent studies have also reported the isolation of a VirB2/VirB5/VirB7 complex and subcomplexes of other VirB proteins, e.g., VirB7/VirB9, VirB6/VirB7/VirB9 and VirB7/VirB9/VirB10 by detergent-solubilization and immunoprecipitation or GST-pull-down assays [19,22,23]. Results from two-hybrid screens complement these findings and further suggest the possibility of other pairwise interactions in vivo [52,65]. Similar studies of the F-like [2,63] and RP4 [62] have identified presumptive complexes among putative channel components.
Taken together with computer-based predictions and topology studies of individual VirB subunits, a general architecture for the VirB/D4 T4SS can be presented (Figs. 4 and 5). Accordingly, the VirD4 CP and the two Mpf ATPases, VirB4 and VirB11 ATPase, are positioned predominantly or exclusively at the cytoplasmic face of the IM. VirB6 is a highly hydrophobic protein predicted to span the IM several times, whereas VirB8 and VirB10 are bitopic proteins with short N-terminal cytoplasmic domains, a TM, and large C-terminal periplasmic domains. VirB2, VirB3, and VirB5 are located in the periplasm, and VirB2, VirB5, and VirB7 also assemble as the extracellular T pilus. VirB7, a small lipoprotein, and VirB9 form a covalently cross-linked dimer and this dimer or more probably a higher-order VirB7–VirB9 multimer assembles at the OM. Among the VirB proteins, VirB9 is the best candidate for forming an oligomeric secretion-like pore for mediating passage of substrates and/or protrusion of the T pilus across the OM (Fig. 4).
Fig. 5.
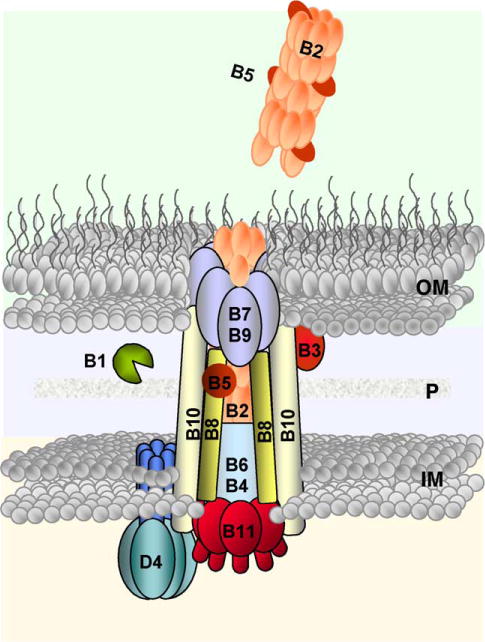
A model depicting the A. tumefaciens VirB/D4 T4SS as a single, supramolecular organelle. The VirD4 CP is a homomultimeric integral membrane complex required for substrate transfer. The VirB proteins assemble as a secretion channel and an extracellular T pilus. VirD4 and the VirB proteins function together to mediate substrate transfer, and the VirB proteins direct pilus assembly. Cells are postulated to shed the adhesive T pilus as a means of promoting aggregation of donor and recipients on solid surfaces. IM, inner membrane; P, periplasm; OM, outer membrane.
6. Energetic components—VirB4 and VirB11
In the next sections, information will be presented about the putative VirB channel subunits and the genetic requirements for substrate passage, focusing first on the energetic components.
VirB11
VirB11 is a member of a large family of ATPases associated with systems dedicated to secretion of macromolecules [66]. Homologs are widely distributed among the Gram-negative bacteria, and they also function in secretion systems of Gram-positive bacteria as well as various species of the Archaea.
VirB11 itself is extremely insoluble and has been difficult to characterize in vitro, whereas soluble forms of VirB11 homologs have been characterized enzymatically and structurally. Purified TrbBRP4, TrwCR388, and H. pylori HP0525Cag were each shown to assemble as homohexameric rings discernible by electron microscopy [67]. Like the TrwBR388 CP, HP0525Cag also presented as a homohexamer by X-ray crystallography [68]. However, quite unlike the CP, HP0525Cag is a double-stacked ring with a central cavity of ~50 Å in diameter. One ring is formed by N-terminal domain interactions, the second by C-terminal domain interactions. The N-terminal domain is unique to HP0525Cag, whereas the C-terminal domain adopts a RecA fold. In the first crystal structure generated, a molecule of ADP is bound at the interface between the two domains [68].
The overall HP0525Cag structure appears to be highly conserved, even among distantly related ATPases associated with other transport or fimbrial biogenesis systems. For example, L. pneumophila DotB of the Dot/Icm T4SS [69], Aquifex aeolicus PilT associated with type IV pilus retraction (these pili are assembled via a type II secretion system (T2SS) and are not related to type IV secretion) [70], and Actinobacillus actinomycetemcomitans TadA associated with Flp pilus biogenesis [71]; each purifies as ring-shaped homohexamers with dimensions similar to those observed for HP0525Cag. A crystal structure of Vibrio cholerae EpsE associated with the Eps T2SS recently was reported [72], and the closest structural homolog is HP0525Cag.
