Inhibition of Nuclear Receptor Signalling by Poly(ADP-Ribose) Polymerase (original) (raw)
Abstract
Mammalian poly(ADP-ribose) polymerase (PARP) is a nuclear chromatin-associated protein with a molecular mass of 114 kDa that catalyzes the transfer of ADP-ribose units from NAD+ to nuclear proteins that are located within chromatin. We report here the identification of a novel property of PARP as a modulator of nuclear receptor signalling. PARP bound directly to retinoid X receptors (RXR) and repressed ligand-dependent transcriptional activities mediated by heterodimers of RXR and thyroid hormone receptor (TR). The interacting surface is located in the DNA binding domain of RXRα. Gel shift assays demonstrated that PARP bound to TR-RXR heterodimers on the response element. Overexpression of wild-type PARP selectively blocked nuclear receptor function in transient transfection experiments, while enzyme-defective mutant PARP did not show significant inhibition, suggesting that the essential role of poly(ADP-ribosyl) enzymatic activity is in gene regulation by nuclear receptors. Furthermore, PARP fused to the Gal4 DNA binding domain suppressed the transcriptional activity of the promoter harboring the Gal4 binding site. Thus, PARP has transcriptional repressor activity when recruited to the promoter. These results indicates that poly(ADP-ribosyl)ation is a negative cofactor in gene transcription, regulating a member of the nuclear receptor superfamily.
Nuclear hormone receptors for steroids, retinoids, thyroid hormone, vitamin D3, and prostanoids comprise a large family of sequence-specific transcription factors. They play diverse roles in development, differentiation, and homeostasis (18) by modulating gene transcription. Retinoid X receptors (RXR) are members of a superfamily of nuclear hormone receptors and heterodimerize with a variety of other family members, including all-_trans_-retinoic acid receptor (RAR), thyroid hormone receptor (TR), and vitamin D receptor (VDR), indicating that RXR play a central role in ligand-dependent transcriptional regulation by nuclear receptors (15, 35, 37). These heterodimers bind to specific DNA sequences and directly regulate transcription of target genes in response to specific ligands. Nuclear receptors are thought to mediate their transcriptional effects in concert with coregulator proteins that modulate receptor interactions with components of the basal transcription machinery (3, 5, 6, 9, 11–14, 16, 30–32).
The mechanism of transcriptional regulation by nuclear receptors has been a focus of intense study. The demonstration of direct interactions of receptors with basal transcription factors, such as TFIIB and TBP (19, 20, 22, 23, 34), suggests that liganded receptors may directly influence the function of the basal transcription machinery. However, these direct interaction models do not explain transcriptional squelching between receptors or the roles of receptor-associated cofactors. Negative transcriptional regulation by TR and RAR is mediated, in part, by their association with a class of silencing mediators termed SMRT and N-CoR (4, 21, 24, 25, 27). In addition, at least three distinct classes of receptor cofactors have been identified, including SRC-1, TRIP1, RIP140/160, and TIF1. A continuing search for cofactors that mediate ligand-dependent transactivation functions of the nuclear hormone receptors led to the finding that CREB binding protein (CBP) and its homolog, p300, can interact with several nuclear receptors and potentiate their transactivation activities. These recent studies suggest that bridging protein factors exist and function to transmit the signal of ligand-induced conformational change to the basal transcription machinery.
Structural change in targeted chromatin has been postulated to be associated with regulation of gene expression. Recent biochemical and genetic studies support the notion that hyperacetylation of core histones is a characteristic of gene activation and that, conversely, histone deacetylation is involved in transcriptional repression. Strikingly, nuclear receptor corepressors SMRT and N-CoR form complexes with Sin3 and histone deacetylase proteins, suggesting that chromatin remodeling by histone deacetylation is a possible mechanism for receptor-mediated repression. It was found that CBP/p300 interact functionally with a human histone acetyltransferase protein, P/CAF (31). Recently, it was also found that CBP and p300 themselves have intrinsic histone acetyltransferase activities (3). Therefore, it appears that CBP/p300 and its associated protein P/CAF play a pivotal role in ligand-dependent transcriptional regulation and function through targeted modification of chromatin structure (2).
