hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci (original) (raw)
Abstract
Proliferating cell nuclear antigen (PCNA) has been implicated in eukaryotic postreplicative mismatch correction, but the nature of its interaction with the repair machinery remained enigmatic. We now show that PCNA binds to the human mismatch binding factors hMutSα and hMutSβ via their hMSH6 and hMSH3 subunits, respectively. The N-terminal domains of both proteins contain the highly conserved PCNA-binding motif Qxx[LI]xx[FF]. A variant of hMutSα, lacking this motif because of deletion of 77 N-terminal residues of the hMSH6 subunit, no longer was able to interact with PCNA in vitro and failed to restore mismatch repair in hMSH6-deficient cells. Colocalization of PCNA and hMSH6 or hMSH3 to replication foci implies an intimate link between replication and mismatch correction. We postulate that PCNA plays a role in repair initiation by guiding the mismatch repair proteins to free termini in the newly replicated DNA strands.
Keywords: PCNA, hMSH6, hMSH3, mismatch repair, DNA repair, replication foci
Mismatches introduced into DNA during replication are addressed by the postreplicative mismatch repair (MMR) system. In Escherichia coli, binding of the mismatch by the MutS protein, which is able to recognize both base–base mismatches (Jiricny et al. 1988; Su et al. 1988) and insertion–deletion loops (IDLs) containing up to four extrahelical nucleotides (Parker and Marinus 1992), triggers an ATP-driven assembly of the MMR repairosome. This contains, in addition to the homodimeric MutS protein, also the strand-discrimination endonuclease MutH and the MutL homodimer, thought to play a bridging role between MutS and MutH. The process also requires the DNA helicase UvrD, single-strand DNA-binding protein Ssb, one of several exonucleases, DNA polymerase III holoenzyme, and DNA ligase. The Dam methylase, which modifies GATC sites in E. coli DNA, plays a key accessory role in the MMR process. Because this enzyme lags behind the replication fork by ∼2 min (Barras and Marinus 1989), the newly synthesized strand remains unmethylated during this time and therefore can be distinguished from the methylated, template DNA. This property is used by the MutH endonuclease, which initiates the repair process by incising the newly synthesized, unmethylated strand of the DNA heteroduplex 5′ from an unmethylated GATC sequence. The MutL protein then loads the helicase–exonuclease complex at this site (Dao and Modrich 1998), and the error-containing strand is degraded until the mispair is eliminated. The repair tract, stabilized by the Ssb protein, is filled in by DNA polymerase III, and ligation of the remaining nick and modification of the hemimethylated DNA by Dam methylase completes the repair process (for review, see Modrich 1989, 1991; Modrich and Lahue 1996).
The MMR process is highly conserved throughout evolution. In eukaryotes, the mismatch recognition function is fulfilled by two heterodimeric factors composed of MutS homologs MSH2 and MSH6 (MutSα) or MSH2 and MSH3 (MutSβ). The former recognizes base–base mismatches and small IDLs (Drummond et al. 1995; Palombo et al. 1995), whereas the latter binds IDLs larger than one extrahelical nucleotide with high efficiency (Palombo et al. 1996; Genschel et al. 1998). The MutL function also is conserved in the form of MutLα, a heterodimer of MLH1 and PMS1 (hMLH1 and hPMS2 in humans) (Kolodner and Marsischky 1999; Jiricny 2000). Interestingly, no MutH homologs have been found to date; this function appears to exist only in gram-negative bacteria. In Streptococcus pneumoniae, mismatch correction is accomplished without methylation direction (Lacks et al. 1982), and a similar situation must exist in organisms that do not methylate their DNA, such as Saccharomyces cerevisiae and Drosophila melanogaster. In extracts of cultured D. melanogaster (Holmes et al. 1990) and human cells (Holmes et al. 1990; Thomas et al. 1991), mismatch-dependent DNA degradation is targeted to either strand of a heteroduplex by pre-existing nicks or gaps, and it has been shown that the repair tracts commence at these strand interruptions (Fang and Modrich 1993). The system appears to lack a mismatch-dependent endonuclease activity, as covalently closed circular substrates are refractory to mismatch repair (Holmes et al. 1990; Thomas et al. 1991).
