Victors: a web-based knowledge base of virulence factors in human and animal pathogens (original) (raw)
Abstract
Virulence factors (VFs) are molecules that allow microbial pathogens to overcome host defense mechanisms and cause disease in a host. It is critical to study VFs for better understanding microbial pathogenesis and host defense mechanisms. Victors (http://www.phidias.us/victors) is a novel, manually curated, web-based integrative knowledge base and analysis resource for VFs of pathogens that cause infectious diseases in human and animals. Currently, Victors contains 5296 VFs obtained via manual annotation from peer-reviewed publications, with 4648, 179, 105 and 364 VFs originating from 51 bacterial, 54 viral, 13 parasitic and 8 fungal species, respectively. Our data analysis identified many VF-specific patterns. Within the global VF pool, cytoplasmic proteins were more common, while adhesins were less common compared to findings on protective vaccine antigens. Many VFs showed homology with host proteins and the human proteins interacting with VFs represented the hubs of human–pathogen interactions. All Victors data are queriable with a user-friendly web interface. The VFs can also be searched by a customized BLAST sequence similarity searching program. These VFs and their interactions with the host are represented in a machine-readable Ontology of Host–Pathogen Interactions. Victors supports the ‘One Health’ research as a vital source of VFs in human and animal pathogens.
INTRODUCTION
Infectious diseases caused by pathogenic microorganisms remain globally a major source of human mortality (1), and veterinary infectious diseases result in significant losses of livestock, pets and wildlife (2). Domestic animals and wild primates are also major sources of human infectious diseases (3). Approximately 60% of all infectious diseases in humans are zoonotic diseases that are transmissible from animals to humans (4); however, there is still a big gap in understanding the underlying mechanism. To more effectively study the zoonotic diseases and achieve better public health outcomes, the emerging ‘One Health’ movement aims to unite human and veterinary medicine and integrate human, animal and environmental health (5). Understanding the molecular mechanisms that enable pathogens to cause infectious diseases is critical to find effective treatment strategies for combating human and animal infectious diseases. In particular, further studies of virulence factors (VFs) that allow microbial pathogens to evade host defense mechanisms and cause disease will improve rational vaccine and drug design for preventing and treating infectious diseases.
Currently, several VF databases are available online. Among them, the Virulence Factors of Pathogenic Bacteria (VFDB) is a database of bacterial VFs with a focus on human bacterial pathogens (http://www.mgc.ac.cn/VFs/) (6). VFDB does not include VFs from parasites, fungi or viruses. VFDB only provides general VF references without the detail of the experimental verification evidences. Another database, PHI-base (http://www.phi-base.org/) contains genes involved in host–pathogen interactions. In addition to its primary focus on plant pathogens, PHI-base also contains medically important pathogens that can cause human diseases, and the database provides links to appropriate publications (7). In addition to VFs, PHI-base also includes pathogen proteins that interact with host without known effect on pathogenesis (7). The PATRIC bacterial bioinformatics database BLASTs all genomes, both public and private, against a database of virulence factors and displays those genes with homology on a page showing information about the genome and on a page for specialty genes of interest (8,9). PATRIC imports the VFs from VFDB and Victors (the database introduced in this paper). In addition, PATRIC has manually curated virulence factors for some bacterial pathogens and provides integrative data annotation and analysis (10).
To address the need for a comprehensive, curated database of human, animal and zoonotic pathogen VFs and to improve our understanding of the complexities of host–pathogen interactions, the Victors database was created. Victors is a manually curated, web-based (http://www.phidias.us/victors) VF database and analysis engine for human and animal pathogens. As an independent program in the PHIDIAS pathogen–host interaction database (11), whose original focus was on integrating existing sequence and disease information from various human and animal pathogens, the Victors VF database has an increased emphasis on the manual annotation and analysis of VFs. Each of the VFs in Victors is associated with manually annotated evidence from at least one peer-reviewed citation. It currently has over 5000 VFs from over 100 bacterial, viral, parasitic and fungal pathogens. The characteristics and functions of these VFs were extracted or analyzed using bioinformatics tools. Furthermore, Victors enabled us to make novel predictions about the interactions between specific proteins of the human host and experimentally verified microbial VFs.
