Origins and Genetic Legacy of Prehistoric Dogs (original) (raw)
. Author manuscript; available in PMC: 2021 Apr 30.
Published in final edited form as: Science. 2020 Oct 29;370(6516):557–564. doi: 10.1126/science.aba9572
Abstract
Dogs were the first domestic animal, but little is known about their population history and to what extent it was linked to humans. We sequenced 27 ancient dog genomes and found that all dogs share a common ancestry distinct from present-day wolves, with limited gene flow from wolves since domestication, but substantial dog-to-wolf gene flow. By 11,000 years ago, at least five major ancestry lineages had diversified, demonstrating a deep genetic history of dogs during the Paleolithic. Co-analysis with human genomes reveals aspects of dog population history that mirror humans, including Levant-related ancestry in Africa and early agricultural Europe. Other aspects differ, including the impacts of steppe pastoralist expansions in West- and East Eurasia, and a complete turnover of Neolithic European dog ancestry.
Wolves were the first animal with which humans formed a mutualistic relationship, eventually giving rise to dogs. While there is little consensus regarding when (1–9), where (2, 8–13), and how many times (1, 8, 9, 14) domestication took place, the archaeological (9, 15) record attests to a long-term and close relationship to humans (9, 16–18). Modern dog genomes have revealed a complex population structure (5, 8, 10, 12, 19, 20), but because only six ancient dog and wolf genomes are currently available (4, 9, 14, 21), the process by which this structure emerged remains largely unknown.
Previous mitochondrial DNA (22–29) and genomic (9, 14, 21) studies have suggested an association between the genetic signatures of dogs and their archeological context. However, dog and human genomes have not been quantitatively co-analyzed to assess the degree to which the population history of dogs was linked to that of humans—or may have been decoupled as a result of trade, human preference for particular types of dogs, variation in infectious disease susceptibility, or dogs moving between human groups.
To reconstruct dog population history we sequenced 27 ancient dog genomes up to 10,900 years old from Europe, the Near East and Siberia (table S1) to a median of 1.5-fold coverage (range 0.1-11X) (Fig. 1A, table S2; (30)). To test the association with human population history, we compiled 17 sets of human genome-wide data (30) that matched the age, geographic location and cultural contexts of the ancient dogs (table S4), and directly compared genetic relationships within the two species.
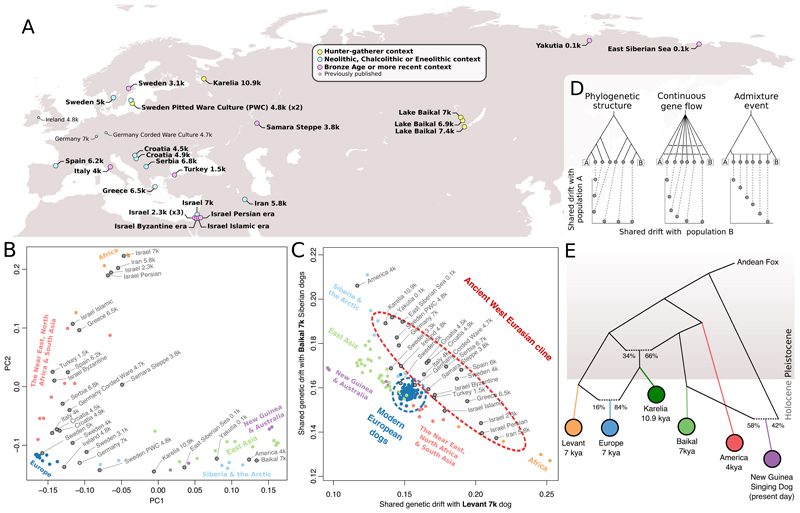
Figure 1. Genomic structure of dogs dates to the Pleistocene.
A) Sampling locations of ancient dogs. B) Principal components analysis on all possible _f4_-statistics among ancient dogs (gray) and a selection of worldwide modern dogs. C) Outgroup f 3-statistics reveal a cline of Levant versus Baikal (horizontal and vertical axes, respectively) related ancestry across ancient west Eurasian dogs, but not among modern European dogs. D) Coalescent simulations demonstrating that a diagonal f 3-cline as in panel C is consistent with an admixture event, but less so with continuous gene flow and not with phylogenetic structure alone. E) An admixture graph that fits all _f4_-statistics between major dog lineages. The European dog was grafted onto the graph identified through exhaustive testing.
Global dog population structure has its origins in the Pleistocene
To characterize the global population structure of ancient and modern dogs, we applied principal component analysis (PCA) to a matrix of all possible f 4-statistics (30), alleviating differences in error rates and missing data. This approach recapitulates a major east-west axis of dog ancestry (PC1) (8, 9, 12), in which the western extreme comprises modern and ancient western Eurasian dogs and modern African dogs (Fig. 1B). The eastern extreme is represented by pre-contact North American dogs (21), three 7 ky dogs from Lake Baikal in Siberia, and modern East Asian dogs including New Guinea Singing Dogs and Australian dingoes. Similar results were obtained through standard model-based clustering (fig. S2).
All ancient and modern European dogs have greater affinity to eastern dog ancestry than ancient Near Eastern dogs have in f 4-tests (fig. S3), despite the overall east-west axis on PC1. Ancient European dogs are also distributed widely across a genetic cline between the East Eurasian and ancient Near Eastern dogs, which furthermore manifests as a linear cline along the diagonal when contrasting shared genetic drift with Baikal dogs and Levantine (Israel, 7kya) dogs using outgroup-f3 statistics (Fig. 1C). Simulations indicate that this linear, diagonal cline is difficult to explain with long-standing continuous gene flow or a tree-like history, but instead suggest that the history of Mesolithic and Neolithic European dogs was marked by a major admixture episode (Fig. 1D) (30).
We modeled the genetic history underlying dog population structure for five populations that represent major ancestries, and tested all 135,285 possible admixture graph models with up to two admixture events (30). One model uniquely fits the data, and features the Mesolithic Karelian dog (10.9 kya) as having received part of its ancestry from a lineage related to eastern dogs, and part from the Levantine lineage (Fig. 1E) (two highly similar models nearly fit, fig. S4). The model can be extended to feature the earliest Neolithic European dog (7 kya)(14) as a mixture of the Karelian and the Levantine branches without loss of fit (fig S5), supporting the dual ancestry model for European dogs suggested by the ancient ancestry cline (Fig. 1C). The observed phylogenetic structure implies that all five ancestry lineages (Neolithic Levant, Mesolithic Karelia, Mesolithic Baikal, ancient America, New Guinea Singing dog) must have existed by 10.9kya (the radiocarbon date of the Karelian dog), and thus most likely prior to the transition from the Pleistocene to the Holocene epoch ~11.6 kya.