In an effort to identify potential coregulators, human RXR fusions with glutathione _S_-transferase (GST) were used to isolate proteins capable of binding the receptor. We report here the selective isolation of poly(ADP-ribose) polymerase (PARP) as a protein that interacts with receptors. We provide in vitro and in vivo evidence that identifies PARP as a repressor for transcriptional activation by nuclear receptors. Our findings strongly suggest that, as well as histone acetylation, poly(ADP-ribosyl)ation of histone or transcriptional factors must have a critical role in nuclear receptor signalling. PARP participates in DNA excision and repair by automodification (7, 26, 33). These properties of PARP, together with the nature of the interactions with nuclear receptors in vitro and vivo, raise the possibility of coupled transcription and DNA repair.
MATERIALS AND METHODS
Isolation of interacting proteins.
A rat GH3 cDNA library was constructed by using T7 expression phage and was screened by full-length human RXRα as a probe. Isolated clones were subcloned into pGEM 3 and sequenced by an ABI 3300 autosequencer. [35S]methionine-labelled peptides were produced with the T7 TNT-coupled system (Promega), and their interactions with RXRα were confirmed by a pull-down experiment using matrix-bound GST-RXRα. About 106 clones were screened, and one clone, which contained amino acids 82 to 220 of PARP [PARP(82-220)], was confirmed as an interacting partner with RXR.
Cell culture and transient transfection and reporter assays.
COS1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 100 μg of penicillin per ml, and 0.25 mg of streptomycin per ml at 37°C in 5% CO2. Transfection was done in COS1 cells by the standard calcium phosphate procedure. Typically, 0.25 μg of DR4- or DR1-driven luciferase reporter was cotransfected with 2 ng of the indicated expression vectors. Cells were incubated for 12 h, and the appropriate ligands were added.
In vitro transcription and translation.
Coupled transcription and translation of PARP(82-220) was carried out with the TNT in vitro transcription/translation kit (Promega), according to the manufacturer’s instructions.
GST pull-down assay and PARP enzymatic assays.
GST-RXRα(1-462) was generated as described previously (3). Ten microliters of GST-Sepharose beads containing 2 to 5 μg of GST recombinant proteins were incubated with [35S]Met-labelled proteins or COS cell extracts for 1 h at 4°C. Complexes were then centrifuged, washed three times in gel shift buffer, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Radiolabelled signals were visualized and quantified with a phosphorimager (Fuji BAS 1500).
Gel retardation assays.
Synthetic oligonucleotides representing each strand of sequences were purified by PAGE, eluted, and annealed. Double-stranded oligonucleotides were radiolabelled with dCTP (>3,300 Ci/mmol) (ICN Biomedicals, Costa Mesa, Calif.) by fill-in reactions with Klenow large-fragment DNA polymerase. Radiolabelled probes (10 fmol, 20,000 to 30,000 cpm) were then incubated with binding proteins in 30 μl of a reaction mixture containing 10 mM KPO4 (pH 8.0) buffer, 1 mM EDTA, 80 mM KCl, 1 μg of poly(dI-dC), 1 mM dithiothreitol (DTT), 0.5 mM MgCl2, 5 μg of bovine serum albumin, and 10% glycerol. Reaction mixtures were incubated for 30 min at room temperature and analyzed on a 5% nondenaturing polyacrylamide gel in Tris-acetate-EDTA buffer. Electrophoresis was performed at a constant 200 V at 4°C in the same buffer.
Expression of recombinant proteins.
To express the fusion protein with GST, PCR-amplified full-length RXRα cDNA or truncated fragments were inserted in-frame into the _Bam_HI and _Eco_RI cloning sites of the pGEX-2TK vector (Pharmacia). Overnight cultures of Escherichia coli BL21 carrying the recombinant GST fusions or GST control plasmid were diluted 100-fold, cultured for 5 to 6 h, and then induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After another 3 h, bacteria were collected and washed with phosphate-buffered saline (PBS). Pellets were suspended in PBS containing 1% (vol/vol) Triton X-100 and sonicated. Debris was removed by centrifugation. The fusion protein or the GST control protein was bound to glutathione-Sepharose (Pharmacia) and extensively washed with PBS containing 1% (vol/vol) Triton X-100. Matrix-bound proteins were used for interaction experiments.