SYSTEM DESIGN, ANNOTATION, ANALYSIS PIPELINE AND STATISTICS
System and database design
Victors is implemented using a three-tier architecture built on two University of Michigan virtual servers that run the Redhat Linux operating system (Redhat Enterprise Linux ES 4). Users can submit database or analysis queries through the web. These queries are then processed using PHP/SQL (middle-tier, application server based on Apache) against a MySQL (version 5.0) relational database (back-end, database server). The result of each query is then presented to the user in the web browser. Two servers are scheduled to back up data every week. Victors is a relatively independent program of the PHIDIAS database and analysis resource (11).
Semi-automatic annotation of VFs
The semi-automatic Victors annotation system was developed by modifying an in-house web-based literature mining and curation system called Limix (11) (Supplementary Figure S1). The interactive Limix data submission and review system: (i) allows a curator to search literature, copy and edit text, and submit data to the database and (ii) provides a data reviewer tools to review, edit and approve the curated data on one comprehensive web interface. The major criterion to classify a VF is the loss or reduction of pathogenicity in the host after the VF gene mutation. Limix also features automated reference tracking and management. Upon approval after critical review by a domain expert (i.e. a scientist who is knowledgeable in the domain and has at least a postdoctoral position), the data will be posted publicly.
VF Statistics
Victors currently contains 5296 VFs from 126 pathogens. Specifically, Victors includes 1160 VFs from 15 Gram-positive pathogens, 3488 VFs from 36 Gram-negative pathogens, 179 VFs from 54 viruses, 105 VFs from 13 parasitic pathogens and 364 VFs from 8 fungal pathogens that primarily infect humans and other animal species (Table 1 and Supplementary Table S1 shows representative statistics). More detailed statistical information is available at http://www.phidias.us/victors/stats.php.
Table 1.
Representative Victors statistics as of 14 August 2018
| # | Pathogen | No. of VFs | No. of human PPIs |
|---|---|---|---|
| G+ Bacteria: out of 1160 VFs from 15 pathogens | |||
| 1 | Bacillus anthracis | 54 | – |
| 2 | Clostridium botulinum | 7 | 470-2* |
| 3 | Listeria monocytogenes | 106 | – |
| 4 | Mycobacterium tuberculosis | 360 | 192-11 |
| 5 | Streptococcus agalactiae | 30 | 1,224-12 |
| 6 | Streptococcus pneumoniae | 413 | 1549-18 |
| 7 | Streptococcus pyogenes | 77 | 2144-12 |
| G- Bacteria: out of 3488VFs from 36 pathogens | |||
| 1 | Brucella spp. | 439 | 2203-221 |
| 2 | Escherichia coli | 569 | 1412-84 |
| 3 | Haemophilus influenzae | 52 | 9-4 |
| 4 | Legionella pneumophila | 68 | 48-9 |
| 5 | Neisseria meningitidis | 175 | 1507-21 |
| 6 | Salmonella spp. | 387 | 1765-16 |
| 7 | Shigella spp. | 349 | 2024-84 |
| Viruses: out of 179 from 54 pathogens | |||
| 1 | Feline infectious peritonitis virus | 5 | – |
| 2 | Herpes simplex virus type 1 and 2 | 9 | – |
| 3 | Pseudorabies virus | 9 | – |
| Parasites: out of 105 from xx pathogens | |||
| 1 | Plasmodium spp. | 11 | – |
| 2 | Toxoplasma gondii | 9 | – |
| Fungi: out of 232 from xx pathogens | |||
| 1 | Candida albicans | 108 | 216-12 |
| 2 | Cryptococcus neoformans | 66 | – |
Bioinformatic analyses
The Clusters of Orthologous Group (COG) categories of VFs were retrieved from the NCBI COG database (12). The protein MW and PI are calculated from the protein sequences using the Bioperl program (https://bioperl.org/) (13). The Vaxign analysis pipeline (14) was used to analyze the subcellular localization, adhesin probability and conserved domains of VFs. Customized BLAST was used for BLAST sequence similarity search (15). For the prediction of host–pathogen protein–protein interactions (PPIs), the orthologs of proteins were defined by employing Inparanoid program (http://inparanoid.cgb.ki.se/) with default parameters (16), and the PPI database HPRD (http://www.hprd.org/) was used to identify the PPIs (17). More detail about PPI prediction is provided in Supplementary Figure S2.