The link of MMR defects with hereditary nonpolyposis colon cancer (HNPCC) (for review, see Kolodner 1995; Modrich and Lahue 1996; Jiricny and Nyström-Lahti 2000) has prompted an intensive search for the missing members of the eukaryotic MMR repairosome. Although the DNA helicase and the 3′-5′ exonuclease(s) remain to be identified, the system could be shown to involve, in addition to MutSα, MutSβ, and MutLα, also the 5′-3′ exonuclease EXOI (Szankasi and Smith 1995; Tishkoff et al. 1997), PCNA (Johnson et al. 1996; Umar et al. 1996; Eissenberg et al. 1997; Gu et al. 1998), RP-A (Lin et al. 1998), and polymerase-δ (Longley et al. 1997; for a recent review, see Kolodner and Marsischky 1999). The participation of PCNA, which acts as a processivity factor for polymerase-δ, was not surprising. What was surprising, however, was that this homotrimeric sliding clamp appeared to be required not only in the gap-filling reaction, but also in the steps preceding the degradation of the error-containing strand (Umar et al. 1996; Gu et al. 1998). Given that PCNA is known to be loaded at DNA termini (for review, see Jonsson and Hubscher 1997) and that the MMR process in human cell extracts commences at such sites, we decided to study the role of this protein in mismatch correction.
The first hint of PCNA involvement in MMR came from Kunkel's laboratory, where screening of yeast and human expression libraries for proteins interacting with MSH2 and MLH1 in two-hybrid assays yielded clones expressing PCNA or its S. cerevisiae homolog encoded by the POL30 gene (Umar et al. 1996). A mutant allele of the yeast PCNA homolog, pol30_–_52 was shown to impart a mutator phenotype and appeared epistatic with MMR gene mutations. Biochemical studies substantiated the requirement for PCNA in MMR inasmuch as the repair process in vitro could be inhibited by the addition of a p21Cip1/WAF1 peptide containing the PCNA-binding motif, known to abrogate the interaction of proteins with PCNA (for review, see Warbrick 1998). Addition of excess PCNA rescued the repair reaction. In a similar study from Prakash's laboratory, the pol30_–_104 allele appeared to be phenotypically similar to pol30_–_52, and PCNA was shown to interact with MutSβ but not with MSH2 in affinity gel filtration experiments (Johnson et al. 1996). Eissenberg and colleagues have examined a series of yeast PCNA mutants and succeeded in identifying two, pol30_–_79 and pol30_–_90, which conferred mutator phenotypes and were epistatic with a Δ_pms1_ mutation, suggesting that the defect was linked with MMR (Eissenberg et al. 1997).
We now show that PCNA interacts with hMSH3 and hMSH6 both in vitro and in vivo, and that the hMSH6–PCNA interaction is required for the correction of base–base mismatches.
Results
hMSH3 and hMSH6 contain PCNA-binding motifs
PCNA appears to interact with other proteins via a conserved motif Qxx[ILM]xx[FH][FY], followed by a nonconserved sequence containing basic amino acids and often also prolines (Warbrick 1998; Zhang et al. 1999). This motif has to date been found in several human proteins involved in cell cycle control and DNA metabolism, such as the cell cycle regulatory protein p21Cip1/WAF1 (hCDN1) (Nakanishi et al. 1995; Warbrick et al. 1995), the flap endonuclease FEN1 (hFEN1) (Gary et al. 1997; Warbrick et al. 1997), the nucleotide excision repair endonuclease XPG (hXPG) (Gary et al. 1997), DNA methyl transferase I (hMTDM) (Chuang et al. 1997), DNA ligase I (hDNL1) (Montecucco et al. 1998), uracil DNA glycosylase (hUNG) (Otterlei et al. 1999), and the small subunit of DNA polymerase-δ (hp66) (Reynolds et al. 2000). Moreover, a partial PCNA-binding motif was found in the large subunit of replication factor C (RFC1) (Montecucco et al. 1998), the catalytic subunit of DNA polymerase-δ (p125) (Zhang et al. 1999), and in the Werner Syndrome gene product (Lebel et al. 1999). For this reason, we sought this motif also in the MMR proteins MLH1 or MSH2, which were reported to interact with PCNA in two-hybrid assays (Umar et al. 1996). To our surprise, these latter polypeptides lacked a recognizable PCNA-binding motif; however, it was present in hMSH6 and hMSH3 (Fig. 1A). With the exception of the S. pombe MSH3 homology, the motif is highly conserved among all known eukaryotic MSH3 and MSH6 homologs (Fig. 1B), in which it appears invariably very close to the N termini (Fig. 1C). We also have aligned the sequences immediately downstream from the PCNA-binding motifs. In the case of p21Cip1/WAF1, this region is involved in the interaction with the interdomain connector loop (IDCL) of PCNA (Gulbis et al. 1996). The known PCNA-binding proteins can be divided into two distinct groups, based on the amino acid sequence of this region: one with a preponderance of basic residues and the other with a high percentage of proline residues (Zhang et al. 1999). As shown in Figure 1B, MSH6 contains the proline-rich region and therefore belongs to the same group as uracil DNA glycosylase and DNA methyl transferase I (Gulbis et al. 1996). In contrast, with the exception of the Arabidopsis sequence, MSH3 does not contain prolines in this region.