Both EpsEVch and HP0525Cag also are structurally similar to members of the AAA ATPase superfamily [72,73]. These structural similarities are intriguing because AAA ATPases display functions compatible with the possible activities of the EpsE_Vch_ and VirB11-like proteins. Many AAA ATPases are energy-dependent unfoldases whose activities are associated with substrate remodeling. In view of the fact that the T4SS are protein exporters, the VirB11-like ATPases might act to unfold these substrates at the channel entrance. Alternatively or in addition, a VirB11 chaperone activity might facilitate assembly of the secretory apparatus and/or the T pilus. Another interesting structural relationship was noted with the p97 AAA ATPase, which is involved in membrane fusion and organelle biogenesis [74,75]. In this context, it is interesting that the VirB11 homolog, TrwDR388, was shown to induce dose-dependent vesicle aggregation and intervesicular mixing of outer monolayer lipids [76,77]. While the investigators suggest this activity might be related to early stages of transporter assembly, it also seems possible that VirB11-like proteins are involved in membrane remodeling during formation of the mating pair junction.
As with TrwBR388 CP, the X-ray structure of the ADP-bound form of HP0525Cag offers only a snapshot view of the VirB11 ATPase structure. More recent comparative studies of the apo-, non-hydrolyzable ATPγS-, and ADP-bound structures, together with analytical ultracentrifugation experiments, showed that one consequence of ATP-binding is a swiveling of the N-terminal domains [75]. This conformational switch converts the open and asymmetric hexamer to a more compact, symmetrical hexamer (Fig. 4). Moreover, a molecule of polyethylene glycol was found bound to the N-terminal domain in the HP0525Cag crystal structure [68]. In vivo, this is probably a molecule of phospholipid, consistent with results of fractionation studies and other experimental findings showing that the VirB11-like ATPases associate tightly but peripherally at the inner face of the IM [78,79]. Interestingly, VirB11 as well as DotB_Lpn_ mutants with Walker A nucleotide-binding motif defects bind the IM more tightly than the wild-type protein, suggestive of an ATP-regulated membrane interaction [69,79].
Finally, genetic studies supplied evidence for the coordinated actions of VirB11 and the VirD4 CP for substrate transfer. A genetic screen resulted in isolation of virB11 alleles displaying dominance in virB11/virB11* merodiploid strains [80]. These alleles were then subclassified as functional or nonfunctional on the basis of whether the mutant protein supports substrate transfer in the absence of native VirB11. Studies of the functional subclass identified one mutant protein that supported substrate transfer (Tra+) but completely blocked biogenesis of the T pilus (Pil−). Conversely, some mutant proteins of the nonfunctional class supported T pilus biogenesis but blocked substrate transfer (Tra−, Pil+). Hence, these were termed “uncoupling” mutations, and their isolation indicates that VirB11 contributes both to pilus biogenesis and to assembly or function of the secretion machine by mechanisms distinguishable by mutation [81]. Such mutations might selectively disrupt VirB11 contacts with VirD4 to block substrate transfer or, alternatively, contacts with other Mpf proteins to disrupt either transfer or pilus biogenesis.
In considering the nature of the structural interaction between the CP and VirB11-like protein hexamers, in principle, the two subunits could form a composite double-stacked ring at the IM. Yet, such a structure is difficult to envision in view of the dimensions of the hexameric structures of the respective homologs and the subcellular dispositions of both subunits, e.g., both homohexamers associate with the IM, one integrally and one peripherally (Fig. 4). A more likely model is that the VirD4 and VirB11 hexamers form independently but localize next to each other at the IM (Fig. 5). These structures would undergo coordinated ATP-dependent conformational switches to mediate substrate recruitment to the CP and delivery to VirB11 for subsequent transfer through the Mpf structure. Consistent with this structural arrangement, Llosa et al. [57] developed a model based on evolutionary considerations that depicts conjugation systems as two separately acting IM translocases: the CP evolved from ring helicases exclusively for T-strand export and the Mpf proteins correspond to an ancestral protein export system. According to this model, the CP still functions as a general recruitment factor for all T4SS substrates. Instead of delivering the entire substrate to the Mpf proteins, however, the CP would translocate the T-strand across the IM while simultaneously transferring the relaxase to VirB11 and the other Mpf proteins for secretion. This interesting model satisfactorily explains many experimental findings. But without invoking significant, dynamic and tightly coordinated changes in CP/VirB11 conformations and oligomeric structures, one still has difficulty envisioning how two translocases sitting next to one another at the membrane coordinate their activities to drive secretion of one substrate—the covalently bound nucleoprotein particle—across the IM.
VirB4
VirB4 possesses two putative membrane-spanning domains, one near the N terminus and a second more centrally located and near the Walker A nucleoside triphosphate binding motif (Fig. 4) [78]. A transmembrane topology is also predicted for VirB4 homologs, e.g., TrbCRP4 [82]. With respect to subunit–subunit contacts, to date, there is biochemical evidence only for a VirB4 self-interaction [83]. A VirB4–VirB11 interaction is suggested by a yeast two-hybrid screen, but not yet confirmed with complementary biochemical or genetic approaches [65].