Database contents
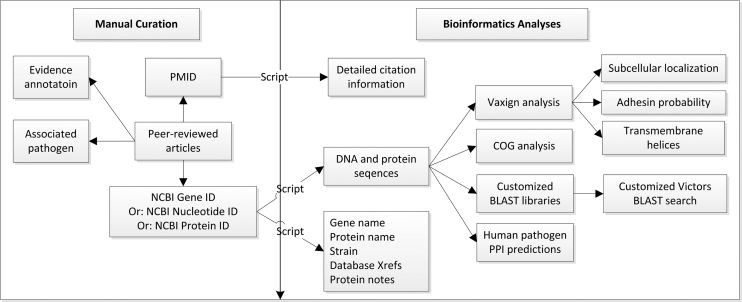
For each specific VF, the Victors database contains the following information: (i) general information on each VF gene symbol, protein name, general gene/protein functions, COG category (if available), and DNA and protein sequences; (ii) manually annotated text from peer-reviewed article that proves the status of VF and (iii) computationally calculated results for each VF, for example, protein weight, protein pI, subcellular localization, adhesin probability, conserved domains of VFs and predicted PPIs (Figure 1). Supplementary Table S2 summarized the COG functional classification of all Victors VFs and Brucella VFs stored in Victors. More analysis results are described below.
Figure 1.
The workflow of the Victors virulence factor annotation and analysis. .
VF DISTINCT PATTERNS COMPARED TO PROTECTIVE ANTIGENS
Previous studies found that protective antigens used for successful vaccine development are typically enriched in the areas of adhesin proteins, extracellular or cell surface bacterial proteins, and do not have sequence similarity to host proteins (18–20). We first compared the proportions of adhesin proteins in VFs and the vaccine immunogen proteins. We found that only 4% of viral VFs are adhesins or adhesin-like proteins (Table 2), while in the remaining types of pathogens adhesins or adhesin-like proteins represent 9–13% of VFs. Using the same Vaxign analysis method (14), it was previously found that over 50% of protective antigens used for development of experimentally verified vaccines are adhesins or adhesin-like proteins (20,21), showing that the VFs and protective antigens exhibit principally distinct molecular patterns.
Table 2.
Vaxign analysis of Victors virulence factors
| Pathogens | G+ bacterium | G- bacterium | Viruses | Parasites | Fungi | Total |
|---|---|---|---|---|---|---|
| Total VFs analyzed | 1160 | 3489 | 179 | 105 | 364 | 5297 |
| Predicted adhesin | 124 (0.11) | 439 (0.13) | 8 (0.04) | 9 (0.09) | 19 (0.05) | 599 (0.11) |
| Similarity to human proteins | 292 (0.25) | 665 (0.19) | 21 (0.12) | 30 (0.29) | 180 (0.49) | 1188 (0.22) |
| Similarity to mouse protein | 293 (0.25) | 670 (0.19) | 21 (0.12) | 31 (0.3) | 181 (0.5) | 1196 (0.23) |
| Extracellular | 117(0.1) | 274 (0.08) | – | – | – | – |
| Cell wall | 73 (0.06) | – | – | – | – | – |
| Outer membrane | – | 282 (0.08) | – | – | – | – |
| Cytoplasmic | 792 (0.68) | 2021 (0.58) | – | – | – | – |
| Cytoplasmic membrane | 282 (0.24) | 560 (0.17) | – | – | – | – |
| Periplasmic | – | 165 (0.05) | – | – | – | – |
| Unknown | 152 (0.13) | 543 (0.16) | – | – | – | – |
We further examined the frequency of VFs that possess human orthologs, which were detected as defined in reference (14), among different groups of pathogens. Out of 4749 proteins, 1188 (22%) are homologous to some human proteins (Table 2). Among them, 30 (29%) out of 109 parasitic proteins and 180 (49%) out of 364 fungal proteins have orthologs in human proteins. In contrast, only 21 (12%) out of 179 viral proteins have human orthologs. There are 665 (19%) and 292 (25%) VFs from Gram-negative and Gram-positive bacteria, respectively, which have homology to human proteins. The results suggest that fungi and parasites, both eukaryotic organisms, could much more frequently use the ‘orthologue strategy’ to interact with the host and as a mechanism of virulence. Similar results were obtained when the VFs were compared with the mouse proteome (Table 2).