No detectable evidence for multiple dog origins or extensive gene flow from wild canids
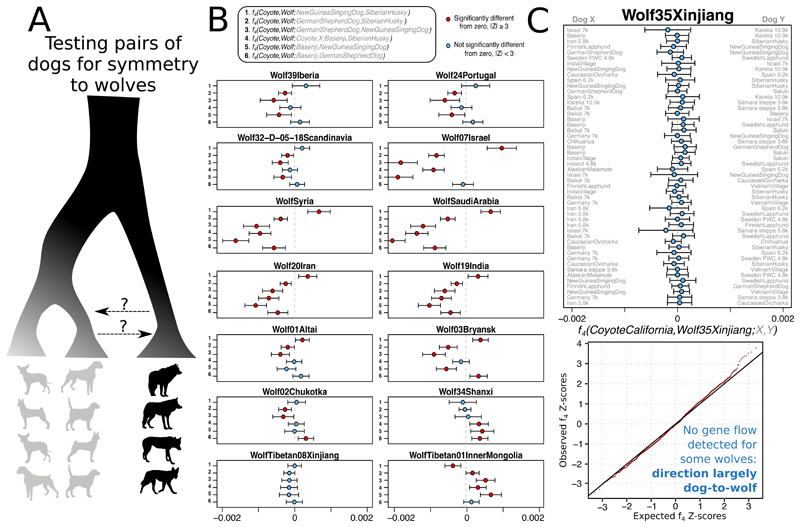
Studies have suggested that wolf populations in Europe (3, 11), the Middle East (12), Central Asia (10), Siberia (31), and East Asia (2, 8), or more than one of these (9), contributed to early dog diversity. One study, however, demonstrated that modern wolves and dogs are reciprocally monophyletic, and suggested bidirectional gene flow (5). We corroborated that gene flow must have occurred by identifying widespread asymmetries between dogs in their affinity to wolves (Fig. 2A,B, fig S7). However, the gene flow was likely largely unidirectional from dogs into wolves, since we also identified some gray wolves that are symmetrically related to all modern and ancient dogs (Fig. 2C). Past gene flow from wolves into specific dog populations would have manifested as an affinity to any member of the modern gray wolf lineage in these tests, so our results suggest that persistent gene flow into dogs has been so limited as to be undetectable at the current resolution of the data. Furthermore, this result is consistent with a scenario in which all dogs derive from a single ancient, now extinct wolf population, or possibly multiple closely related wolf populations. While it is still possible that other, thus far unsampled ancient wolf populations were independently involved in early domestication (3, 9, 31), our data indicate that they did not contribute substantially to later dogs.
Fig. 2. All detectable gene flow is consistent with being unidirectional from dogs into wolf populations.
A) Illustration of asymmetry tests (f 4-statistics) comparing 35 Eurasian gray wolves to all pairs of 66 ancient and modern dogs. B) Selected results using Coyote as outgroup. C) A wolf from Xinjiang, western China, is not closer to some dog populations than to others, as the test statistics are consistent with being normally distributed around 0 (the quantile-quantile plot includes all 66 dogs). If there was a substantial gene flow from some wolf population into some dog population, we would expect all wolf individuals to display asymmetric relationships.
In contrast to the lack of wolf admixture into dogs, we identified dog admixture into almost all analyzed present-day wolves (Fig. 2B), with the strongest signals typically coming from dogs into geographically proximate wolf populations in Europe, the Near East and East Asia (fig S7). We also replicated affinities between ancient American dogs and Coyotes (21), and between African dogs and African Golden Wolves (32), though the direction of gene flow in both cases is unclear, and the small magnitude is unlikely to impact most analyses of dog relationships (table S5). We did not find genome-wide evidence for gene flow from Tibetan wolves into Tibetan dogs, despite evidence for wolf ancestry locally around the EPAS1 gene associated with adaptation to altitude (33, 34). Dogs thus do not show similar evidence of wild introgression that has been found in pigs, goats, horses, sheep and cattle (35–40).
Assessing the relationship between dog and human population histories
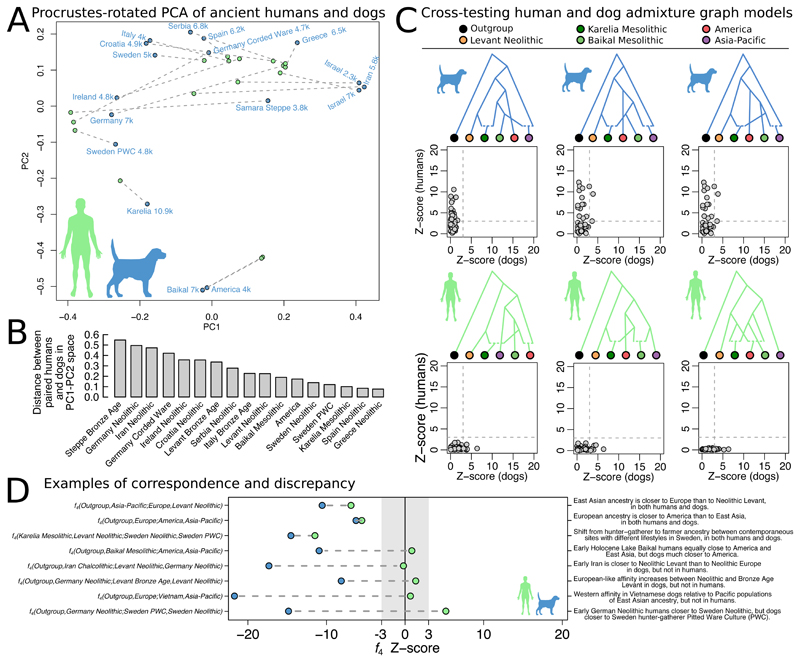
We next quantitatively compared the population relationships observed in dogs with those of humans. First, using Procrustes rotation to align f 4-PCA results obtained on dog and human genomes matched in time and space (Fig. 3A; (30)), we find that the population structures of the two species resemble each other (Procrustes correlation = 0.48, p = 0.043). However, there are also several cases where the matched dogs and humans cluster in different parts of the PCA space. The greatest differences (Fig. 3B) are observed for Chalcolithic Iran, in which the human population is different from the Neolithic Levant (41, 42) but the dogs in the two regions are similar. In Neolithic Germany and Ireland, the humans are more shifted towards the Levant (43, 44) but the dogs are shifted towards Northern European hunter-gatherer contexts. In the Bronze Age Steppe and in Corded Ware Germany, the humans are shifted away from the Neolithic European cluster (45, 46) in a manner not seen in dogs.
Figure 3. Quantitative comparisons between dog and human population genomic structure.
A) Principal components analysis on all possible f 4-statistics on ancient dogs (blue), overlaid through Procrustes transformation by the corresponding analysis performed on ancient humans matched in time, space, and cultural context to the dogs (green). Dashed lines connect each matched pair. B) Euclidian residuals between the Procrustes-rotated human and dog coordinates. C) The three admixture graphs that fit for one species and provide the smallest error for the other. Scatter plots show absolute Z-scores for the difference between observed and predicted _f4_-statistics. D) Examples of _f4_-statistics that reveal similarities and differences between humans and dogs (far right text).
Second, we evaluated if the admixture graph topologies that best fit the data for one species could also explain population relationships of the other. Though we found no graphs that fit the data perfectly for both species, graphs that fit, or nearly fit dogs rank among the 0.8-2.8% top scoring graphs in the human search, and graphs that fit humans rank among the 0.007-1.2% top scoring graphs in the dog search (Fig. 3C, fig. S9). However, we note that this analysis does not take into account the different time depth of the two species’ population histories: the >40kya divergence of human East- and West Eurasian ancestries (47) is significantly older than the earliest appearance of dog morphology in the fossil record, conservatively dated to 14.5kya (48) though older (3, 31), disputed specimens (49, 50), have been claimed.
Third, we found that the sign (positive or negative) of f 4-statistics in dogs match the sign in humans in 71% of 31,878 tests (null expectation 50%) across 24 matched dog-human pairs, although this decreases to 58% when restricted to dogs and humans from Europe. We identified specific f 4-statistics that exemplify both concordance and discrepancy between the species (Fig. 3D). While it is not known what degree of concordance would be expected between the histories of two species based on biogeographical factors alone, the results of these three analyses demonstrate that ancestry relationships in dogs and humans share overall features, but are not identical over space and time, and there are several cases where they must have been decoupled.
Recurrent population histories
One notable example of concordance is that both humans and dogs in East Asia are closer to European than to Near Eastern populations, which in both humans (43) and our best-fitting graph (Fig. 1E) is best modelled by European ancestry being a mixture of ancestry related to the Near East and East Asia. However, the divergence of Near Eastern 'Basal Eurasian' ancestry in humans was likely >45 kya (43), suggesting that dog population dynamics may have mimicked earlier processes in humans. A second example is that all European dogs have a stronger affinity towards American and Siberian dogs than they have to New Guinea singing dogs, which likely represent a type of unadmixed East Asian dog ancestry, mirroring a circumpolar affinity between humans in Europe and the Americas (51) (Fig. 3D). Human groups at Lake Baikal 24-18kya had western Eurasian origins and contributed to Native American ancestry (51), but were largely replaced by the Holocene (52). Though the dogs at Lake Baikal dated to 7kya constitute a similar link between the Americas and Europe (Fig. 1C,E), they do so >10ky later (Fig. 3D). Thus, shared circumpolar ancestry through northern Eurasia is an important feature of both human and dog population structures, though this did likely not result from the same migration episodes.