ATP hydrolysis has not been convincingly shown for purified VirB4-type proteins, but these proteins require intact Walker A motifs to mediate substrate export [82,84]. Intriguingly, several years ago it was reported that the presence of a subset of VirB proteins in agrobacterial recipient cells leads to a significant increase in the efficiency of plasmid acquisition in matings with agrobacterial donors [85]. These findings led to a proposal that these VirB proteins assemble as a complex that stabilizes mating junctions or perhaps directly facilitate DNA uptake across the recipient cell envelope. VirB4 is one of the VirB proteins that contribute to this stimulatory effect [19]. However, it was also shown that a Walker A mutation does not alter the capacity of VirB4 to stimulate plasmid acquisition by recipients. Therefore, VirB4 probably contributes structural information necessary for substrate transfer—in either direction—across the cell envelope, but an intact ATP-binding domain is necessary for configuring this T4SS specifically for substrate export [83]. Clearly, the VirD4, VirB11, and VirB4 proteins must interact in complex and dynamic ways—most probably via ATP-driven conformational changes—to energize substrate transfer to and across the IM (Figs. 4 and 5).
7. IM proteins—VirB6, VirB8, and VirB10
In contrast to the phylogenetically wide distribution of the VirB11-like ATPases, the VirB6, VirB8, and VirB10 IM proteins or functional orthologs are found mainly as components of T4SS elaborated by Gram-negative bacteria [1,2,66].
VirB6
VirB6 is a highly hydrophobic protein with five, six or nine possible TMs predicted by the TmPred, TMHMM, or TopPred2 algorithm, respectively (Fig. 4). Polytopic proteins are common components of secretion or transport systems, e.g., SecY, SecE, and SecG of the GSP [86] and TatC of the twin-arginine translocation (TAT) system [87]. Thus, early on it was postulated that VirB6 is a channel constituent. A virB6 deletion mutation results in diminished levels of several VirB proteins, consistent with a role in maintaining the structural integrity of this T4SS [20,22,88]. Additionally, Krall et al. [64] showed that VirB6 co-fractionates with other VirB proteins as a high-molecular-weight complex(es) in detergent-solubilized cell extracts, further suggestive of interactions with other VirB proteins as a stable supramolecular structure. Protein–protein interaction studies also have supplied evidence for VirB6 contacts with the VirB7 and VirB9 OM proteins. These interactions influence the formation of a disulfide cross-linked VirB7 homodimer as well as higher-order VirB9 multimers detectable by electrophoresis through protein gels under nonreducing conditions. Moreover, several four-residue insertion mutations affecting VirB9 multimerization mapped to a central, periplasmic loop of VirB6. The VirB6 interaction with the VirB7 and VirB9 proteins might constitute a contact between IM and OM channel subunits of this T4SS (see below) [22].
VirB8 and VirB10
VirB8 and VirB10 are both bitopic IM proteins with N-terminal proximal TMs and large C-terminal periplasmic domains (Fig. 4) [46]. VirB8 and VirB10 each were shown to self-associate and also to interact with each other and with VirB9 [23,52,89]. Mutations affecting these interactions abolished virulence, confirming that VirB8, VirB9, VirB10 complex formation is essential for machine function [52,89]. As noted above, foci composed of VirB8, VirB9, and VirB10 were identified around the cell periphery by immunofluorescence and electron microscopy [89]. On examination of various A. tumefaciens mutants, it was further shown that VirB8 production is necessary for focal formation of VirB9 and VirB10. The investigators proposed that VirB8 nucleates assembly of the VirB8, VirB9, VirB10 complex [89]. Consistent with the notion that these proteins form a stable complex, these three proteins co-fractionate in blue native gels and during gel filtration [64]. These findings plus the demonstrations of interactions between homologs of VirB10 and VirD4 [54,55] suggest that the VirB10 proteins function as structural scaffolds, linking CPs to their cognate T4SS and also IM subcomplexes to OM subcomplexes of the T4SS. Consistent with such a transmembrane ‘bridging’ function, VirB10 proteins possess Pro- or Glyrich central domains suggestive of a rigid, extended structure in the periplasm.
8. Periplasmic/OM proteins—VirB2, VirB7 and VirB9, VirB3 and VirB5, VirB1 and AcvB/VirJ
Six VirB proteins—VirB1, VirB2, VirB3, VirB5, VirB7, and VirB9—carry characteristic GSP signal sequences at their N termini. All of these proteins localize predominantly in the periplasm or OM, whereas VirB2, VirB5, and VirB7 also polymerize to form the extracellular T pilus (Fig. 4). The contributions of the cell-associated forms of these proteins to substrate transfer are discussed below.
VirB2
VirB2 is the pilin subunit of the T pilus (Fig. 4) [11]. The overall pilus biogenesis pathway is poorly understood (see below), but several of the processing reactions leading to mature pilin are well characterized for this and other conjugation systems. The propilin is delivered into or across the IM by the GSP. In the periplasm, cleavage of the signal sequence, most probably by leader peptidase LepB, is followed by one or more modification reactions. For example, the F TraA-like pilins are N-terminally acetylated, RP4 TrbCRP4 is proteolytically cleaved near its C terminus, and both TrbCRP4 and A. tumefaciens VirB2 undergo a novel head-to-tail cyclization reaction that covalently links N- and C-terminal residues making a closed circular polypeptide [2,90–93]. In all systems, mature pilin is integrated into the inner membrane; for F TraA, integration requires TraQ [2]. The integrated pilin is thought to comprise a pool of pilin monomers accessible for polymerization as the T pilus upon receipt of an exogenous signal from recipient cells.