Interaction experiments.
In vitro-translated 35S-labelled proteins (1 to 2 μl) were incubated for 20 min at room temperature with glutathione-Sepharose (10 μl) preloaded with GST fusion or GST control protein in 250 μl of binding buffer (20 mM Tris-Cl, pH 7.8–100 mM NaCl–10% glycerol–1 mM DTT–1 mM EDTA–1 mM phenylmethylsulfonyl fluoride–1 mM leupeptin–1 mM pepstatin) in the presence or absence of 10−6 M T3 and/or 9-_cis_-RA. After extensive washing with binding buffer, bound proteins were eluted in 25 μl of Laemmli sample buffer, boiled for 10 min, and resolved by SDS–10% PAGE followed by autoradiography. The results of in vitro reactions and amounts of 35S-labeled protein bound by GST or GST-RXRα were visualized and quantified with a phosphorimager (Fuji BAS 1500).
RESULTS
Isolation of PARP as an interacting partner with RXR.
We employed biochemical methods to identify the protein interacting with RXR. A rat GH3 cell cDNA library was screened with a GST fusion containing full-length human RXRα as a probe. Positive clones were transcribed by T7 RNA polymerase, translated into 35S-labelled peptides with [35S]methionine, and used for pull-down experiments with GST-RXRα to confirm the interactions. One positive clone was identified and found to be a fragment (amino acid residues 82 to 220) of rat PARP cDNA (Fig. 1a).
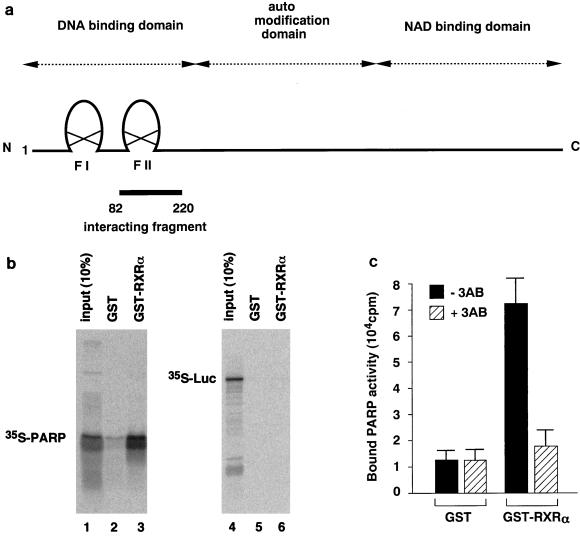
FIG. 1.
PARP interacts with nuclear receptors in vitro. (a) Schematic diagram of the domain structure of full-length rat PARP. The isolated interacting fragment (amino acids 82 to 220) is indicated. (b) PARP interacts with RXR. 35S-labelled PARP(82-220) or luciferase was synthesized by in vitro translation and incubated separately with GST (lanes 2 and 5) or GST-RXR (lanes 3 and 6) bound to glutathione-Sepharose beads. Ten percent of input 35S-labelled proteins are indicated (lanes 1 and 4). (c) Endogenous PARP interacts with hormone receptors. Crude COS1 cell extracts were allowed to interact with immobilized RXR. COS1 cell extracts were incubated with matrix-bound GST-RXR or GST. After a wash, PARP activity which associated with beads was measured in the absence or presence of 3-aminobenzamide (3AB). The initial velocity of [32P]NAD incorporation into acid-insoluble acceptors was measured at 25°C for 1 min. The results represent averages and standard deviations from three independent experiments.
PARP interacts with nuclear receptors.