Neolithic expansion into Europe
Ancient human genomes have revealed a major ancestry transformation associated with the expansion of Neolithic agriculturalists from the Near East into Europe (43, 45, 53), and a study of ancient dog mitochondria suggested they were accompanied by dogs (27). We hypothesized that the genomic ancestry cline we observe across ancient European dogs (Fig. 1C) could be, at least in part, due to admixture between dogs associated with Mesolithic hunter-gatherers and incoming Neolithic farmers. Three observations support this: first, the hypothesized hunter-gatherer end of the cline is occupied by the 10.9kBP Mesolithic Karelian dog, and dogs from a 4.8kBP hunter-gatherer Pitted Ware Culture site in Sweden. Second, relative to the Swedish hunter-gatherer dogs, a contemporaneous dog from a Swedish Neolithic agricultural context is shifted towards the Levantine end of the cline, mirroring humans at the same sites (41, 53, 54) (Fig. 3A,D; fig. S10D). Third, Neolithic Levantine affinity increases towards the south (p=0.0196, linear regression), consistent with a range expansion alongside Neolithic human groups. While dogs clearly associated with Mesolithic continental 'Western hunter-gatherer' (43) human groups have yet to be identified, our results suggest that such dogs would have strong affinity towards the Siberian end of the European cline. Overall, these results indicate that the Neolithic expansion of farmers into Europe was also associated with an ancestry transformation for dogs.
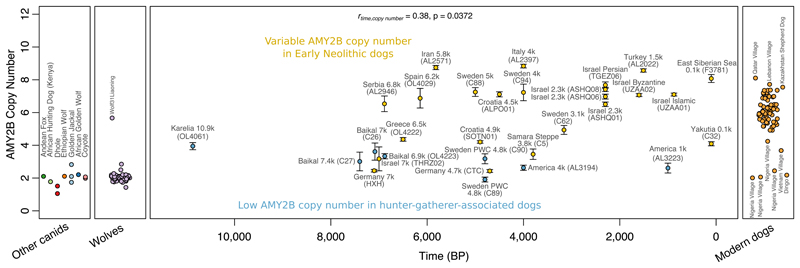
Increased copy number of the AMY2B gene, involved in starch digestion, has been linked to dietary adaptations of dogs during the agricultural transition (6, 55, 56). The paralogous AMY1 gene has been under adaptive evolution in humans (57), though this does not seem clearly linked to agriculture (58). We observe low copy numbers in dogs from human hunter-gatherer contexts (Fig. 4), although the Mesolithic Karelian dog may already have possessed an elevated number relative to wolves. Several Neolithic dogs have as many copies as present-day dogs, as early as in 5.8 ky old Iranian and 6 ky old Spanish dogs, but others display low numbers (14, 56), e.g. the 7 ky Levantine individual. These results suggest that selection for increased AMY2B copy number did not take place during the early stages of domestication, and in contrast to humans (58) was not advanced in Mesolithic hunter-gatherer contexts, but was variable in early agricultural populations and did not become widespread until several thousand years after the first appearance of starch-rich agricultural lifestyles.
Figure 4. Expansion of copy number in the AMY2B pancreatic amylase gene largely occurred after the transition to agriculture.
Ancient dogs are plotted against their age, with blue color indicating dogs from likely hunter-gatherer human contexts. Bars denote 95% binomial confidence intervals around the ratio of the number of reads mapping to the copy number variable region to those mapping to control regions throughout the genome.
Africa and the Near East
The clustering of modern African dogs with ancient dogs from the Levant and Iran, especially the oldest individual dating to 7 kya, suggests a Near Eastern origin (Fig. 1B,C, fig. S2). Western (Anatolia and the Levant) and eastern (Zagros mountains of Iran) human groups in the Fertile Crescent were highly genetically differentiated (41), and the western groups were the primary source of gene flow into Europe and Africa (41, 59) during the Neolithic. A source of African dog ancestry from the Levant (7kya) is a better fit than Iran (5.8kya) (Fig. 5A), mirroring the human history, as well as that of cattle (40). In contrast, we are unable to distinguish whether the Levant or Iran is the better source for Neolithic dog ancestry in Europe. Our results suggest a single origin of sub-Saharan African dogs from the Levant (Fig. 5B), with limited gene flow from outside the continent until the past few hundred years.
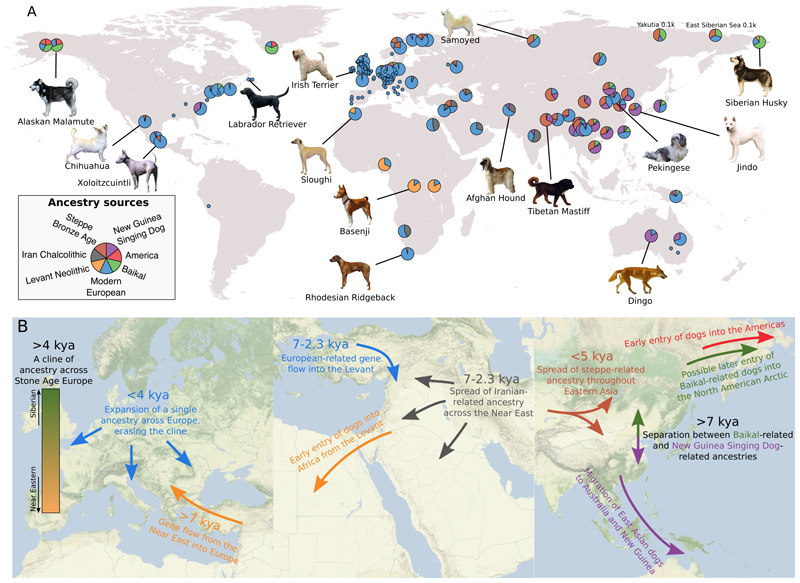
Figure 5. Ancestry of global dogs today.
A) For each present-day population, the ancestry proportions estimated by the best-fitting qpAdm model, restricted to models containing up to four of seven selected sources, are displayed. Populations for which a single component accounts for ≥98% of the ancestry are collapsed to smaller circles. Dog pictures were obtained from Wikimedia under the CC BY-SA 3.0 license (https://commons.wikimedia.org/wiki/Special:ListFiles/Desaix83). B) Illustrations of inferred population histories in three regions of the world.
In contrast to Africa, the 7kya Neolithic Levantine population does not appear to have contributed much, if any, ancestry to present-day dogs in the Near East. Instead, 2.3 ky old dogs in the Levant can be modelled as having 81% Iran-related and 19% Neolithic Europe-related ancestry (Data S1). By this time in the Levant, there was also human gene flow from Iran (41) and transient gene flow from Europe (60). However, our results suggest a more complete replacement of dog ancestry in the Levant by 2.3 kya (Fig. 5B). Later, modern Near Eastern dogs are best modelled as mixtures of the 2.3 ky Levantine and modern European sources (Data S1).
Steppe pastoralist expansions
Expansions of steppe pastoralists associated with the Yamnaya and Corded Ware cultures into Late Neolithic and Bronze Age Europe transformed the ancestry of human populations (43, 45, 46). To test if dog ancestry was similarly affected, we analyzed a 3.8 ky old dog from the eastern European steppe associated with the Bronze Age Srubnaya culture. While its ancestry resembles that of western European dogs (Fig. 1C, fig. S10), it is an outlier in the center of PC1-PC2 space (Fig. 1B). A Corded Ware-associated dog (4.7kya) from Germany, hypothesized to have steppe ancestry (14), can be modelled as deriving 51% of its ancestry from a source related to the Srubnaya steppe dog, and the rest from a Neolithic European source ((30); Data S1). We obtain similar results for a Bronze Age Swedish dog (45%; 3.1kya), but not a Bronze Age Italian dog (4kya).