The cell-associated VirB2 might be used exclusively for pilus biogenesis on demand, but one line of study suggests at least a portion of the pilin alternatively assembles as a structural subunit of the secretion channel in the periplasm (Fig. 4). We asked whether the Tra+, Pil− mutant strains described above are still capable of translocating substrate in the absence of the VirB2 pilin protein. We deleted the virB2 gene from strains synthesizing the virB9 or virB11 “uncoupling” alleles, and then we tested whether such Pil− cells retained the capacity to translocate substrates. The virB2 gene deletion abolished substrate transfer, establishing that VirB2 pilin is essential for substrate transfer (S.J. Jakubowski, unpublished data). Thus, the VirB/D4 T4SS can transfer DNA intercellularly in the absence of detectable T pilus, but not in the absence of pilin protein. The simplest explanation for these findings is that the pilin protein is a structural subunit of the secretion channel in the periplasm or at the OM.
VirB3 and VirB5
VirB3 and VirB5 are required for substrate transfer. Both proteins are exported across the IM where they sort to the periplasm and the OM (Fig. 4). Both proteins also are thought to play a role in biogenesis of the T pilus and, indeed, VirB5 has been shown to interact with the cell-associated VirB2 as well as with the extracellular T pilus [64,93,94]. With respect to VirB3, presently there are no reports of interactions with other VirB proteins, but a finding that production of VirB6 enhances steady-state levels of VirB5 and VirB3 suggests the possibility of stabilizing interactions among these VirB proteins [88]. Both proteins might thus contribute to assembly or function of the secretion channel in the periplasm.
Very recently, a crystal structure of a VirB5 homolog, TraC of pKM101, was presented [95]. This is a single domain protein with a mostly α-helical, elongated structure. Analyses of TraCKM101 mutants identified regions likely to be involved in protein–protein interactions. A couple of mutant proteins conferred altered conjugation frequencies as well as defects in infection by bacteriophages PRD1 and Ike. The investigators have proposed that the VirB5-like proteins bind along the pilus, including the pilus tip (the site of Ike attachment) and base (the site of PRD1 attachment); these sites of contact are thought to promote pilus biogenesis as well as adherence to recipient cells (Figs. 4 and 5) [95]. At this point, however, the TraCKM101 structure does not provide much insight into how this family of pilus-associated proteins contributes to channel formation or activity. One possibility is that VirB5 interacts with cell-associated VirB2 pilin in the periplasm and modulates VirB2 interactions with the VirB9 OM protein or the VirB6, VirB8, and VirB10 proteins in the periplasm or at the IM.
VirB7 and VirB9
VirB9 has nine possible β-sheet OM-spanning segments according to the Schirmer-Cowan algorithm [96] (S.J. Jakubowski, unpublished data). VirB9 assembles as a disulfide cross-linked heterodimer with the VirB7 lipoprotein (Fig. 4) [97–99]. Consistent with an OM localization of this dimer, the VirB7 sequence at the N-terminal lipid-modification site (CQTN) is devoid of the Asp2+ IM-retention signal [100]. Mutations of the reactive Cys residues involved in disulfide bridge formation destabilize VirB7 and VirB9 and several other VirB proteins, and confer avirulence unless VirB9 is significantly overproduced [96–99,101]. Thus, the heterodimer or, more probably, a higher-order multimer of dimers represents the physiologically relevant form of these subunits.
VirB9 also has sequence similarities with pore-forming domains of secretin proteins associated with type II (PulD) [102] and III (YscC) [103] secretion, filamentous bacteriophage f1 (pIV) [104], and Pap pilus biogenesis (PapC) [105] [2; S.J. Jakubowski, unpublished data]. Intriguing parallels also exist with a subset of secretins, namely those that form a stabilizing interaction with a cognate OM lipoprotein (e.g., PulS–PulD) [102]. These findings suggest the interesting possibility that VirB9, with a structural contribution of VirB7 lipoprotein, assembles as a secretin for substrate passage and, possibly, protrusion of the T pilus across the OM. At this time, however, a secretin-like function remains an enticing, but experimentally unproven, model.
VirB7 also forms a disulfide cross-linked homodimer that co-partitions with the IM and OM fractions and, curiously, is released into the extracellular milieu at abundant levels [61,90]. In pilus-producing cells, the VirB7 homodimer associates with T pili that are released by sloughing or shearing from the cell surface (Fig. 4) [90]. In cells, the lipoprotein is also found in this extracellular fraction. The physiological relevance of the cell-bound and extracellular VirB7 homodimer is not known, but these novel findings suggest this small lipoprotein contributes in several ways to assembly or function of this T4SS.
VirB1 and AcvB/VirJ
Three periplasmic proteins were postulated to contribute to T-strand transfer through effects on T4SS biogenesis or function. VirB1 carries a trans-glycosylase motif and is thought to “punch” holes in the peptidoglycan to facilitate T4SS assembly [106]. VirB1-related transglycosylases contribute to conjugal DNA trans fer as most extensively shown for the P19 transglycosylase of plasmid R1 [107]. Additionally, transglycosylases are important for assembly of other T4SS, PtlE for B. pertussis Ptl [108], and AtlA for an F-like Tra system carried on the gonococcal genetic island of N. gonorrhoeae that mediates DNA release into the milieu [4]. VirB1 transglycosylase activity is important for biogenesis of the A. tumefaciens T pilus, but not for substrate transfer, as shown by the capacity of the ΔvirB1 mutant to incite tumor formation on plants [19].