The interaction between PARP(82-220) and RXR is shown in Fig. 1b. A matrix-bound fusion protein of GST with RXR (GST-RXR), but not GST alone, retains [35S]methionine-labelled PARP(82-220) (lanes 2 and 3), while [35S]methionine-labelled luciferase binds to neither GST-RXR nor GST alone (lanes 5 and 6). Addition of RXR ligand 9-_cis_-RA during incubation did not alter the binding of 35S-labelled PARP (data not shown). The interaction between cellular PARP and nuclear receptors was further analyzed by incubating COS1 cell nuclear extracts with matrix-bound GST-RXR. Evidence for the interaction between RXR and cellular PARP is shown in Fig. 1c. Equivalent amounts of COS1 cell extracts were incubated with matrix-bound GST or GST-RXR, and after washing, bound proteins were eluted by the addition of 1 mM glutathione. The PARP activity of affinity-selected fractions shows that RXR can associate with PARP in the presence of a full complement of nuclear proteins.
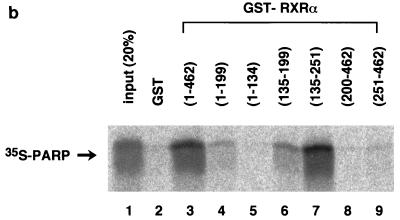
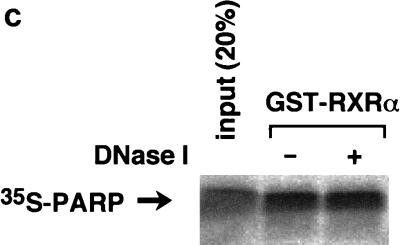
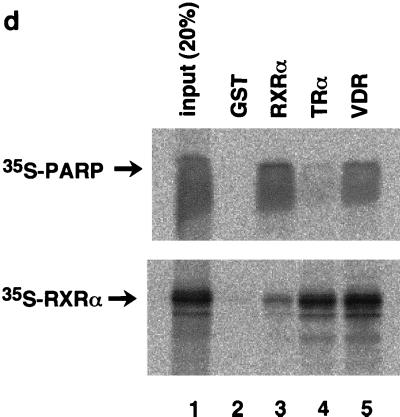
To map the specific domains in RXR that mediate interactions with PARP, a series of GST fusion proteins representing overlapping portions of RXR were expressed in bacteria, purified (Fig. 2a), and used to bind 35S-labelled PARP(82-220) (Fig. 2b). The DNA binding domain (C domain) of RXR is required for interaction. The C domain itself possessed only weak binding activity to PARP (Fig. 2b, lanes 4 and 6), and an additional hinge domain (D domain) is necessary for full interaction (Fig. 2b, lane 7), although the D domain itself has no binding activity (Fig. 2b, lane 8). Because the regions identified as putative interaction surfaces correspond to the DNA binding domains of both proteins, there is the possibility that the interaction may be mediated by nonspecific binding to DNA fragments present in the assays. To exclude the above possibility, we performed the in vitro pull-down experiment in the presence of DNase I. As shown in Fig. 2c, DNase I treatment did not alter the interaction, indicating that the interaction is direct protein-protein binding. It is not surprising that the highly conserved DNA binding domain of nuclear receptors could also serve as a site for coregulator proteins. We next investigated the binding of PARP to other nuclear receptors. 35S-labelled PARP significantly interacted with GST-VDR but not with GST-TRα1, while GST-VDR and GST-TRα1 retained equal abilities to interact with 35S-labelled RXRα (Fig. 2d).
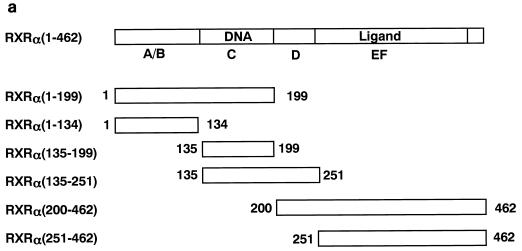
FIG. 2.