Despite this potential link between the steppe and the Corded Ware dog, most later European dogs display no particular affinity to the Srubnaya dog. Modern European dogs instead cluster with Neolithic European dogs (Fig. 1B), and do not mirror the lasting ancestry shift seen in humans after the pastoralist expansion (Fig. 3A). While earlier and additional steppe dog genomes are needed to better understand this process, the relative continuity between Neolithic and present-day individuals suggests that the arrival of steppe pastoralists did not result in persistent large-scale shifts in the ancestry of European dogs.
Although steppe pastoralists also expanded east, they do not appear to have contributed much ancestry to present-day people in East Asia (46, 52). Many modern Chinese dogs display unambiguous evidence (negative f 3 tests (30)) of being the product of admixture between a population related to the New Guinea Singing Dog (and the Australian Dingo) and a West Eurasian-related population (table S6). A recent study also found a mitochondrial turnover in Chinese dogs in the last few thousand years (61). The best-fitting models involve ancestry from modern European breeds, but also substantial contributions from the 3.8k BP Srubnaya steppe dog (Fig. 5A, Data S1). Some populations, especially those in Siberia, additionally require a fourth source related to the 7ky old Lake Baikal dogs, but no or minimal New Guinea Singing Dog-related ancestry. Our results thus raise the possibility that the eastward migrations of steppe pastoralists had a more substantial impact on the ancestry of dogs than humans in East Asia (Fig. 5B).
Later homogenization of dog ancestry in Europe
The extensive range of ancestry diversity among early European dogs is not preserved today, as modern European dogs are all symmetrically related to the ancient dogs in our dataset (Fig. 1C, fig. S13, Data S1, (30)). This suggests little to no contribution of most local Mesolithic and Neolithic populations to present-day diversity in Europe. Instead, we found that a single dog from a Neolithic megalithic context dated to 5 kya at the Frälsegården site in southwestern Sweden can be modelled as a single-source proxy for 90-100% of the ancestry of most modern European dogs, to the exclusion of all other ancient dogs (fig. S13, Data S1). This implies that a population with ancestry similar to this individual, but not necessarily originating in Scandinavia, replaced other populations and erased the continent-wide genetic cline (Fig 5B). This ancestry was in the middle of the cline (Fig 1C), such that present-day European dogs can be modelled as about equal proportions of Karelian and Levantine-related ancestries (54% and 46% respectively, for German Shepherd using the admixture graph (Fig 1E)).
The Frälsegården dog is also favored as a partial ancestry source for a 4ky old Bronze Age dog from Italy, a 1.5ky old dog from Turkey and Byzantine and Medieval, but not earlier dogs in the Levant (Data S1), providing some constraints on the timing of this ancestry expansion. However, the circumstances that initiated or facilitated the homogenization of dog ancestry in Europe from a narrow subset of that present in the European Neolithic, including the phenomenal phenotypic diversity and genetic differentiation of modern breeds (12, 19, 20) (Fig. 1C), remain unknown.
More recently, this modern European ancestry has dispersed globally, and today is a major component of most dog populations worldwide (Fig. 5A). Our ancestry models, however, reveal that some pre-colonial ancestry does survive in breeds such as the Mexican Chihuahua (~4%) and Xoloitzcuintli (~3%), and the South African Rhodesian Ridgeback (~4%) (Data S1).
Discussion
The diversification of at least five dog ancestry lineages by the onset of the Holocene was followed by a dynamic population history that in many ways tracked that of humans, likely reflecting how dogs migrated alongside human groups. However, in several instances, these histories do not align, suggesting that humans also dispersed without dogs, dogs moved between human groups, or that dogs were cultural and/or economic trade commodities.
Certain aspects of genetic relationships between dog populations, such as an east-west Eurasian differentiation, circumpolar connections, and possible basal lineages in the Near East, resemble features of human population history that were established before the earliest estimated dates of dog domestication. This superficial mirroring between the species may therefore instead point to recurrent population dynamics, due to biogeographic or anthropological factors that remain to be understood. A key question is how dogs spread across Eurasia and the Americas by the Holocene, since no major human population movements have been identified after the initial out-of-Africa expansion that could have driven this global dispersal.
We find that the modern and ancient genomic data are consistent with a single origin for dogs, though a scenario involving multiple closely related wolf populations remains possible. However, in our view, the geographical origin of dogs remains unknown. Previously suggested points of origin based upon present-day patterns of genomic diversity (2, 8, 10) or affinities to modern wolf populations (12) are sensitive to the obscuring effects of more recent population dynamics and gene flow. Ultimately, integrating DNA from dogs and wolves even older than those analyzed here with archaeology, anthropology, ethology and other disciplines, is needed to determine where, and in which environmental and cultural context the first dogs originated.
Supplementary Material
Data S1
Supplementary material
One Sentence Summary.
Ancient dog genomes reveal no evidence for multiple origins but an early diversification, followed by a genetic history that both mirrors and differs from humans.
Acknowledgements
We thank S. Charlton, I. Lazaridis, A. Manin and I. Mathieson for comments on the manuscript, G.-D. Wang and C. Marsden for help with data access, and GORDAILUA (the Gipuzkoa Centre for Heritage Collections), S. San José, C. Olaetxea, M. Urteaga, A. Sampson, A.R. Sardari Zarchi and M. Abdollahi (ICHHTO, Iran) for facilitating sample access.
Funding
Ancient genome sequencing was supported by SciLifeLab National Projects and the Erik Philip Sörensen Foundation (to P.S.). A.B., T.D., and P.S. were supported by the Francis Crick Institute core funding (FC001595) from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust. P.S. was also supported by the European Research Council (grant no. 852558) and Wellcome Trust Investigator award (217223/Z/19/Z). R.L. was supported by the Social Sciences and Humanities Research Council of Canada (#SSHRC IG 435-2014-0075). Y. K. was supported by State Assignment of the Sobolev Institute of Geology and Mineralogy. M.S. was supported by ZIN RAS (state assignment no. АААА-А19-119032590102-7). A.T.L. was supported by the Smithsonian’s Peter Buck Postdoctoral Fellowship. Archaeological work in Serbia was supported by AHRC grant AH/J001406/1. Computations were supported by SNIC-UPPMAX (b2016004), and the UOXF ARC facility. L.A.F.F. was supported by the Wellcome Trust (Grant 210119/Z/18/Z) and by Wolfson College (University of Oxford). G.L. was supported by the ERC (Grant ERC-2013-StG-337574-UNDEAD). G.L. and K.D. were supported by the Natural Environmental Research Council (Grants NE/K005243/1 and NE/K003259/1). Dating was supported by the NERC Radiocarbon Facility (NF/2016/2/4).
Footnotes
Author contributions: GL and PS initiated the study. JS, K-GS, DA, EA, SA, GB-O, VIB, JB, DB, SF, IF, DF, MG, LH, LJ, JK-C, YK, RJL, DLD, MM, MN, VO, DO, MP, MR, DR, BR, MS, IS, AT, KT, IU, AV, PW, AG, and LD contributed material and archaeological information. RS, EE, OL, LG-F, JH, AJ, HR and AL did ancient DNA molecular work, supervised by AG, LD, RP, GL and PS. AB, LF, AC, TD, EKI-P and PS processed the genome sequence data, supervised by LF and PS. AB did population genomic analyses, supervised by PS. ATL did mtDNA analyses, supervised by GL. AB, LF, GL and PS wrote the paper with input from RP, KD and all other authors.
Competing interests: Authors declare no competing interests.
Data and materials availability
The generated DNA sequencing data will be made available in the European Nucleotide Archive (ENA) under study accession PRJEB38079.