Figure 1.
Consensus PCNA-binding motif. (A) Alignment of the conserved PCNA-binding motifs of hMSH6, hMSH3, p21Cip1/WAF1 (hCDN1), flap endonuclease FEN1 (hFEN1), DNA methyltransferase I (hMTDM), XPG endonuclease (hXPG), DNA ligase I (hDNL1), uracil DNA glycosylase (hUNG), and the small subunit of DNA polymerase-δ (hp66). (B) Evolutionary conservation of the putative PCNA-binding motifs of hMSH6 and hMSH3 homologs from human (Homo sapiens), mouse (Mus musculus), budding yeast (S. cerevisiae), fission yeast (Schizosaccharomyces pombe), and Arabidopsis thaliana. The residues important for binding are shown in boldface. Basic residues in the flanking sequences are in gray, and proline residues are underlined. The numbers in brackets denote the amino acid residues of the respective PCNA-binding motifs. (C) Schematic representation of hMSH6, hMSH6Δ77 (which lacks 77 N-terminal amino acid residues of hMSH6), and hMSH3, showing location of the PCNA-binding motifs, the proposed sites of interaction with hMSH2, as well as the positions of the mismatch-, ATP- and magnesium-binding sites. The numbers represent amino acid residues; numbers in brackets above hMSH6Δ77 refer to the corresponding amino acid residues in full-length hMSH6.
We wanted to see whether PCNA does indeed interact with MSH6 and MSH3 and whether this motif is essential for the interaction.
PCNA interacts with hMSH6 and hMSH3 in far-Western assays
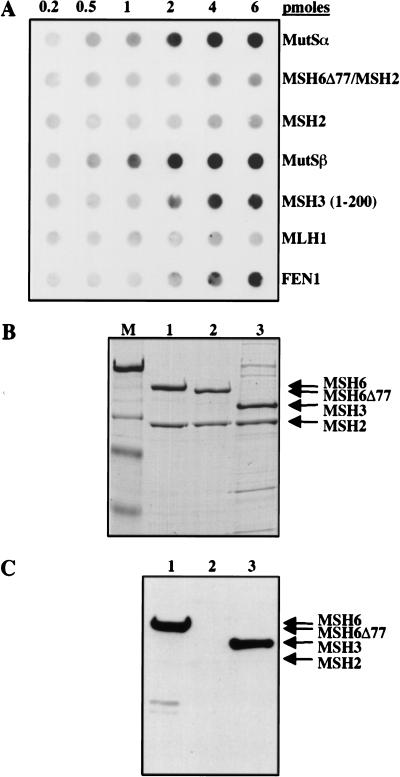
In the following experiments, we used human MutS and MutL homologs expressed in the Baculovirus system as described previously (Iaccarino et al. 1998; Räschle et al. 1999). The exception was an N-terminal fragment of hMSH3, which was overexpressed in E. coli (Marra et al. 1998). The proteins were spotted directly on the membrane, which then was blocked with skim milk, washed, allowed to hybridize with recombinant PCNA, washed again, and allowed to hybridize with the anti-PCNA antibody 19F4. As shown in Figure 2A, PCNA remained bound to hMutSα or hMutSβ, as well as to the N-terminal 200 amino acids of hMSH3, but no significant interaction was detected with hMSH2. The truncated variant of hMutSα, MSH6Δ77/MSH2, which lacks 77 N-terminal amino acid residues of hMSH6 and thus also the consensus PCNA interaction motif (Fig. 1C), also was unable to interact with PCNA in this assay. Similarly, no interaction was detected with hMLH1 (Fig. 2A) or with the hMLH1/hPMS2 heterodimer hMutLα (data not shown). Although this assay is only semiquantitative, the interaction of hMutSα or hMutSβ with PCNA appeared to be stronger that that of the sliding clamp with the flap endonuclease FEN1, shown to be mediated via the conserved motif QGRLDDFF near its C terminus (Warbrick et al. 1997).