Domains within nuclear receptors required for PARP interactions. (a) Series of N- and C-terminal deletions of RXR used in pull-down experiments. (b) In vitro interaction of PARP(82-220) with RXR. Bacterially produced GST-RXR deletions or GST alone was bound to glutathione-Sepharose beads and incubated with equivalent amounts of 35S-labelled PARP(82-220) produced by in vitro translation. Associated proteins were analyzed by SDS–15% PAGE and visualized by BAS 1500 (Fuji, Tokyo, Japan). (c) 35S-labelled PARP(82-220) was incubated with matrix-bound GST-RXRα in the absence or presence of 1 U of DNase I, and associated proteins were analyzed by SDS–15% PAGE. (d) Differential binding of PARP to nuclear receptors. 35S-labelled PARP(82-220) or 35S-labelled RXRα was incubated with GST alone (lane 2), GST-RXRα (lane 3), GST-TRα (lane 4), or GST-VDR (lane 5). Associated proteins were analyzed by SDS–15% PAGE and visualized.
These results led us to evaluate the effect of DNA binding on the RXR-PARP interaction. Inclusion of 10 molar excesses of RXRE relative to RXR during incubation had little effect on RXR binding to PARP, and RXR strongly binds RXRE under such conditions (data not shown). Thus, DNA binding does not appear to block the interaction. We can now identify the nuclear protein PARP as a high-affinity binding protein for the CD regions of the RXR.
PARP associates with receptors on the hormone response element.
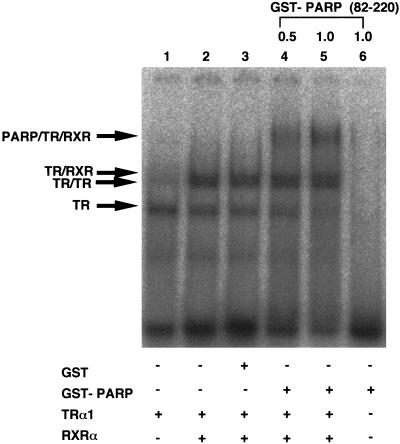
The above results were of interest because the DNA binding domain of RXR has been reported to be involved in the formation of TR-RXR heterodimers on direct repeat DNA elements. To characterize further interaction between PARP and receptor heterodimers, we have performed gel mobility shift assays with bacterially expressed and purified proteins. The results shown in Fig. 3 indicate that TR-RXR heterodimers were further shifted by addition of bacterially expressed and purified GST-PARP. Addition of GST alone did not alter the migration of the TR-RXR heterocomplex, and GST-PARP alone did not show the retarded bands. It should be noted that recruitment of PARP to receptor-DNA complexes does not incur significant change in the affinity of receptors to hormone response elements, suggesting that PARP probably does not function through altering receptor DNA binding.
FIG. 3.
PARP associates with receptors on hormone response elements. Shown is the interaction of PARP(82-220) with an RXR-TR heterodimer on a DR4 element comprising an AGGTCA direct repeat spaced by four nucleotides in a gel retardation assay. Bacterially expressed and purified RXR and TR were incubated with radiolabelled DR4 probe in the presence or absence of GST-PARP(82-220) or GST alone. The RXR-TR heterocomplex and PARP-RXR-TR ternary complex are indicated by arrows.
Expression of PARP specifically inhibits ligand-dependent transcriptional activation by TR.
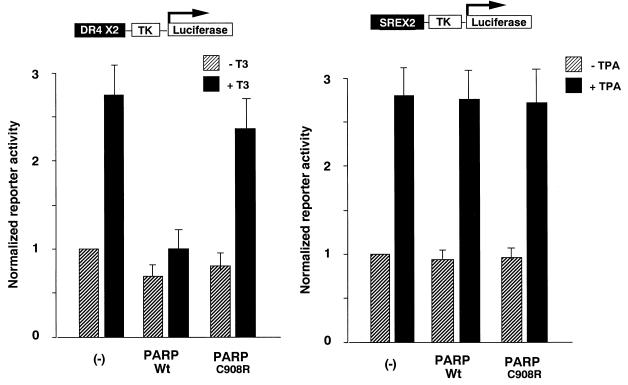
Our results with the interaction between PARP and nuclear receptors in vitro prompted us to test the physiological relevance of the interaction. To evaluate the function of PARP in nuclear receptor signalling, full-length PARP was coexpressed in transient transfection assays. As shown in Fig. 4, full-length PARP specifically blocked the ligand-dependent transcriptional activity by TR but had no effect on a promoter under control of the cytomegalovirus promoter (data not shown) or TPA (12-_O_-tetradecanoylphorbol-13-acetate)-stimulated serum response element (SRE) thymidine kinase (TK) promoter activity. Moreover, the PARP enzyme-defective mutant PARP(C908R) (23) did not alter the nuclear receptor signalling, suggesting that recruitment of PARP enzyme activity plays an inhibitory role in nuclear receptor-dependent transcription.