Finally, we examined the subcellular locations of VFs among different groups of pathogens, including distinctions between Gram+ and Gram- bacterial VFs. Table 2 also shows the statistics of the cellular locations of virulence factors in Gram+ and Gram- bacterial VFs. Interestingly, 82% and 75% of Gram+ and Gram- bacterial VFs are located in the cytoplasmic and cytoplasmic membrane areas. However, the portions of extracellular or cell wall (or outer membrane) proteins are relatively low, representing <20% of the total bacterial VFs. These results indicate that key bacterial VFs are ‘designed’ to work inside the bacterial cells rather than on the surface or as secreted molecules. These results contrast with the patterns of bacterial protective antigens, where over 50% protective antigens are located on the cell surface or secreted out (20). Therefore, similar to the adhesin and ortholog studies reported above, the VFs and protective antigens also follow differential patterns of subcellular localization.
PREDICTION OF HOST–PATHOGEN INTERACTIONS BASED ON VICTORS
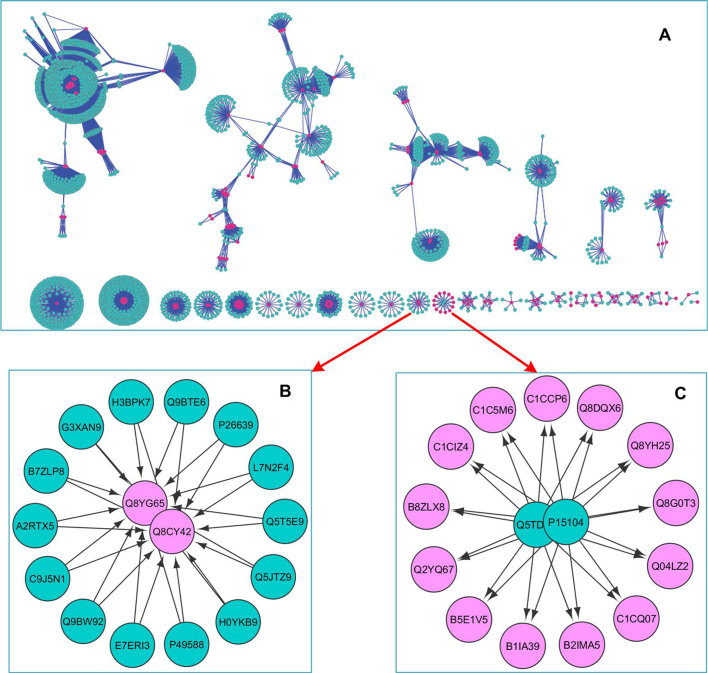
Our next goal was to assess the value of Victors in predicting pathogen–human PPIs based on the analyses of protein domains and phylogeny. Using all of the VFs experimentally verified in the human system, we searched for potential interactions among the host proteins. As detailed in Supplementary Figure S2, candidate PPIs were generated using ‘domain inferred’ and ‘ortholog inferred’ predictions (22–24). The ‘domain inferred prediction’ was used to infer inter-species PPIs based on the domain–domain interactions (DDI). Such DDI inference is theoretically supported by the conservation and importance of domains in proteins (24). If one domain in the pathogen protein A interacted with another domain in the human protein B (domain interacts with domain), we inferred these two proteins could potentially interact (protein A interacts with protein B). The ‘ortholog inferred prediction’ infers the inter-species PPIs based on their orthologs. If a specific protein A interacts with another protein B and has an ortholog protein C, we inferred that protein B and C interact with each other. The interactions generated using ‘domain inferred’ and ‘ortholog inferred’ methods were then combined into a unique human–pathogen interactome (Figure 2 and Supplementary Figure S2).