The Ti plasmid-encoded VirJ protein and its chromosomal homolog, AcvB, were shown to localize in the periplasm [109] and evidence was also presented for a VirJ interaction with the VirB/D4 T4SS as well as the VirE2 effector and VirD2 relaxase [110]. A virJ/acvB mutant is avirulent [109], leading to a proposal that these proteins function as periplasmic chaperones to mediate substrate transfer through the periplasm [110]. At this time, however, it is premature to conclude that AcvB and VirJ contribute directly to substrate transfer through the VirB/D4 T4SS.
9. T pilus and secretion channel: anatomical and spatial relationships
The genetic requirements for assembly of the secretion channel closely match those for T pilus biogenesis (Fig. 4). All of the VirB proteins are needed for both processes, with the exception of VirB1 transglycosylase, which as noted above is required for pilus production but not for substrate transfer. Thus, it is tempting to depict channel assembly and pilus outgrowth as two interlocked reaction pathways that culminate in assembly of a single, supramolecular organelle, the secretion channel/T-pilus. Accordingly, such a structure would consist of a set of IM channel proteins and a pilus structure extending through the periplasm and a (VirB9?) secretin-like pore at the OM (Fig. 5). Consistent with such a model, in other conjugation systems it has been shown that pilin subunits are recruited from the IM and added to the base of the pilus [2].
A prediction of this model is that the lumen of the pilus serves as a conduit for type IV secretion substrates. In fact, however, there is no strong experimental support for substrate transfer through an intact T pilus. To the contrary, at least two lines of evidence argue strongly against the conduit model, at least for the VirB/D4 system. First, a nonpolar null mutation of virB1 fails to assemble the T pilus, yet the ΔvirB1 mutant still translocates the oncogenic T-DNA to plant cells. This shows that the secretory apparatus is intact in Pil− cells [20]. Second, we have now isolated “uncoupling” mutations in several VirB proteins—VirB11 [10], VirB6 [22], and VirB9 (S.J. Jakubowski, unpublished data). As with the ΔvirB1 derivative, strains producing these mutant proteins transfer substrate in the absence of detectable T pilus production. The isolation of such mutations firmly establishes that the wild-type T pilus is not an obligatory organelle for T4SS-mediated substrate transfer.
The T pilus filament extending from the cell surface thus probably functions predominantly or exclusively to bring cells into close apposition for formation of the mating junction. As shown in Fig. 6, this can be achieved in two ways. Conjugative pili elaborated by the F T4SS and related systems are thought to use an extension–retraction mechanism. This mechanism is reminiscent of that described for type IV pili produced by T2SS (as noted above, in spite of the nomenclature, type IV pili are evolutionarily unrelated to conjugal pili) [2,111]. The T pili assemble and establish contact with recipient cells and then retract to bring the cells together, most probably by an energy-dependent process. During retraction, the pilin monomers might reinsert into the donor cell membrane, forming a pool for use in a new round of pilus biogenesis, or be shunted to a degradation pathway. Once cell membranes establish direct contact, formation of the stable mating junction triggers opening of the secretion channel for substrate passage across the OM.
Fig. 6.
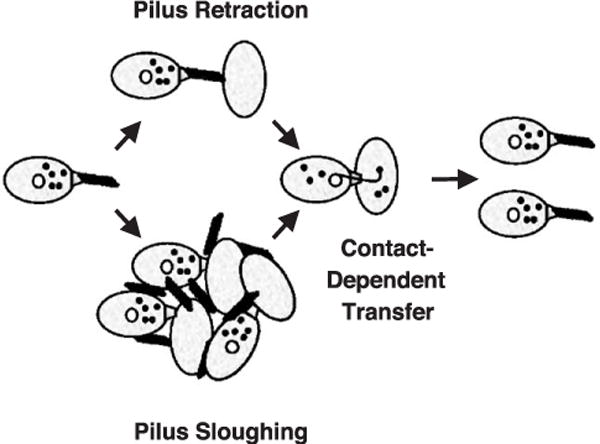
Conjugative pili likely function by a retraction mechanism, e.g., F pilus, or a sloughing mechanism, e.g., RP4, R388, VirB/D4 pili. Transfer systems utilizing the latter mechanism shed their pili presumably to induce aggregation of donor and recipient cells. Both mechanisms bring donor and recipient cell membranes into close apposition for formation of mating junctions. Contact between donor and recipient membranes stimulates channel opening and substrate transfer. Following transfer, the mating junction disassembles by an unknown mechanism(s). “Uncoupling” mutations of the VirB/D4 T4SS permit substrate transfer on solid surfaces in the absence of detectable pilus production [80,81].