Figure 2.
Far-Western analysis of PCNA interactions with MMR proteins and their variants. (A) Increasing amounts (top line) of the purified recombinant proteins listed on the right were spotted onto a nitrocellulose membrane, which then was hybridized with PCNA. The membrane subsequently was probed with the anti-PCNA monoclonal antibody 19F4 (see Materials and Methods). [MSH3 (1–200)] 200 N-terminal amino acid residues of hMSH3 expressed in E. coli; (FEN1) flap endonuclease used as a positive control. (B) hMutSα (lane 1), hMSH6Δ77/hMSH2 (lane 2), and hMutSβ (lane 3) subunits separated by SDS-PAGE and visualized by staining with Coomassie blue. (M) Molecular weight markers (from top to bottom: 200, 116, 97, and 66 kD). (C) The denatured proteins from B were transferred onto a nitrocellulose membrane, which was treated as in A above (see Materials and Methods). A and C are autoradiographs of the membranes, where the protein bands were visualized by ECL (see Materials and Methods).
As no significant interaction could be detected between PCNA and hMSH2 or hMSH6Δ77/hMSH2, we postulated that the observed interaction of PCNA with hMutSα and hMutSβ must be mediated by hMSH6 and hMSH3, respectively. However, we wanted to exclude the possibility that PCNA recognized conformational epitopes of the heterodimeric factors, rather than single peptidic motifs. We therefore conducted a second far-Western experiment, in which the constituent polypeptides of hMutSα, hMutSβ, and hMSH6Δ77/hMSH2 were separated before incubation with PCNA by denaturing polyacrylamide gel electrophoresis on SDS-PAGE (Fig. 2B). The proteins were electrotransferred onto a membrane, which subsequently was blocked with skim milk and then hybridized with PCNA. After extensive washing, the membrane was incubated with anti-PCNA antibodies, washed, and exposed to film as above. As shown in Figure 2C, PCNA could be seen to interact exclusively with the full-length hMSH3 and hMSH6 polypeptides. This experiment shows that the interaction between PCNA and the latter proteins is not mediated by conformational epitopes present either in the heterodimer or in its individual subunits, as the proteins in this assay are fully denatured. This finding also was supported by the fact that the N-terminal fragment of hMSH3 appeared to bind PCNA with high affinity in the absence of hMSH2 (Fig. 2A).
PCNA coimmunoprecipitates with hMutSα and hMutSβ
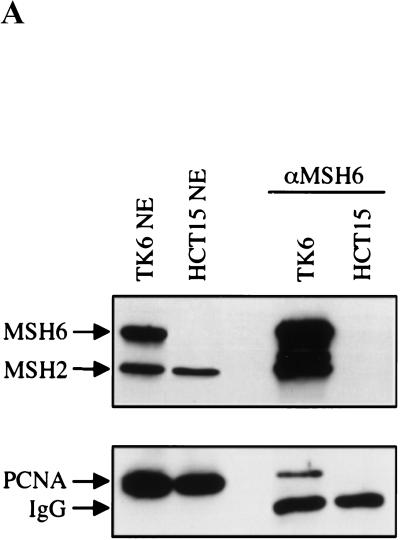
The interaction of hMSH6 and PCNA in vivo could be substantiated by coimmunoprecipitation experiments (Fig. 3A), in which the anti-hMSH6 monoclonal antibody 66H6, which recognizes the native polypeptide, was used to precipitate its cognate protein from human cell extracts. PCNA and hMSH2 were coimmunoprecipitated with hMSH6 from extracts of the MMR proficient TK6 cells, but not from extracts of HCT15 cells, which lack full-length hMSH6 (Papadopoulos et al. 1995). Unfortunately, the inverse experiment could not be conducted, as the anti-PCNA antibody available to us binds an epitope in the PCNA protein–protein interaction region and therefore does not function in coimmunoprecipitations.