References and notes
- 1.Vilà C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 2.Savolainen P, Zhang Y, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 3.Germonpré M, Sablin MV, Stevens RE, Hedges REM, Hofreiter M, Stiller M, Després VR. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J Archaeol Sci. 2009;36:473–490. [Google Scholar]
- 4.Skoglund P, Ersmark E, Palkopoulou E, Dalén L. Ancient Wolf Genome Reveals an Early Divergence of Domestic Dog Ancestors and Admixture into High-Latitude Breeds. Curr Biol. 2015;25:1515–1519. doi: 10.1016/j.cub.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B, Beale H, et al. Genome Sequencing Highlights the Dynamic Early History of Dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelsson E, Ratnakumar A, Arendt M-L, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar A, Lindblad-Toh K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 7.Skoglund P, Götherström A, Jakobsson M. Estimation of population divergence times from non-overlapping genomic sequences: examples from dogs and wolves. Mol Biol Evol. 2011;28:1505–1517. doi: 10.1093/molbev/msq342. [DOI] [PubMed] [Google Scholar]
- 8.Wang G-D, Zhai W, Yang H-C, Wang L, Zhong L, Liu Y-H, Fan R-X, Yin T-T, Zhu C-L, Poyarkov AD, Irwin DM, et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 2016;26:21–33. doi: 10.1038/cr.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frantz LAF, Mullin VE, Pionnier-Capitan M, Lebrasseur O, Ollivier M, Perri A, Linderholm A, Mattiangeli V, Teasdale MD, Dimopoulos EA, Tresset A, et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science. 2016;352:1228–1231. doi: 10.1126/science.aaf3161. [DOI] [PubMed] [Google Scholar]
- 10.Shannon LM, Boyko RH, Castelhano M, Corey E, Hayward JJ, McLean C, White ME, Said MAbi, Anita BA, Bondjengo NI, Calero J, et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proceedings of the National Academy of Sciences. 2015;112:13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thalmann O, Shapiro B, Cui P, Schuenemann VJ, Sawyer SK, Greenfield DL, Germonpré MB, Sablin MV, López-Giráldez F, Domingo-Roura X, Napierala H, et al. Complete Mitochondrial Genomes of Ancient Canids Suggest a European Origin of Domestic Dogs. Science. 2013;342:871–874. doi: 10.1126/science.1243650. [DOI] [PubMed] [Google Scholar]
- 12.vonHoldt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pang J-F, Kluetsch C, Zou X-J, Zhang A-B, Luo L-Y, Angleby H, Ardalan A, Ekström C, Sköllermo A, Lundeberg J, Matsamura S, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botigué LR, Song S, Scheu A, Gopalan S, Pendleton AL, Oetjens M, Taravella AM, Seregély T, Zeeb-Lanz A, Arbogast R-M, Bobo D, et al. Ancient European dog genomes reveal continuity since the Early Neolithic. Nat Commun. 2017;8 doi: 10.1038/ncomms16082. 16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morey DF. The Early Evolution of the Domestic Dog. Am Sci. 1994;82:336–347. [Google Scholar]
- 16.Clutton-Brock J. Man-made dogs. Science. 1977;197:1340–1342. doi: 10.1126/science.197.4311.1340. [DOI] [PubMed] [Google Scholar]
- 17.Davis SJM, Valla FR. Evidence for domestication of the dog 12,000 years ago in the Natufian of Israel. Nature. 1978;276:608–610. [Google Scholar]
- 18.Sablin M, Khlopachev G. The Earliest Ice Age Dogs: Evidence from Eliseevichi 11. Curr Anthropol. 2002;43:795–799. [Google Scholar]
- 19.Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 20.Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, Ostrander EA. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development. Cell Rep. 2017;19:697–708. doi: 10.1016/j.celrep.2017.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leathlobhair MNí, Perri AR, Irving-Pease EK, Witt KE, Linderholm A, Haile J, Lebrasseur O, Ameen C, Blick J, Boyko AR, Brace S, et al. The evolutionary history of dogs in the Americas. Science. 2018;361:81–85. doi: 10.1126/science.aao4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Asch B, Zhang A-B, Oskarsson MCR, Klütsch CFC, Amorim A, Savolainen P. Pre-Columbian origins of Native American dog breeds, with only limited replacement by European dogs, confirmed by mtDNA analysis. Proceedings of the Royal Society B: Biological Sciences. 2013;280 doi: 10.1098/rspb.2013.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard JA, Wayne RK, Wheeler J, Valadez R, Guillen S, Vila C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- 24.Castroviejo-Fisher S, Skoglund P, Valadez R, Vila C, Leonard J. Vanishing native American dog lineages. BMC Evol Biol. 2011;11:73. doi: 10.1186/1471-2148-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greig K, Gosling A, Collins CJ, Boocock J, McDonald K, Addison DJ, Allen MS, David B, Gibbs M, Higham CFW, Liu F, et al. Complex history of dog (Canis familiaris) origins and translocations in the Pacific revealed by ancient mitogenomes. Sci Rep. 2018;8 doi: 10.1038/s41598-018-27363-8. 9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E, Lundeberg J. A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proc Natl Acad Sci U S A. 2004;101:12387–12390. doi: 10.1073/pnas.0401814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ollivier M, Tresset A, Frantz LAF, Bréhard S, Bălăşescu A, Mashkour M, Boroneanţ A, Pionnier-Capitan M, Lebrasseur O, Arbogast R-M, et al. Dogs accompanied humans during the Neolithic expansion into Europe. Biol Lett. 2018;14 doi: 10.1098/rsbl.2018.0286. 20180286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ameen C, Feuerborn T, Linderholm A, Brown S, Hulme-Beaman A, Lebrasseur O, Sinding M-HS, Lounsberry ZT, Lin A, Appelt M, et al. Specialised sledge dogs accompanied Inuit dispersal across the North American Arctic. Proceedings of the Royal Society B: Biological Sciences. 2019 doi: 10.1098/rspb.2019.1929. available at https://abdn.pure.elsevier.com/en/publications/specialised-sledge-dogs-accompanied-inuit-dispersal-across-the-no. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmström H, Vilà C, Gilbert MTP, Storå J, Willerslev E, Holmlund G, Götherström A. Barking up the wrong tree: modern northern European dogs fail to explain their origin. BMC Evol Biol. 2008;8:71. doi: 10.1186/1471-2148-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materials and methods are available as supplementary materials. [Google Scholar]
- 31.Ovodov ND, Crockford SJ, Kuzmin YV, Higham TFG, Hodgins GWL, van der Plicht J. A 33,000-Year-Old Incipient Dog from the Altai Mountains of Siberia: Evidence of the Earliest Domestication Disrupted by the Last Glacial Maximum. PLoS One. 2011;6:e22821. doi: 10.1371/journal.pone.0022821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y-H, Wang L, Xu T, Guo X, Li Y, Yin T-T, Yang H-C, Hu Y, Adeola AC, Sanke OJ, Otecko NO, et al. Whole-Genome Sequencing of African Dogs Provides Insights into Adaptations against Tropical Parasites. Mol Biol Evol. 2018;35:287–298. doi: 10.1093/molbev/msx258. [DOI] [PubMed] [Google Scholar]
- 33.Miao B, Wang Z, Li Y. Genomic Analysis Reveals Hypoxia Adaptation in the Tibetan Mastiff by Introgression of the Gray Wolf from the Tibetan Plateau. Mol Biol Evol. 2017;34:734–743. doi: 10.1093/molbev/msw274. [DOI] [PubMed] [Google Scholar]