Figure 2.
Landscapes of predicted bacteria pathogen–human PPIs. (A) All 15 334 PPIs. (B) This figure shows that two pathogen proteins (GPT pyrophosphokinase rsh) from Brucella suis (Q8CY42) and Brucella melitensis (Q8YG65) interact with a group of human proteins (blue dots) on tRNA level, such as t-RNA editing proteins, tRNA ligase proteins. The GO enrichment analysis reveals that this group of proteins is enriched in tRNA aminoacylation (A2RTX5|Q9BW92|Q5JTZ9|P49588|P26639), corrected _P_-value 9.5487E-12. (C) This figure shows that two human proteins lengsin (Q5TDP6) and Glutamine synthetase (P15104) interact with a group of pathogen proteins from different species, such as Streptococcus pneumonia and Brucella suis, Brucella melitensis. Interestingly, all pathogen proteins are Glutamine synthetase. Nodes in red color are pathogen proteins, and nodes in blue are human proteins.
Our initial candidate PPI predictions based on the ortholog and domain interfaces used all the proteins from individual pathogens. However, the results included many false positives since some pathogen proteins may not interact with host proteins due to various reasons (e.g. physical barriers). In order to increase the quality of the PPI predictions, we utilized experimentally verified VFs in Victors to screen candidate pairs. Based on our literature annotation, 553 VFs from 30 pathogen species were confirmed experimentally using human-specific analysis approaches (e.g. human cells). The PPI pairs that had no Victors’ VFs were removed. Finally, we inferred 15 334 human–pathogen interactions comprised of 185 VFs from 19 pathogen species and 2502 human proteins (Table 1 and Supplementary Table S3).
Bacterial VFs interact with host (e.g. human) proteins in the complex process of bacterium–host interactions. Our approach predicted 15 334 bacteria–human PPIs from 11 bacteria (Supplementary Figure S3). For each of the 15 334 PPIs, we calculated the degree of proteins and then plotted the density curves (Supplementary Figure S3), which uncovered higher degree of VFs interactions with human proteins, compared to that of the entire bacterial proteome. These findings point out that relative to the non-VF bacterial proteins, VFs are much more likely to interact with human proteins.
To visualize the landscape of predicted bacteria–human PPIs, Cytoscape (25) was utilized to display the interaction networks and perform GO enrichment analyses (Figure 2). The results revealed that most of the interactions were well-connected and formed 31 component networks in 7 specific network patterns (Figure 2). Interestingly, many proteins are found at the nexus of ‘linkages’ or ‘hubs’ of large numbers of interactions. To further explore these multiple targeted proteins, we calculated the number of interacting bacterial VFs for each human protein (Supplementary Table S3). For example, each of two cell adhesion functional proteins, P13611 (Versican core protein) and Q96GW7 (Brevican core protein), connects to 17 VFs, consistent with the process of attaching to human cells being required for bacterial virulence.
DATABASE QUERY AND ACCESS
Database query and Blast analysis
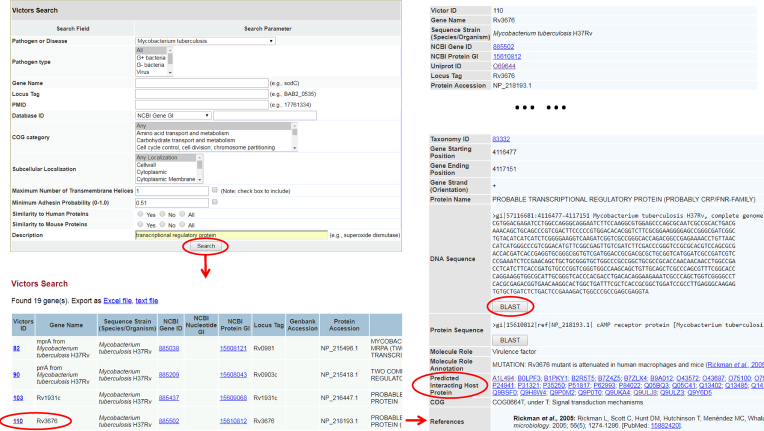
The manually curated and pre-computed VFs data can be efficiently queried specifying one or multiple criteria: (i) gene or protein name, (ii) pathogen species or strain, (iii) locus tag of a VF, (iv) other existing database identifiers, such as NCBI Gene ID, NCBI nucleotide or protein GI, (v) keywords search with Boolean mode support, (vi) COG category, (vii) subcellular localization, (viii) four limiting factors, including the maximum number of transmembrane helices, the minimum adhesin probability and protein sequence similarity to human proteins, and protein sequence similarity to mouse proteins. User-friendly web interfaces are provided to display the results (Figure 3).