Figure 3.
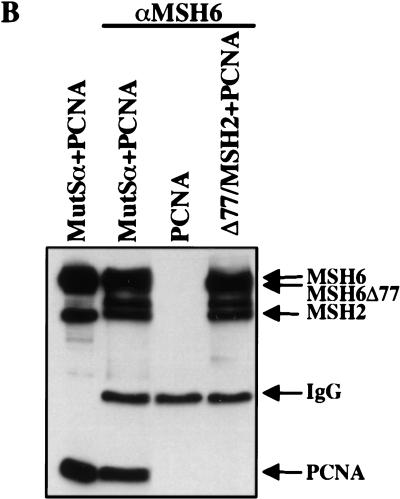
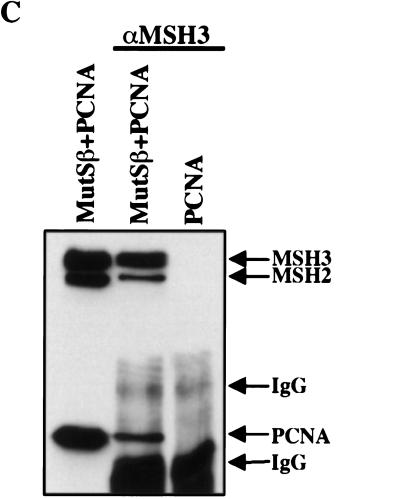
Coimmunoprecipitation of hMSH6 or hMSH3 with PCNA. (A) Two hundred micrograms of nuclear extracts of TK6 or HCT15 cells was incubated with the anti-hMSH6 monoclonal antibody 66H6, and the immunoprecipitates were loaded onto a denaturing SDS–polyacrylamide gel adjacent to lanes containing 20 μg of the untreated extracts of the same lines. The proteins then were electrotransferred onto a nitrocellulose membrane, which was hybridized with antibodies against hMSH6 and hMSH2 (top) or PCNA (bottom). PCNA can be seen to coimmunoprecipitate with hMSH6 in TK6 extracts (lane TK6). No PCNA signal was detectable in coimmunoprecipitates from HCT15 extracts (lane HCT15), which lack full-length hMSH6 (lane HCT15 NE). (IgG) Immunoglobulin light chain. (B) Two picomoles of recombinant hMutSα or hMSH6Δ77/hMSH2 was mixed with 6 pmoles of recombinant PCNA, and the immunoprecipitation was performed with the anti-hMSH6 monoclonal antibody as described in Materials and Methods. No PCNA was coimmunoprecipitated from a mixture with hMSH6Δ77/hMSH2 or when hMutSα was omitted (lane PCNA). (IgG) Immunoglobulin heavy chain. Left lane contains the input proteins (1 pmole hMutSα and 3 pmoles PCNA) loaded on the gel directly without immunoprecipitation. (C) Same as B except that hMutSβ was used in place of hMutSα, and the immunoprecipitations were conducted with a polyclonal anti-hMSH3 antiserum (see Materials and Methods). Left lane contains the reference proteins (1 pmole hMutSβ and 3 pmoles PCNA) loaded directly on the gel without immunoprecipitation. The figures are autoradiographs of the membranes, where the proteins were visualized by ECL (see Materials and Methods).
Because of the low amounts of hMSH3 in human cells (Genschel et al. 1998; Marra et al. 1998), the interaction of this latter polypeptide with PCNA contained in cell extracts also could not be shown. For this reason, we examined the interaction of the recombinant proteins in the coimmunoprecipitation assays. The polyclonal anti-hMSH3 antiserum coimmunoprecipitated hMSH3 with hMSH2 and PCNA (Fig. 3C). As anticipated, the anti-hMSH6 antibody 66H6 coimmunoprecipitated hMSH6 with hMSH2 and PCNA, but only hMSH2 could be co-immunoprecipitated from mixtures containing PCNA together with the truncated variant hMSH6Δ77/hMSH2 (Fig. 3B).