- 34.vonHoldt B, Fan Z, Ortega-Del Vecchyo D, Wayne RK. EPAS1 variants in high altitude Tibetan wolves were selectively introgressed into highland dogs. PeerJ. 2017;5:e3522. doi: 10.7717/peerj.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frantz LAF, Haile J, Lin AT, Scheu A. Ancient pigs reveal a near-complete genomic turnover following their introduction to Europe. Proceedings of the. 2019 doi: 10.1073/pnas.1901169116. available at https://www.pnas.org/content/116/35/17231.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daly KG, Delser PMaisano, Mullin VE, Scheu A, Mattiangeli V, Teasdale MD, Hare AJ, Burger J, Verdugo MP, Collins MJ, Kehati R, et al. Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science. 2018;361:85–88. doi: 10.1126/science.aas9411. [DOI] [PubMed] [Google Scholar]
- 37.Fages A, Hanghøj K, Khan N, Gaunitz C, Seguin-Orlando A, Leonardi M, Constantz McCroryC, Gamba C, Al-Rasheid KAS, Albizuri S, Alfarhan AH, et al. Tracking Five Millennia of Horse Management with Extensive Ancient Genome Time Series. Cell. 2019;177:1419–1435.e31. doi: 10.1016/j.cell.2019.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbato M, Hailer F, Orozco-terWengel P, Kijas J, Mereu P, Cabras P, Mazza R, Pirastru M, Bruford MW. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci Rep. 2017;7 doi: 10.1038/s41598-017-07382-7. 7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SDE, Magee DA, McGettigan PA, Teasdale MD, Edwards CJ, Lohan AJ, Murphy A, Braud M, Donoghue MT, Liu Y, Chamberlain AT, et al. Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biol. 2015;16:234. doi: 10.1186/s13059-015-0790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdugo MP, Mullin VE, Scheu A, Mattiangeli V, Daly KG, Delser PMaisano, Hare AJ, Burger J, Collins MJ, Kehati R, Hesse P, et al. Ancient cattle genomics, origins, and rapid turnover in the Fertile Crescent. Science. 2019;365:173–176. doi: 10.1126/science.aav1002. [DOI] [PubMed] [Google Scholar]
- 41.Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, Connell S, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broushaki F, Thomas MG, Link V, López S, van Dorp L, Kirsanow K, Hofmanová Z, Diekmann Y, Cassidy LM, Díez-del-Molino D, Kousathanas A, et al. Early Neolithic genomes from the eastern Fertile Crescent. Science. 2016;353:499. doi: 10.1126/science.aaf7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, Berger B, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cassidy LM, Martiniano R, Murphy EM, Teasdale MD, Mallory J, Hartwell B, Bradley DG. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proceedings of the National Academy of Sciences. 2016;113:368–373. doi: 10.1073/pnas.1518445113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, Fu Q, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allentoft ME, Sikora M, Sjogren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlstrom T, Vinner L, Malaspinas A-S, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 47.Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Pääbo S. DNA analysis of an early modern human from Tianyuan Cave, China. Proceedings of the National Academy of Sciences. 2013;110:2223–2227. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janssens L, Giemsch L, Schmitz R, Street M, Van Dongen S, Crombé P. A new look at an old dog: Bonn-Oberkassel reconsidered. J Archaeol Sci. 2018;92:126–138. [Google Scholar]
- 49.Perri A. A wolf in dog’s clothing: Initial dog domestication and Pleistocene wolf variation. J Archaeol Sci. 2016;68:1–4. [Google Scholar]
- 50.Morey DF. In search of Paleolithic dogs: a quest with mixed results. J Archaeol Sci. 2014;52:300–307. [Google Scholar]
- 51.Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Jr, Orlando L, Metspalu E, Karmin M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018;557:369–374. doi: 10.1038/s41586-018-0094-2. [DOI] [PubMed] [Google Scholar]
- 53.Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MTP, Götherström A, Jakobsson M. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 54.Skoglund P, Malmström H, Omrak A, Raghavan M, Valdiosera C, Günther T, Hall P, Tambets K, Parik J, Sjögren K-G, Apel J, et al. Genomic Diversity and Admixture Differs for Stone-Age Scandinavian Foragers and Farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 55.Arendt M, Cairns KM, Ballard JWO, Savolainen P, Axelsson E. Diet adaptation in dog reflects spread of prehistoric agriculture. Heredity. 2016;117:301–306. doi: 10.1038/hdy.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ollivier M, Tresset A, Bastian F, Lagoutte L, Axelsson E, Arendt M-L, Bălăşescu A, Marshour M, Sablin MV, Salanova L, Vigne J-D, et al. Amy2B copy number variation reveals starch diet adaptations in ancient European dogs. R Soc Open Sci. 2016;3 doi: 10.1098/rsos.160449. 160449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathieson S, Mathieson I. FADS1 and the Timing of Human Adaptation to Agriculture. Mol Biol Evol. 2018;35:2957–2970. doi: 10.1093/molbev/msy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skoglund P, Thompson JC, Prendergast ME, Mittnik A, Sirak K, Hajdinjak M, Salie T, Rohland N, Mallick S, Peltzer A, Heinze A, et al. Reconstructing Prehistoric African Population Structure. Cell. 2017;171:59–71.e21. doi: 10.1016/j.cell.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feldman M, Master DM, Bianco RA, Burri M, Stockhammer PW, Mittnik A, Aja AJ, Jeong C, Krause J. Ancient DNA sheds light on the genetic origins of early Iron Age Philistines. Sci Adv. 2019;5:eaax0061. doi: 10.1126/sciadv.aax0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Sun G, Ren L, Yuan H, Dong G, Zhang L, Liu F, Cao P, Ko AM-S, Yang MA, Hu S, et al. Ancient DNA Evidence from China Reveals the Expansion of Pacific Dogs. Mol Biol Evol. 2020;37:1462–1469. doi: 10.1093/molbev/msz311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sampson A, editor. Skoteini, Tharrounia. The cave, the settlement and the cemetery. Athens, Ephorate of Paleoanthropology - Speleology. 1993 Σκοτεινή Θαρροσνίων. To ο σπήλαιο. O οικισμός και το νεκροταυείο. [Google Scholar]
- 63.Kotjabopoulou E, Trantalidou K. Faunal analysis of the Skoteini cave. Sampson A, editor. Skoteini, Tharrounia: the Cave, the Settlement and the Cemetery. 1993:392–434. [Google Scholar]
- 64.Abdollahi M, Sardari ZA. EASTERN CENTRAL ZAGROS DURING THE NEOLITHIC PERIOD: BASED ON THE EXCAVATION AT TAPPEH QELA GAP. PAZHOHESH-HA-YE BASTANSHENASI IRAN. 2013;3:117–138. [Google Scholar]
- 65.Amiri S, Mashkour M, Mohaseb A, Tengberg M, Abdolahi M, Sardari A. The Subsistence Economy of Qela Gap; Lurestan, Iran: From the Late Neolithic to the Iron Age. Archaeology. 2019:125–132. [Google Scholar]
- 66.Amiri S, Mashkour M, Mohaseb A, Tengberg M, Abdolahi M, Sardari A. H AKM, Hkanipour M, Naseri R, editors. Proceeding of the International Conference of Young Archaeologists, The Faculty of Litterature and Humanities and the Cultural Division of University of Tehran. 2014:597–626. [Google Scholar]
- 67.de Mazzorin JG, Tagliacozzo A. Dogs through time: an archaeological perspective. Oxford: Archaeopress; 2000. Morphological and osteological changes in the dog from the Neolithic to the Roman period in Italy; pp. 141–161. [Google Scholar]
- 68.Onar V, Çakırlar C, Janeczek M, Kızıltan Z. Skull Typology of Byzantine Dogs from the Theodosius Harbour at Yenikapı, Istanbul. Anat Histol Embryol. 2012;41:341–352. doi: 10.1111/j.1439-0264.2012.01143.x. [DOI] [PubMed] [Google Scholar]
- 69.Onar V, Pazvant G, Alpak H, Ince NG, Armutak A, Kiziltan ZS. Animal skeletal remains of the Theodosius harbor: general overview. Turkish Journal of Veterinary and Animal Sciences. 2013;37:81–85. [Google Scholar]