Figure 3.
Example of Victors data query and BLAST sequence similarity analysis. In this example, a user selected ‘_Mycobacterium tuberculosis_’ in the ‘Pathogen or Disease’ menu bar and typed ‘transcriptional regulatory protein’. After clicking the ‘Search’ button, a list of 19 genes was shown up in the results page. A click on the Victors ID ‘110’ (the forth on the list) led to another page providing details about the VF (e.g. the gene mutation phenotype information and predicted interacting host proteins). A BLAST search was initiated by clicking ‘BLAST’.
To facilitate antigen comparison and dynamical sequence analysis, two customized BLAST libraries have been generated. One library contains the protein sequences of all VFs in Victors, and the other contains all of the DNA sequences from all VFs. Victors users are able to perform BLAST sequence similarity searches using different BLAST tools.
Data transfer and download
Several methods are provided in Victors to facilitate data exchange and transfer. First, a Victors web page (http://www.phidias.us/victors/download.php) is available for users to freely download the DNA and protein sequences of all VFs. In addition, after a web query from the Victors query website, a user can export queried VF information into a Microsoft Excel document.
We have also created an Ontology of Host–Pathogen Interactions (OHPI; https://github.com/OHPI/ohpi/). OHPI is developed by following the Open Biomedical Ontology (OBO) Foundry principles (e.g. openness and collaboration) (26). OHPI represents the VFs and how the mutants of VFs in Victors become less virulence inside a host organism or host cells. Currently, OHPI includes over 6700 terms. OHPI reuses existing ontologies (27). For example, OHPI reuses terms from the Gene Ontology (GO) (28), Ontology of Genes and Genomes (OGG) (29) and Cell Line Ontology (CLO) (30) to represent cellular components, genes and cell lines, respectively. OHPI includes object properties to semantically represent the relations between VFs and host entities (Supplementary Figure S4A). For example, the OHPI object property ‘gene mutant attenuated in host cell’ represents a relation between a gene and a host cell where the microbial mutant lacking the gene is attenuated in the host cell compared to the wild-type microbe. Such an object property can be used to represent a VF and its interaction in a host cell, for example, an oxidative stress response gene ahpC of M. tuberculosis strain H37Rv and mouse macrophage cell line J774 cell line (Supplementary Figure S4B), where the ahpC mutant of strain H37Rv is attenuated in J774 cells (31). Using the machine-readable Web Ontology Language (OWL) format (http://www.w3.org/TR/owl2-quick-reference), OWL-based software programs can be developed to parse and extract the information from the ontology and offer advanced data analysis. For example, using an OHPI SPARQL program (http://www.phidias.us/ohpi/sparql/), we could generate a simple SPARQL query script to identify 386 VFs whose mutants are attenuated in macrophages (Supplementary Figure S5A) and extract detailed information about the gene names, OGG IDs and VF annotations of these VFs (Supplementary Figure S5B).
Availability and requirements
Victors is freely available for usage on the website http://www.phidias.us/victors. The Victors system can be accessed using a web browser.
DISCUSSION
This article is the first formal introduction of the Victors database, a comprehensive knowledge base focused on experimentally verified VFs for human and veterinary pathogens. In addition to all the manually curated VFs data, Victors also provides the OHPI ontology representation and computational analysis of the data and user-friendly web interfaces for querying and analyzing the results. Its usage will support researchers in the areas of microbiology, vaccinology and immunology with curated data and bioinformatics tools.