FIG. 4.
Overexpression of PARP inhibits ligand-dependent transactivation by nuclear receptors. Luciferase reporter gene activities under control of the DR4-TK or SREX2-TK promoter were measured from extracts of COS1 cells after the cells were transiently transfected by the corresponding reporter and expression plasmids. Relative luciferase activities in the presence (solid bars) or absence (hatched bars) of cognate ligands after being normalized by the internal control for β-galactosidase activities are presented. The results represent averages and standard deviations from at least three independent experiments.
PARP has transcriptional repression activity.
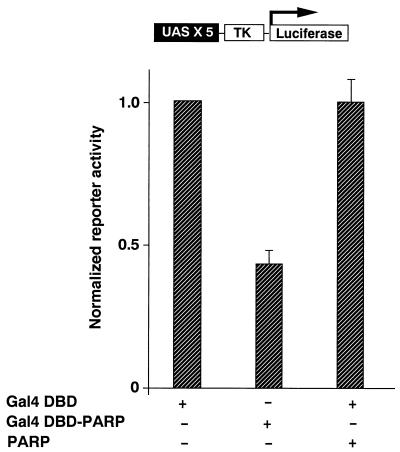
In order to elucidate whether PARP possesses transcriptional repression activity, the pM-PARP eukaryotic expression construct encoding a Gal4 DNA binding domain-PARP fusion protein was cotransfected with a luciferase reporter plasmid containing five Gal4 binding sites into COS1 cells. Compared with cells cotransfected with the reporter construct and only the Gal4 DNA binding domain, luciferase activity was lower in cells cotransfected with pM-PARP (Fig. 5). The C908R mutation, which results in a loss of poly(ADP-ribosyl) enzyme activity, eliminated the repressor activity of the protein. There was a dose-dependent repression of luciferase activity when full-length PARP was coexpressed as a Gal4 fusion protein with the reporter (data not shown).
FIG. 5.
PARP has transcriptional repression activity. The pM-PARP eukaryotic expression construct encoding a Gal4 DNA binding domain-PARP fusion protein was cotransfected into COS1 cells with a luciferase reporter plasmid containing five Gal4 binding sites. Compared with cells cotransfected with the reporter construct and Gal4 DNA binding domain, luciferase activity was lower in cells cotransfected with pM-PARP. The C908R mutation, which results in loss of poly(ADP-ribosyl) enzyme activity, eliminated the repressor activity of the protein. All luciferase activity was corrected for transfection efficiency by measuring β-galactosidase activity of cells transfected together. The results represent averages and standard deviations from three independent experiments.
DISCUSSION
In this study, we demonstrated that PARP is a novel partner of nuclear receptors and inhibits nuclear receptor signalling. The identification of PARP as a potential inhibitor of nuclear receptor function is an important extension of the cellular roles of ADP-ribosylation, raising questions regarding the role of poly(ADP-ribosyl)ation in transcriptional regulation. Poly(ADP-ribosyl)ation of nuclear proteins has been reported to occur during the processes of DNA repair, DNA replication, and DNA transcription (22). It was reported that histones are the predominant poly(ADP-ribose) binding species in mammals, Xenopus sp., and yeasts (20, 30). The polymer binding site is confined specifically to the histone domains responsible for DNA condensation, i.e., histone tails (23). Remodeling of chromatin structure and nucleosome positioning by targeting ADP-ribosylation has been linked to gene regulation.