- 70.Onar V, Janeczek M, Pazvant G, Ince NGezer, Alpak H, Armutak A, Chrószcz A, Kızıltan Z. Estimating the body weight of Byzantine dogs from the Theodosius Harbour at Yenikapı, Istanbul. Kafkas Üniversitesi Veteriner Fakültesi Dergisi. 2015;21:55–59. [Google Scholar]
- 71.Oshibkina SV. The Mesolithic culture Veret’ye. Chronology and periods. Rossiiskaya Arkheologiya. 2004:100–110. [Google Scholar]
- 72.Oshibkina SV. Mezolit Vostochnogo Prionezhya. Kultura Veretye. Moscow: Institute of Archaeology, Russian Academy of Sciences. 2006 [Google Scholar]
- 73.Sjögren KG. In: Aspects of Neolithic burial practices: I. Brink K, Hydén S, Jennbert K, Larsson L, Olausson D, editors. Neolithic Diversities. Perspectives from a conference in Lund; Sweden Lund: 2015. pp. 200–212. [Google Scholar]
- 74.Ullén I. Horse and dog in the Swedish Bronze Age: A close-up study of the relation of horse and dog to man in the Bronze Age settlement of Apalle. Archäologisches Korrespondenzblatt. 1996;26:145–166. [Google Scholar]
- 75.Ullén I. Lager och hus. Bronsåldersboplatsen vid Apalle i Uppland. Arkeologi på väg--undersökningar för E18. 1997:22–75. [Google Scholar]
- 76.Losey RJ, Garvie-Lok S, Leonard JA, Katzenberg MAnne, Germonpré M, Nomokonova T, Sablin MV, Goriunova OI, Berdnikova NE, Savel’ev NA. Burying Dogs in Ancient Cis-Baikal, Siberia: Temporal Trends and Relationships with Human Diet and Subsistence Practices. PLoS One. 2013;8:e63740. doi: 10.1371/journal.pone.0063740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Losey RJ, Bazaliiskii VI, Garvie-Lok S, Germonpré M, Leonard JA, Allen AL, Katzenberg MAnne, Sablin MV. Canids as persons: Early Neolithic dog and wolf burials, Cis-Baikal, Siberia. Journal of Anthropological Archaeology. 2011;30:174–189. [Google Scholar]
- 78.Cava A. El depósito arqueológico de la cueva de Marizulo (Guipúzcoa) Munibe. 1978;30:155–172. [Google Scholar]
- 79.Basabe JM. Restos humanos del yacimiento de Marizulo. Munibe. 1971;23:104–124. [Google Scholar]
- 80.Altuna J. Fauna de mamíferos del yacimiento prehistórico de Marizulo (Urnieta), Guipúzcoa. Sociedad de Ciencias Naturales Aranzadi; 1967. [Google Scholar]
- 81.Bulatović J. Arheozoološki aspekti društvenih i kulturnih promena na Centralnom Balkanu u petom milenijumu pre nove ere. PhD thesis, Faculty of Philosophy; University of Belgrade: 2018. [Google Scholar]
- 82.Radivojević M, Roberts BW, Marić M, Kuzmanović-Cvetković J, Rehren T. The Rise of Metallurgy in Eurasia: The Archaeology of Early Metallurgy and Society in the Central Balkans. UCL Press; London: 2018. [Google Scholar]
- 83.Garašanin MV. Hronologija vinčanske grupe. Ljubljana: Univerza v Ljubljana. 1951 [Google Scholar]
- 84.Radivojevic M, Kuzmanovic-Cvetkovic J. Copper minerals and archaeometallurgical materials from the Vinča culture sites of Belovode and Pločnik: Overview of the evidence and new data. Starinar. 2014;2014:7–30. [Google Scholar]
- 85.Stalio B. Pločnik-Prokuplje-naselje. Arheološki pregled. 1960;2 [Google Scholar]
- 86.Šljivar D, Kuzmanović-Cvetković J. Pločnik kod Prokuplja, istraživanja u 1997. Glasnik Srpskog arheološkog društva. 1998;14:79–85. [Google Scholar]
- 87.Whittle A, Bayliss A, Barclay A, Gaydarska B, Bánffy E, Borić D, Draşovean F, Jakucs J, Marić M, Orton DC, Pantović I, et al. A Vinča potscape:: formal chronological models for the use and development of Vinča ceramics in south-east Europe. Documenta Praehistorica. 2016;60 [Google Scholar]
- 88.Anthony DW, Brown DR. The dogs of war: A Bronze Age initiation ritual in the Russian steppes. Journal of Anthropological Archaeology. 2017;48:134–148. [Google Scholar]
- 89.Dayan T, Galili E. A preliminary look at some new domesticated dogs from submerged Neolithic sites off the Carmel coast. BAR INTERNATIONAL SERIES. 2000;889:29–34. [Google Scholar]
- 90.Horwitz LK, Wolff SR, Ortiz S, Lev-Tov J, Gilbert A, Hesse P. The Context and Biometry of Iron Age II and Hellenistic Period Dog “Burials” from Tel Gezer Compared to Those from Other Sites in the Region. The Wide Lens in Archaeology: Honoring Brian Hesse’s Contributions to Anthropological Archaeology. 2017:297–333. [Google Scholar]
- 91.Dever WG, Lance HD, Wright GE. Gezer I: Preliminary Report of the 1964-66 Seasons. Jerusalem: Hebrew Union College Biblical and Archaeological School in Jerusalem. 1970;1 [Google Scholar]
- 92.Ortiz SM, Wolff SR. Tel Gezer excavations 2006--2015: the transformation of a border city. The Shephelah during the Iron Age: Recent Archaeological Studies. 2017:61–102. [Google Scholar]
- 93.Getzov N, Gorni DAvshalom, Gorin-Rosen Y, Stern EJ, Syon D, Tatcher A. Horbat ‘Uza: The 1991 Excavations, ii. The Late Periods. 2009 [Google Scholar]
- 94.Master DM, Schloen JD, Stager LE. Ashkelon 1: Introduction and overview (1985-2006) Eisenbrauns; 2008. [Google Scholar]
- 95.Stager LE. Why were hundreds of dogs buried at Ashkelon? Biblical Archaeology Review. 1991;17:26–42. [Google Scholar]
- 96.Wapnish P, Hesse B. Pampered Pooches or Plain Pariahs? The Ashkelon Dog Burials. The Biblical Archaeologist. 1993;56:55–80. [Google Scholar]
- 97.Hansen HB, Damgaard PB, Margaryan A, Stenderup J, Lynnerup N, Willerslev E, Allentoft ME. Comparing Ancient DNA Preservation in Petrous Bone and Tooth Cementum. PLoS One. 2017;12:e0170940. doi: 10.1371/journal.pone.0170940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;6 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 99.Rodríguez-Varela R, Günther T, Krzewińska M, Storå J, Gillingwater TH, MacCallum M, Arsuaga JL, Dobney K, Valdiosera C, Jakobsson M, Götherström A, et al. Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans. Curr Biol. 2017;27:3396–3402.e5. doi: 10.1016/j.cub.2017.09.059. [DOI] [PubMed] [Google Scholar]
- 100.Günther T, Malmström H, Svensson EM, Omrak A, Sánchez-Quinto F, Kılınç GM, Krzewińska M, Eriksson G, Fraser M, Edlund H, Munters AR, et al. Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 2018;16:e2003703. doi: 10.1371/journal.pbio.2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ersmark E, Orlando L, Sandoval-Castellanos E, Barnes I, Barnett R, Stuart A, Lister A, Dalén L. Population demography and genetic diversity in the Pleistocene cave lion. Open Quaternary. 2015;1:1–14. [Google Scholar]
- 102.Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Pääbo S. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 2010;38:e87. doi: 10.1093/nar/gkp1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci U S A. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, Whittle A, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nature Communications. 2014;5 doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, Sudmant PH, et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Skoglund P, Northoff BH, Shunkov MV. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proceedings of the. 2014 doi: 10.1073/pnas.1318934111. available at https://www.pnas.org/content/111/6/2229/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pečnerová P, Díez-Del-Molino D, Dussex N, Feuerborn T, von Seth J, van der Plicht J, Nikolskiy P, Tikhonov A, Vartanyan S, Dalén L. Genome-Based Sexing Provides Clues about Behavior and Social Structure in the Woolly Mammoth. Curr Biol. 2017;27:3505–3510.e3. doi: 10.1016/j.cub.2017.09.064. [DOI] [PubMed] [Google Scholar]
- 109.Gower G, Fenderson LE, Salis AT, Helgen KM, van Loenen AL, Heiniger H, Hofman-Kamińska E, Kowalczyk R, Mitchell KJ, Llamas B, Cooper A. Widespread male sex bias in mammal fossil and museum collections. Proc Natl Acad Sci U S A. 2019;116:19019–19024. doi: 10.1073/pnas.1903275116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shi N-N, Fan L, Yao Y-G, Peng M-S, Zhang Y-P. Mitochondrial genomes of domestic animals need scrutiny. Mol Ecol. 2014;23:5393–5397. doi: 10.1111/mec.12955. [DOI] [PubMed] [Google Scholar]