Alternatively, conjugative pili encoded by plasmids of the IncP (e.g., RP4), IncN (pKM1010), or IncW (R388) incompatibility groups probably do not retract, but instead are sloughed from the cell surface (Fig. 6) [2,59]. Sloughing can be so extensive that cell-bound pili are difficult to detect by microscopy, as is the case for the RP4 pilus. It is not known what signals induce pilus sloughing, but the abundant accumulation of adhesive filaments in the milieu probably induces formation of cell aggregates and, consequently, mating junctions. As might be expected, cells elaborating F-like retracting pili transfer substrates efficiently in broth and on solid surfaces, whereas those dependent on pilus sloughing systems transfer substrates efficiently only on solid surfaces. The A. tumefaciens VirB/D4 T4SS likely use the pilus sloughing mechanism (Figs. 5 and 6), as suggested by extensive sequence relationships with the IncN, IncP, and IncW conjugation systems, efficient DNA transfer only on solid surfaces, and studies showing that T pili accumulate at abundant levels in the extracellular milieu [81,93].
Finally, it is interesting to consider why the VirB/D4 T4SS system—pilus and secretion channel—assembles at the A. tumefaciens poles (Fig. 3). With respect to the mechanics of secretion (Fig. 5) or formation of the mating junction (Fig. 6), it is not immediately obvious why such a polar localization would be advantageous. However, a recent study showed that the Ti plasmid, the source of the oncogenic T-DNA substrate, partitions to A. tumefaciens poles during cell division [112]. This finding raises the interesting, though completely uncharacterized, general question of how cells coordinate two metabolic reactions—plasmid partitioning and conjugation—that clearly cannot proceed simultaneously. It is conceivable that A. tumefaciens positioned the VirB/D4 T4SS at the poles in order to achieve temporal and spatial control of Ti plasmid segregation to daughter cells and T-DNA transfer to plant cells. Intriguingly, the A. tumefaciens VirC1 protein, which is thought to function as a Dtr protein by binding adjacent to the T-DNA border repeats, is a ParA homolog. ParA proteins function in segregation of chromosome and plasmid DNA to daughter cells during cell division [13]. Thus, VirC1 might function as part of a global regulatory mechanism operating to control the timing of T-DNA transmission during the division cycle.
10. Summary and future directions
Conjugation machines are remarkably dynamic surface organelles. Detailed structure–function studies of the A. tumefaciens VirB/D4 T4SS and other paradigmatic conjugation systems have shown that the conjugation systems are composed of three protein subcomplexes, Dtr, CP, and Mpf, that act sequentially to drive three distinct reactions, substrate processing, recruitment, and transfer. Two especially noteworthy recent advances include: (i) crystallographic structures of three T4SS subunits, homologs of the VirD4 CP, pilus-associated VirB5, and the VirB11 ATPase; and (ii) definition of subunit–subunit contacts among machine components. The X-ray structures and accompanying biochemical findings suggest the energetic components undergo cycles of ATP-dependent conformational changes that affect intersubunit contacts. The TrwBR388 and HP0525Cag are ring-shaped homohexamers structurally similar to proteins that move along DNA and AAA ATPase unfoldase proteins, respectively. It is tempting to suggest that ATP-binding regulates T-strand-relaxase substrate interactions and associated translocase or unfoldase activities. Our knowledge of the CP and VirB11-like proteins is now sufficient to apply structure-based mutagenesis strategies to define specific contributions of individual residues or domains to machine biogenesis and substrate transfer.
Correspondingly, studies defining topologies and subunit–subunit contacts among the channel components are yielding a structural view of a secretory apparatus that so far has eluded detection by electron microscopy. The findings have identified several interactions of interest, for example, between energetic components, the putative IM channel subunits VirB6, VirB8, and VirB10, and the periplasmic/OM proteins VirB2, VirB7, and VirB9. Further studies should address the following questions:
- How do the three putative ATPase subunits, VirD4, VirB11, and VirB4, coordinate their activities to recruit and drive substrate transfer across the IM?
- What is/are the IM translocase—VirD4, VirB11, or VirB6/VirB8/VirB10—or a combination of these components?
- What is the role of the VirB10 IM-OM “bridge” in mediating T4SS assembly or function?
- What is the structure–function relationship between the T-pilus and the secretion channel?
- What gating mechanisms regulate channel opening and closure at both the IM and OM?
Over the course of the next few years, studies of these and other questions will supply many examples—at an increasingly atomic level of resolution—of the dynamic nature of these fascinating translocation machines.
Acknowledgments
I thank members of my laboratory for helpful discussions and contributions to development of the figures. I thank Alain Bernadac for the electron micrograph showing T pili at the A. tumefaciens cell pole. Work in my laboratory is supported by NIH grant GM47846.