Nuclear receptor corepressors, N-CoR and the related factor SMRT, which were initially discovered through their ability to bind to unliganded nuclear receptors, recruit histone deacetylase (mSin3 and mRPD3), resulting in condensation of the chromatin structure to repress basal transcription (4, 12, 20, 27). We hypothesize that PARP which is recruited to DNA-bound nuclear receptors may also participate in the remodeling of nucleosome structure in concert with histone acetylation or deacetylation, resulting in transcriptional regulation in response to ligands.
Studies with a chimeric protein of PARP fused to the glucocorticoid receptor revealed that PARP activity can be targeted to specific DNA sequences and repress gene expression (24). Recently, Oei et al. demonstrated that poly(ADP-ribosyl)ation might prevent polymerase II-dependent transcription (21). Here, we also demonstrate that a chimeric protein of PARP fused to the Gal4 DNA binding domain represses gene expression of a promoter harboring the Gal4 binding site. These results suggest that PARP has repressor activity when targeted to the promoter. PARP might facilitate recovery from DNA damage by stimulating DNA repair and silencing transcription. The association of DNA repair factors with the RNA polymerase II complex was reported (1, 25). TFIIH has dual roles in transcription and DNA nucleotide excision repair (7, 26). It seems that transcription and DNA repair are closely related. Future experiments could help to elucidate whether PARP interacts with basal transcriptional machinery or functionally distinct components, such as DNA repair proteins, that have been reported to associate with the basal machinery (25). The results presented here demonstrate that PARP participates with both transcriptional regulators and components with roles in DNA repair. It was reported that mice lacking PARP develop normally but are susceptible to skin disease (33). Evaluation of hormonal responsiveness in these transgenic mice will further elucidate the physiological importance of PARP in vivo.
Although we demonstrated that PARP inhibits nuclear receptor-induced transcription, there is evidence that PARP enhances nonnuclear receptor signaling. For example, inducible nitric oxide synthase expression has been shown to be inhibited by PARP inhibitors (8, 10) and in PARP knockout cells (29). Furthermore, expression of prolactin has been shown to be regulated by poly(ADP-ribosyl)ation in a similar manner (28). Together with these data, our results provide an interesting corollary, namely, how differently nuclear and nonnuclear receptor signal transduction events are regulated. In order to elucidate the functional consequences of this kind of modification, it would be necessary to search for substrates of PARP recruited to promoters.
ACKNOWLEDGMENTS
We thank R. M. Evans for providing RXRα cDNA and UAS × 4 reporter and V. Rolli and G. Murcia for wild-type and mutant (C908R) PARP cDNA.
REFERENCES
- 1.Bardwell A J, Bardwell L, Iyer N, Svejstrup J Q, Feaver W J, Kornberg R D, Friedberg E C. Yeast nucleotide excision repair proteins Rad2 and Rad4 interact with RNA polymerase II basal transcription factor b (TFIIH) Mol Cell Biol. 1994;14:3569–3576. doi: 10.1128/mcb.14.6.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evan R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen J D, Umesono K, Evans R M. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng X, Reginato M J, Andrews N C, Lazar M A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowell P, Ishmael J E, Avram D, Peterson V J, Nevrivy D J, Leid M. p300 functions as a coactivator for the peroxisome proliferator-activated receptor alpha. J Biol Chem. 1997;272:33435–33443. doi: 10.1074/jbc.272.52.33435. [DOI] [PubMed] [Google Scholar]
- 7.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 8.Fujimura M, Tominaga T, Yoshimoto T. Nicotinamide inhibits inducible nitric oxide synthase mRNA in primary rat glial cells. Neurosci Lett. 1997;228:107–110. doi: 10.1016/s0304-3940(97)00373-x. [DOI] [PubMed] [Google Scholar]