- 111.Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, Derevianko A, Pääbo S. A Complete mtDNA Genome of an Early Modern Human from Kostenki, Russia. Curr Biol. 2010;20:231–236. doi: 10.1016/j.cub.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 112.Plassais J, Kim J, Davis BW, Karyadi DM, Hogan AN, Harris AC, Decker B, Parker HG, Ostrander EA. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09373-w. 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang G-D, Zhai W, Yang H-C, Fan R-X, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng L-G, Poyarkov AD, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4 doi: 10.1038/ncomms2814. 1860. [DOI] [PubMed] [Google Scholar]
- 114.Kardos M, Åkesson M, Fountain T, Flagstad Ø, Liberg O, Olason P, Sand H, Wabakken P, Wikenros C, Ellegren H. Genomic consequences of intensive inbreeding in an isolated wolf population. Nat Ecol Evol. 2018;2:124–131. doi: 10.1038/s41559-017-0375-4. [DOI] [PubMed] [Google Scholar]
- 115.Sinding M-HS, Gopalakrishan S, Vieira FG, Castruita JASamaniego, Raundrup K, Jørgensen MPHeide, Meldgaard M, Petersen B, Sicheritz-Ponten T, Mikkelsen JB, Marquard-Petersen U, et al. Population genomics of grey wolves and wolf-like canids in North America. PLoS Genet. 2018;14:e1007745. doi: 10.1371/journal.pgen.1007745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gopalakrishnan S, Sinding M-HS, Ramos-Madrigal J, Niemann J, Castruita JASamaniego, Vieira FG, Carøe C, de Montero MM, Kuderna L, Serres A, González-Basallote VM, et al. Interspecific Gene Flow Shaped the Evolution of the Genus Canis. Curr Biol. 2018;28:3441–3449.e5. doi: 10.1016/j.cub.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 [q-bio.GN]. available at http://arxiv.org/abs/1303.3997. [Google Scholar]
- 118.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Barros Damgaard P, Martiniano R, Kamm J, Moreno-Mayar JV, Kroonen G, Peyrot M, Barjamovic G, Rasmussen S, Zacho C, Baimukhanov N, Zaibert V, et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science. 2018;360 doi: 10.1126/science.aar7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haber M, Doumet-Serhal C, Scheib C, Xue Y, Danecek P, Mezzavilla M, Youhanna S, Martiniano R, Prado-Martinez J, Szpak M, Matisoo-Smith E, et al. Continuity and Admixture in the Last Five Millennia of Levantine History from Ancient Canaanite and Present-Day Lebanese Genome Sequences. Am J Hum Genet. 2017;101:274–282. doi: 10.1016/j.ajhg.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scheib CL, Li H, Desai T, Link V, Kendall C, Dewar G, Griffith PW, Mörseburg A, Johnson JR, Potter A, Kerr SL, et al. Ancient human parallel lineages within North America contributed to a coastal expansion. Science. 2018;360:1024–1027. doi: 10.1126/science.aar6851. [DOI] [PubMed] [Google Scholar]
- 123.Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, Sirak K, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mathieson I, Alpaslan-Roodenberg S, Posth C, Szécsényi-Nagy A, Rohland N, Mallick S, Olalde I, Broomandkhoshbacht N, Candilio F, Cheronet O, Fernandes D, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hofmanová Z, Kreutzer S, Hellenthal G, Sell C, Diekmann Y, Díez-Del-Molino D, van Dorp L, López S, Kousathanas A, Link V, Kirsanow K, et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc Natl Acad Sci U S A. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Olalde I, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szécsényi-Nagy A, Mittnik A, Altena E, et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, Skoglund P, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oksanen J, Blanchet FG, Kindt R, Legendre P, O’hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: community ecology package. R package version 1.17-4. 2010;23 2010. https://cran.r-project.org/ [Google Scholar]
- 129.Kunsch HR. The jackknife and the bootstrap for general stationary observations. Ann Stat. 1989;17:1217–1241. [Google Scholar]
- 130.Lazaridis I, Belfer-Cohen A, Mallick S, Patterson N. Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry. bioRxiv. 2018 available at https://www.biorxiv.org/content/10.1101/423079v1.abstract. [Google Scholar]
- 131.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leppälä K, Nielsen SV, Mailund T. admixturegraph: an R package for admixture graph manipulation and fitting. Bioinformatics. 2017;33:1738–1740. doi: 10.1093/bioinformatics/btx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 135.Skoglund P, Mallick S, Bortolini MC, Chennagiri N, Hünemeier T, Petzl-Erler ML, Salzano FM, Patterson N, Reich D. Genetic evidence for two founding populations of the Americas. Nature. 2015 doi: 10.1038/nature14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baranowska I, Jäderlund KH, Nennesmo I, Holmqvist E, Heidrich N, Larsson N-G, Andersson G, Wagner EGH, Hedhammar Å, Wibom R, Andersson L. Sensory Ataxic Neuropathy in Golden Retriever Dogs Is Caused by a Deletion in the Mitochondrial tRNATyr Gene. PLoS Genetics. 2009;5:e1000499. doi: 10.1371/journal.pgen.1000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Björnerfeldt S, Webster MT, Vilà C. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 2006;16:990–994. doi: 10.1101/gr.5117706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim KS, Lee SE, Jeong HW, Ha JH. The complete nucleotide sequence of the domestic dog (Canis familiaris) mitochondrial genome. Mol Phylogenet Evol. 1998;10:210–220. doi: 10.1006/mpev.1998.0513. [DOI] [PubMed] [Google Scholar]
- 139.Strakova A, Leathlobhair MNí, Wang G-D, Yin T-T, Airikkala-Otter I, Allen JL, Allum KM, Bansse-Issa L, Bisson JL, Domracheva ACastillo, de Castro KF, et al. Mitochondrial genetic diversity, selection and recombination in a canine transmissible cancer. Elife. 2016;5 doi: 10.7554/eLife.14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Webb KM, Allard MW. Mitochondrial genome DNA analysis of the domestic dog: identifying informative SNPs outside of the control region. J Forensic Sci. 2009;54:275–288. doi: 10.1111/j.1556-4029.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- 141.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Molak M, Suchard MA, Ho SYW, Beilman DW, Shapiro B. Empirical calibrated radiocarbon sampler: a tool for incorporating radiocarbon-date and calibration error into Bayesian phylogenetic analyses of ancient DNA. Molecular Ecology Resources. 2015;15:81–86. doi: 10.1111/1755-0998.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ciccarelli FD. Toward Automatic Reconstruction of a Highly Resolved Tree of Life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 148.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, et al. Ensembl 2020. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bai B, Zhao W-M, Tang B-X, Wang Y-Q, Wang L, Zhang Z, Yang H-C, Liu Y-H, Zhu J-W, Irwin DM, Wang G-D, et al. DoGSD: the dog and wolf genome SNP database. Nucleic Acids Res. 2015;43:D777–83. doi: 10.1093/nar/gku1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci. 2013;40:4477–4482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1
Supplementary material
Data Availability Statement
The generated DNA sequencing data will be made available in the European Nucleotide Archive (ENA) under study accession PRJEB38079.