Victors differs from existing VF databases in several significant ways. In addition to bacterial VFs as shown in VFDB (6), Victors includes VFs from viruses, parasites and fungi. While VFDB lists general references for a set of genes, Victors provides references for individual VFs and records the details on the experimental verification. Compared to PHI-base, which focuses on plant pathogens (7), Victors focuses on human and veterinary pathogens. The system design and data types for each VF among Victors, VFDB and PHI-base are also different. The Victors VF records are also semantically represented in the OHPI ontology. Data in the Victors database have been incorporated for many years by the PATRIC Specialty Genes resource (http://patricbrc.org/portal/portal/patric/SpecialtyGenes), a well-recognized bacterial bioinformatics database and analysis resource (9,10). While PATRIC can display Victors data, PATRIC does not provide all the details for each VF and is not responsible for the original annotation.
The predicted interactions between human proteins and VFs provided by Victors have improved our understanding of the pathogenesis of these pathogens. We found that among all pathogen proteins, VFs have higher tendency to interact with human proteins (Supplementary Figure S3). There are two possible explanations for this phenomenon. First, pathogens tend to use the VFs to hijack host defense pathways for the benefit of microbial pathogenesis. Second, human proteins tend to interact with the VFs in order to fight against the pathogen virulence mechanism. Using human–pathogen PPIs, Dyer et al. demonstrated that pathogens preferentially interact with human proteins that are hubs and bottlenecks in human interaction pathways (32,33). However, these studies by Dyer et al. do not differentiate pathogen VFs with other pathogen proteins that are not VFs and thus do not typically significantly contribute to microbial pathogenesis to a significant degree. In contrast, our study provides the evidence that compared to other pathogen proteins, pathogen VFs are more likely to interact with human proteins. Moreover, the human–pathogen interactomes can be used to identify which biological pathways are possibly utilized by the pathogens to invade the host and to develop candidates for future experimental validation.
We intend to keep adding new information to the resource. Our semi-automatic annotation pipeline makes the annotation process efficient. The manual curation and verification process (Supplementary Figure S1) ensures that every VF collected on the website has solid experimental evidence. Meanwhile, we are also testing new literature mining methods to ensure faster VF identification from literature. We also plan to mine and add more experimental conditions used for VF identification in the reported publications. As another future work, in contrast to the correlations between various factors, we will further add their direct associations or causal relations (i.e. network) to Victors by data-driven network methods (34).
Victors is expected to become a central and vital source of VFs and their associated data and support the One Health initiative. It is estimated that ∼75 percent of emerging human pathogens in the past 25 years have originated in animals, and the risk of zoonotic transmission is likely to increase in the future (35). In recent years, the One Health concept has gained well recognition in the public health and animal health communities. Its usage will support researchers in the areas of microbiology, vaccinology and immunology with curated data and bioinformatics tools.
Supplementary Material
Supplementary Data
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH-NIAID R01 [R01AI081062]; University of Michigan startup fund (to Y.H.); Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040700 to L.C.]; National Program on Key Basic Research Project of China [2017YFA0505500, 2014CB910504]; National Natural Science Foundation of China (NSFC) [31771476, 91529303, 81471047]; Innovation Program of Shanghai Municipal Education Commission [13ZZ072]; Shanghai Pujiang Program [13PJD032]; VA Merit Grant [I01BX000656 to M.A.O.]; VA Research Career Scientist Award [1IK6BX003615-01 to M.A.O.]. Funding for open access charge: Dr. Yongqun He bridge fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Becker K., Hu Y., Biller-Andorno N.. Infectious diseases - a global challenge. Int. J. Med. Microbiol. 2006; 296:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller R.S., Farnsworth M.L., Malmberg J.L.. Diseases at the livestock-wildlife interface: status, challenges, and opportunities in the United States. Prev. Vet. Med. 2013; 110:119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfe N.D., Dunavan C.P., Diamond J.. Origins of major human infectious diseases. Nature. 2007; 447:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse M.E., Gowtage-Sequeria S.. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005; 11:1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atlas R.M. One Health: its origins and future. Curr. Top. Microbiol. Immunol. 2013; 365:1–13. [DOI] [PubMed] [Google Scholar]