References
- 1.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 3.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 5.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 6.Ding Z, Atmakuri K, Christie PJ. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 2003;11:527–535. doi: 10.1016/j.tim.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–383. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 8.Baron C, O’Callaghan D, Lanka E. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol Microbiol. 2002;43:1359–1365. doi: 10.1046/j.1365-2958.2002.02816.x. [DOI] [PubMed] [Google Scholar]
- 9.Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O’Callaghan D. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A. 2002;99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie PJ. The Agrobacterium T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai EM, Chesnokova O, Banta LM, Kado CI. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–3716. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, von Bodman SB, Lanka E, Ream W, Farrand SK. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CH, Winans SC. Resection and mutagenesis of the acid pH-inducible P2 promoter of the Agrobacterium tumefaciens virG gene. J Bacteriol. 1996;178:4717–4720. doi: 10.1128/jb.178.15.4717-4720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachman MA, Swanson MS. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol. 2001;40:1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 18.Rouot B, Alvarez-Martinez MT, Marius C, Menanteau P, Guilloteau L, Boigegrain RA, Zumbihl R, O’Callaghan D, Domke N, Baron C. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect Immun. 2003;71:1075–1082. doi: 10.1128/IAI.71.3.1075-1082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Binns AN. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J Bacteriol. 2003;185:3259–3269. doi: 10.1128/JB.185.11.3259-3269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez D, Spudich GM, Zhou X-R, Christie PJ. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakubowski SJ, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J Bacteriol. 2003;185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaupré CE, Bohne J, Dale EM, Binns AN. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clewell DB, Helinski DR. Supercoiled circular DNA–protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci U S A. 1969;62:1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 26.Howard EA, Zupan JR, Citovsky V, Zambryski PC. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 27.Bravo-Angel AM, Gloeckler V, Hohn B, Tinland B. Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J Bacteriol. 1999;181:5758–5765. doi: 10.1128/jb.181.18.5758-5765.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees CED, Wilkins BM. Protein transfer into the recipient cell during bacterial conjugation: studies with F and RP4. Mol Microbiol. 1990;4:1199–1205. doi: 10.1111/j.1365-2958.1990.tb00695.x. [DOI] [PubMed] [Google Scholar]
- 29.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins BM, Thomas AT. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol Microbiol. 2000;38:650–657. doi: 10.1046/j.1365-2958.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- 31.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jacome E, Hooykaas PJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergunst AC, Van Lier MC, den Dulk-Ras A, Hooykaas PJ. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 2003;133:978–988. doi: 10.1104/pp.103.029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simone M, McCullen CA, Stahl LE, Binns AN. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol. 2001;41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 34.Luo Z-Q, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at the cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49:1699–1733. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol. 2002;43:1523–1532. doi: 10.1046/j.1365-2958.2002.02829.x. [DOI] [PubMed] [Google Scholar]
- 37.Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:1–6. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 38.Cabezon E, Lanka E, de la Cruz F. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J Bacteriol. 1994;176:4455–4558. doi: 10.1128/jb.176.14.4455-4458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 40.Sastre JI, Cabezon E, de la Cruz F. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J Bacteriol. 1998;180:6039–6042. doi: 10.1128/jb.180.22.6039-6042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moncalian G, Cabezon E, Alkorta I, Valle M, Moro F, Valpuesta JM, Goni FM, de La Cruz F. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J Biol Chem. 1999;274:36117–36124. doi: 10.1074/jbc.274.51.36117. [DOI] [PubMed] [Google Scholar]
- 42.Hormaeche I, Alkorta I, Moro F, Valpuesta JM, Goni FM, de La Cruz F. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J Biol Chem. 2002;277:46456–46462. doi: 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- 43.Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szpirer CY, Faelen M, Couturier M. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol Microbiol. 2000;37:1283–1292. doi: 10.1046/j.1365-2958.2000.02077.x. [DOI] [PubMed] [Google Scholar]
- 45.Disque-Kochem C, Dreiseikelmann B. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol. 1997;179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das A, Xie YH. Construction of transposon Tn3PhoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–414. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- 47.Lee MH, Kosuk N, Bailey J, Traxler B, Manoil C. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J Bacteriol. 1999;181:6108–6113. doi: 10.1128/jb.181.19.6108-6113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 49.Gomis-Ruth FX, Moncalian G, de la Cruz F, Coll M. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J Biol Chem. 2002;277:7556–7566. doi: 10.1074/jbc.M110462200. [DOI] [PubMed] [Google Scholar]
- 50.Gomis-Ruth FX, de la Cruz F, Coll M. Structure and role of coupling proteins in conjugal DNA transfer. Res Microbiol. 2002;153:199–204. doi: 10.1016/s0923-2508(02)01313-x. [DOI] [PubMed] [Google Scholar]
- 51.Schroder G, Lanka E. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388) J Bacteriol. 2003;185:4371–4381. doi: 10.1128/JB.185.15.4371-4381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilmour MW, Lawley TD, Rooker MM, Newnham PJ, Taylor DE. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol Microbiol. 2001;42:705–715. doi: 10.1046/j.1365-2958.2001.02682.x. [DOI] [PubMed] [Google Scholar]
- 54.Gilmour MW, Gunton JE, Lawley TD, Taylor DE. Interaction between the IncHI1plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol Microbiol. 2003;49:105–116. doi: 10.1046/j.1365-2958.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- 55.Llosa M, Zunzunegui S, Alfonso C, Rivas G, de la Cruz F. Protein TrwB interacts with protein TrwE and with the relaxosome, undercoring its coupling role in bacterial conjugation. Proc Natl Acad Sci U S A. 2003;100:10465–10470. [Google Scholar]
- 56.Errington J, Bath J, Wu LJ. DNA transport in bacteria. Nat Rev, Mol Cell Biol. 2001;2:538–545. doi: 10.1038/35080005. [DOI] [PubMed] [Google Scholar]