- 9.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 10.Hauschildt S, Scheipers P, Bessler W G. Inhibitors of poly (ADP-ribose) polymerase suppress lipopolysaccharide-induced nitrite formation in macrophages. Biochem Biophys Res Commun. 1991;179:865–871. doi: 10.1016/0006-291x(91)91898-m. [DOI] [PubMed] [Google Scholar]
- 11.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 13.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Glass B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 15.Kliewer S A, Umesono K, Mangelsdorf D J, Evans R M. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 17.Leong G M, Wang K S, Marton M J, Blanco J C, Wang I M, Rolfes R J, Ozato K, Segars J H. Interaction between the retinoid X receptor and transcription factor IIB is ligand-dependent in vivo. J Biol Chem. 1998;273:2296–2305. doi: 10.1074/jbc.273.4.2296. [DOI] [PubMed] [Google Scholar]
- 18.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meisterernst M, Stelzer G, Roeder R G. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 21.Oei S L, Griesenbeck J, Ziegler M, Schweiger M. A novel function of poly(ADP-ribosyl)ation: silencing of RNA polymerase II-dependent transcription. Biochemistry. 1998;37:1465–1469. doi: 10.1021/bi9727390. [DOI] [PubMed] [Google Scholar]
- 22.Panzeter P L, Zweifel B, Malanga M, Waser S H, Richard M, Althaus F R. Targeting of histone tails by poly(ADP-ribose) J Biol Chem. 1993;268:17662–17664. [PubMed] [Google Scholar]
- 23.Rolli V, O’Farrel M, Murcia J M, Murcia G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997;36:12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal D, Hong T, Zhang S, Shima T, Danielsen M, Smulson M. Expression and characterization of a fusion protein between the catalytic domain of poly(ADP-ribose) polymerase and the DNA binding domain of glucocorticoid receptor. Biochem Biophys Res Commun. 1994;202:880–887. doi: 10.1006/bbrc.1994.2012. [DOI] [PubMed] [Google Scholar]
- 25.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H, Egly J M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 26.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H, Chambon P, Egly J M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 27.Soderstrom M, Vo A, Heinzel T, Lavinsky R M, Yang W M, Seto E, Peterson D A, Rosenfeld M G, Glass C K. Differential effects of nuclear receptor corepressor (N-CoR) expression levels on retinoic acid receptor-mediated repression support the existence of dynamically regulated corepressor complexes. Mol Endocrinol. 1997;11:682–692. doi: 10.1210/mend.11.6.0018. [DOI] [PubMed] [Google Scholar]
- 28.Suganuma N, Kikkawa F, Seo H, Matsui N, Tomoda Y. Poly (adenosine diphosphate-ribose) synthesis in the anterior pituitary of the female rat throughout the estrous cycle: study of possible relation to cell proliferation and prolactin gene expression. J Endocrinol Investig. 1993;16:475–480. doi: 10.1007/BF03348885. [DOI] [PubMed] [Google Scholar]
- 29.Szabo C, Virag L, Cuzzocrea S, Scott G S, Hake P, O’Connor M P, Zingarelli B, Salzman A, Kun E. Protection against peroxynitrite-induced fibroblast injury and arthritis development by inhibition of poly(ADP-ribose) synthase. Proc Natl Acad Sci USA. 1998;95:3867–3872. doi: 10.1073/pnas.95.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thenot S, Henriquet C, Rochefort H, Cavailles V. Differential interaction of nuclear receptors with the putative human transcriptional coactivator hTIF1. J Biol Chem. 1997;272:12062–12068. doi: 10.1074/jbc.272.18.12062. [DOI] [PubMed] [Google Scholar]
- 31.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 32.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 34.Wesierska G J, Sauermann G. The effect of poly(ADP-ribose) on interactions of DNA with histones H1, H3 and H4. Eur J Biochem. 1988;173:675–679. doi: 10.1111/j.1432-1033.1988.tb14051.x. [DOI] [PubMed] [Google Scholar]
- 35.Yu V C, Delsert C, Andersen B, Holloway J M, Devary O V, Naar A M, Kim S Y, Boutin J M, Glass C K, Rosenfeld M G. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- 36.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X K, Lehmann J, Hoffmann B, Dawson M I, Cameron J, Graupner G, Hermann T, Tran P, Pfahl M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature. 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]