- 6.Chen L., Zheng D., Liu B., Yang J., Jin Q.. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016; 44:D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urban M., Cuzick A., Rutherford K., Irvine A., Pedro H., Pant R., Sadanadan V., Khamari L., Billal S., Mohanty S. et al. PHI-base: a new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017; 45:D604–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wattam A.R., Abraham D., Dalay O., Disz T.L., Driscoll T., Gabbard J.L., Gillespie J.J., Gough R., Hix D., Kenyon R. et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014; 42:D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L. et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017; 45:D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao C., Abraham D., Wattam A.R., Wilson M.J., Shukla M., Yoo H.S., Sobral B.W.. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics. 2015; 31:252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Z., Tian Y., He Y.. PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol. 2007; 8:R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V.. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015; 43:D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle S.M. BBP: Brucella genome annotation with literature mining and curation. Infect. Immun. 2006; 7:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y., Xiang Z., Mobley H.L.. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J. Biomed. Biotechnol. 2010; 2010:297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y. et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostlund G., Schmitt T., Forslund K., Kostler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L.. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010; 38:D196–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009; 37:D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y., Rappuoli R., De Groot A.S., Chen R.T.. Emerging vaccine informatics. J. Biomed. Biotechnol. 2010; 2010:218590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B., Sayers S., Xiang Z., He Y.. Protegen: a web-based protective antigen database and analysis system. Nucleic Acids Res. 2011; 39:D1073–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Y., Xiang Z.. Bioinformatics analysis of bacterial protective antigens in manually curated Protegen database. Procedia Vaccinol. 2012; 6:3–9. [Google Scholar]
- 21.Ong E., Wong M.U., He Y.. Identification of new features from known bacterial protective vaccine antigens enhances rational vaccine design. Front. Immunol. 2017; 8:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sapkota A., Liu X., Zhao X.M., Cao Y., Liu J., Liu Z.P., Chen L.. DIPOS: database of interacting proteins in Oryza sativa. Mol. Biosyst. 2011; 7:2615–2621. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X.M., Chen L., Aihara K.. A discriminative approach for identifying domain-domain interactions from protein-protein interactions. Proteins. 2010; 78:1243–1253. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X.M., Zhang X.W., Tang W.H., Chen L.. FPPI: Fusarium graminearum protein-protein interaction database. J. Proteome Res. 2009; 8:4714–4721. [DOI] [PubMed] [Google Scholar]
- 25.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T.. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011; 27:431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith B., Ashburner M., Rosse C., Bard J., Bug W., Ceusters W., Goldberg L.J., Eilbeck K., Ireland A., Mungall C.J. et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007; 25:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y., Xiang Z., Zheng J., Lin Y., Overton J.A., Ong E.. The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability. J. Biomed. Seman. 2018; 9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y., Liu Y., Zhao B.. The 2014 International Conference on Biomedical Ontologies (ICBO 2014). 2014; 1327:Houston: CEUR Workshop Proceedings; 13–20. [Google Scholar]
- 30.Sarntivijai S., Lin Y., Xiang Z., Meehan T.F., Diehl A.D., Vempati U.D., Schürer T.C., Pang C., Malone J., Parkinson H. et al. CLO: The Cell Line Ontology. J. Biomed. Seman. 2014; 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Master S.S., Springer B., Sander P., Boettger E.C., Deretic V., Timmins G.S.. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology. 2002; 148:3139–3144. [DOI] [PubMed] [Google Scholar]
- 32.Dyer M.D., Murali T.M., Sobral B.W.. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008; 4:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyer M.D., Neff C., Dufford M., Rivera C.G., Shattuck D., Bassaganya-Riera J., Murali T.M., Sobral B.W.. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One. 2010; 5:e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Zhou Y., Zhang X., Chen L.. Part mutual information for quantifying direct associations in networks. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5130–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King D.A., Peckham C., Waage J.K., Brownlie J., Woolhouse M.E.. Epidemiology. Infectious diseases: preparing for the future. Science. 2006; 313:1392–1393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data