hMSH3 and hMSH6 colocalize with PCNA to replication foci
PCNA is an abundant protein that is distributed throughout the nucleus. However, during replication, PCNA forms foci of aggregated protein (Leonhardt et al. 2000) that have been shown to colocalize with newly replicated DNA labeled with 5-bromodeoxyuridine (BrdU). Immunofluorescence studies conducted with the MMR-proficient HeLa cells showed that in cells undergoing DNA replication, both hMSH6 and hMSH3 colocalize with PCNA to these foci. Figure 4A shows that the overlap of the PCNA and hMSH6 signals is essentially complete (yellow spots), even though there is some free excess hMSH6 (green) visible in the merged image. In the case of hMSH3, the overlap is complete, and no free protein is visible in the S phase nuclei (Fig. 4B). This is consistent with the significantly greater abundance of hMSH6 as compared with hMSH3 (Genschel et al. 1998; Marra et al. 1998). In a control experiment, we wanted to show that the hMSH6 and hMSH3 proteins did indeed colocalize with newly replicated DNA. To this end, we labeled the cells with BrdU shortly before fixation. As can be seen in Figure 4C,D, the BrdU signals completely overlap with those due to the two MMR proteins.
Figure 4.
Colocalization of hMSH6 or hMSH3 and PCNA to replication foci in HeLa cells in vivo. (A) Indirect immunofluorescence of HeLa nuclei stained with antibodies against hMSH6 (green) or PCNA (red). Superimposition of the red and green signals gives yellow color, which is indicative of colocalization of the two proteins. (B) As in A except that the cells were stained with a polyclonal anti-hMSH3 antibody (green). (C) As in A except that cells were labeled with BrdU for 1 h before fixation, and the nuclei then were stained with anti-BrdU antibodies (green). (D) As in C except that the nuclei were stained with anti-hMSH3 polyclonal antibody. Note that the MMR protein signals in A and B are green, whereas those in C and D are red.
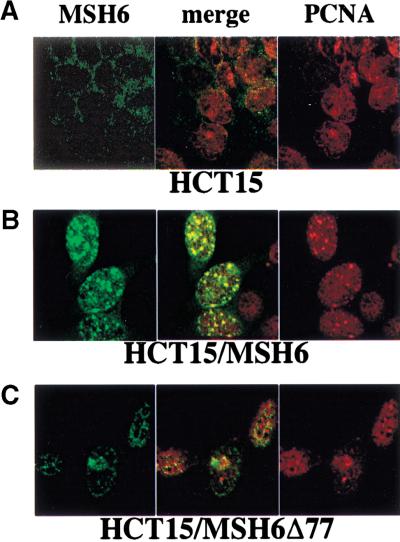
The N-terminal 77 amino acids of hMSH6 are required for localization to replication foci
The PCNA consensus binding motif of hMSH6 is located close to its N terminus (Fig. 1B,C), and we have shown above that a truncated polypeptide lacking the N-terminal 77 amino acids was no longer able to interact with PCNA, either alone or within the context of hMutSα in coimmunoprecipitation assays (Figs. 2 and 3). We wanted to see whether the same was also true in vivo. To answer this question, we made use of hMSH6-deficient HCT15 cells, either untransfected, or stably transfected with expression vectors carrying either full-length hMSH6 cDNA (Lettieri et al. 1999) or the truncated version hMSH6Δ77 (T. Lettieri et al., unpubl.). Indirect immunofluorescence experiments showed that whereas the signal due to hMSH6 was absent from the nuclei of untransfected HCT15 cells (Fig. 5A), because of truncating mutations in both alleles of the hMSH6 gene (Papadopoulos et al. 1995), in the HCT15/MSH6 transfectant, the stably expressed full-length hMSH6 colocalized with PCNA to the replication foci (Fig. 5B). In contrast, the truncated hMSH6 polypeptide in the HCT15/MSH6Δ77 clone was localized in the nuclei but failed to concentrate in replication foci during S phase (Fig. 5C, left), unlike PCNA, which formed distinct foci in the same nuclei (Fig. 5C, right). These results imply the N-terminal 77 amino acid residues of hMSH6 are responsible for targeting hMSH6 to the replication foci in S phase cells in vivo. It is tempting to suggest that the targeting is mediated by PCNA and that it involves its conserved interaction motif in the IDCL. However, we cannot formally exclude the possibility that the N-terminal region of hMSH6 contains other sequence motifs responsible for targeting the protein to replication foci. As the truncated polypeptide is defective in mismatch repair, it is conceivable that it fails to interact with other members of the MMR machinery and that this is the real reason for its staying out of the replication foci. This doubt could only be answered by expressing in HCT15 cells a mutant of hMSH6 that is deficient in MMR, but which can interact with PCNA. Unfortunately, such a mutant is not available at present. However, we believe, based on the study of interactions of PCNA with hMSH6 and hMutSα in vitro (see above), that the PCNA–hMSH6 interaction plays a key role in the localization of the latter protein to the replication foci.