- 57.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 58.Durrenberger MB, Villiger W, Bachi T. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 59.Samuels AL, Lanka E, Davies JE. Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol. 2000;182:2709–2715. doi: 10.1128/jb.182.10.2709-2715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thorstenson Y, Kuldau G, Zambryski P. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993;175:5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez D, Dang TAT, Spudich GM, Zhou X-R, Berger BR, Christie PJ. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grahn AM, Haase J, Bamford DH, Lanka E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: Implications for related macromolecule transport systems. J Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris RL, Hombs V, Silverman PM. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol Microbiol. 2001;42:757–766. doi: 10.1046/j.1365-2958.2001.02667.x. [DOI] [PubMed] [Google Scholar]
- 64.Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 2002;99:11405–11410. doi: 10.1073/pnas.172390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward DV, Draper O, Zupan JR, Zambryski PC. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci U S A. 2002;99:11493–11500. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krause S, Barcena M, Panseqrau W, Lurz R, Carazo J, Lanka E. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc Natl Acad Sci U S A. 2000;97:3067–3072. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeo H-J, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 69.Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol. 2004;186:1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herdendorf TJ, McCaslin DR, Forest KT. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J Bacteriol. 2002;184:6465–6471. doi: 10.1128/JB.184.23.6465-6471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J Mol Biol. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 73.Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- 74.Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- 75.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Machon C, Rivas S, Albert A, Goni FM, de la Cruz F. TrwD, the hexameric traffic ATPase encoded by plasmid R388, induces membrane destabilization and hemifusion of lipid vesicles. J Bacteriol. 2002;184:1661–1668. doi: 10.1128/JB.184.6.1661-1668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goni FM. Non-permanent proteins in membranes: when proteins come as visitors. Mol Membr Biol. 2002;19:237–245. doi: 10.1080/0968768021000035078. [DOI] [PubMed] [Google Scholar]
- 78.Dang TAT, Christie PJ. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rashkova S, Spudich GM, Christie PJ. Mutational analysis of the Agrobacterium tumefaciens VirB11 ATPase: identification of functional domains and evidence for multimerization. J Bacteriol. 1997;179:583–589. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou X-R, Christie PJ. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sagulenko Y, Sagulenko V, Chen J, Christie PJ. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol. 2001;183:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rabel C, Grahn AM, Lurz R, Lanka E. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J Bacteriol. 2003;185:1045–1058. doi: 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dang TAT, Zhou X-R, Graf B, Christie PJ. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol Microbiol. 1999;32:1239–1253. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berger BR, Christie PJ. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bohne J, Yim A, Binns AN. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc Natl Acad Sci U S A. 1998;95:7062–8057. doi: 10.1073/pnas.95.12.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stathopoulos C, Hendrixson DR, Thanassi DG, Hultgren SJ, St Geme JW, III, Curtiss R., III Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2000;2:1061–1072. doi: 10.1016/s1286-4579(00)01260-0. [DOI] [PubMed] [Google Scholar]
- 87.Buchanan G, Leeuw Ed E, Stanley NR, Wexler M, Berks BC, Sargent F, Palmer T. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol Microbiol. 2002;43:1457–1470. doi: 10.1046/j.1365-2958.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 88.Hapfelmeier S, Domke N, Zambryski PC, Baron C. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J Bacteriol. 2000;182:4505–4511. doi: 10.1128/jb.182.16.4505-4511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar RB, Das A. Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8. J Bacteriol. 2001;183:3636–3641. doi: 10.1128/JB.183.12.3636-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie PJ. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J Bacteriol. 2001;183:3642–3651. doi: 10.1128/JB.183.12.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 92.Eisenbrandt R, Kalkum M, Lurz R, Lanka E. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J Bacteriol. 2000;182:6751–6761. doi: 10.1128/jb.182.23.6751-6761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lai EM, Eisenbrandt R, Kalkum M, Lanka E, Kado CI. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J Bacteriol. 2002;184:327–330. doi: 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Baron PC, Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeo H-J, Yuan Q, Beck MR, Baron C, Waksman G. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci U S A. 2003;100:15947–15952. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cao TB, Saier MH., Jr Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology. 2001;147:3201–3214. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]
- 97.Spudich GM, Fernandez D, Zhou XR, Christie PJ. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci U S A. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anderson LB, Hertzel AV, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci U S A. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baron C, Thorstenson YR, Zambryski PC. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 101.Das A, Anderson LB, Xie YH. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol. 1997;179:3404–3409. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley AP. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci U S A. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis GR, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocoliticaforms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 104.Linderoth NA, Simon MN, Russel M. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science. 1997;278:1635–1638. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 105.Thanassi DG, Saulino ET, Lombardo MJ, Roth R, Heuser J, Hultgren SJ. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci U S A. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Llosa M, Zupan J, Baron C, Zambryski P. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J Bacteriol. 2000;182:3437–3445. doi: 10.1128/jb.182.12.3437-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Hogenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rambow-Larsen A, Weiss AA. The PtlE proteins of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J Bacteriol. 2002;184:2863–2869. doi: 10.1128/JB.184.11.2863-2869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kalogeraki VS, Winans SC. The octopine-type Ti plasmid pTiA6 of Agrobacterium tumefaciens contains a gene homologous to the chromosomal virulence gene acvB. J, Bacteriology. 1995;177:892–897. doi: 10.1128/jb.177.4.892-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 111.McBride MJ. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 112.Kahng LS, Shapiro L. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J Bacteriol. 2003;185:3384–3391. doi: 10.1128/JB.185.11.3384-3391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]