Figure 5.
Colocalization of hMSH6 and PCNA to replication foci in HCT15 cells either untransfected (A) or stably transfected with a vector expressing hMSH6 (B) or the truncated variant hMSH6Δ77 (C). Indirect immunofluorescence of cell nuclei stained with antibodies against hMSH6 (green) or PCNA (red). Superimposition of the red and green signals gives yellow color, which is indicative of colocalization of the two proteins. See text for details.
Loss of hMSH6 N terminus attenuates mismatch repair efficiency
The experiments described above showed that the N terminus of hMSH6, which contains the consensus PCNA-binding motif, is necessary for interaction of the two proteins in vitro, as well as for targeting of hMSH6 to replication foci in vivo. We therefore wanted to know how the absence of this motif affected the efficiency of mismatch correction. In previous experiments, extracts of cells deficient in hMutSα could be complemented to mismatch repair proficiency by the addition of the mismatch binding factor, either purified from human cells (Drummond et al. 1995) or expressed in the Baculovirus system (Iaccarino et al. 1998). As shown in the Western blot in Figure 6A, extracts of HCT15 cells lack hMSH6 and, correspondingly, are deficient in mismatch correction (Fig. 6B). Addition of recombinant hMutSα restored the MMR proficiency of these extracts to near wild-type levels (cf. HeLa extract, Fig. 6B), but an equal amount of hMSH6Δ77/hMSH2 was substantially less effective, restoring the repair efficiency to only 20%. Similarly, extracts of the HCT15/MSH6 cells, in which the MMR defect was corrected by the stable expression of full-length hMSH6, were largely MMR proficient, whereas extracts of HCT15/MSH6Δ77 cells, expressing the truncated hMSH6 polypeptide, were MMR deficient (Fig. 6B). In the HCT15/MSH6Δ77 cells, the amount of the truncated hMSH6 polypeptide expressed from the stably transfected vector, as well as that of hMSH2, was noticeably lower than that present in MMR-proficient HeLa cells (Fig. 6A). It therefore could be argued that the MMR deficiency of the HCT15/MSH6Δ77 line might be due to the low level of the MSH6Δ77 polypeptide rather than to its inability to interact with PCNA. We cannot discount this possibility at present. However, expression of a similar amount of the full-length hMSH6 in HCT15/MSH6 cells (Fig. 6A) rescued the MMR-deficient phenotype of these cells, both in vivo and in vitro (Fig. 6B; Lettieri et al. 1999), and it is therefore likely that the repair defect in the HCT15/MSH6Δ77 cells is indeed due to the lack of interaction between the truncated hMSH6 and PCNA.
Figure 6.
Complementation of the MMR defect of HCT15 cells in vitro and in vivo. (A) Western blot of cytoplasmic extracts used in MMR assays shown in B. (Lanes 1,2) 50 μg HeLa and HCT15, respectively; (lane 3) HCT15 cell extract (50 μg) supplemented with 20 ng of recombinant hMutSα; (lane 4) HCT15 cell extract (50 μg) supplemented with 20 ng of recombinant hMSH6Δ77/hMSH2; (lane 5) extract of HCT15 cells stably transfected with hMSH6 cDNA (50 μg); (lane 6) extract of HCT15 cells stably transfected with hMSH6Δ77 cDNA (50 μg). (B) Mismatch repair efficiencies of extracts shown in A; lane number correspond with those in A. The light gray column indicates the repair efficiency of HCT15 extract supplemented with 200 ng of recombinant hMSH6Δ77/hMSH2.
In the above experiments, we complemented the deficient HCT15 extracts with amounts of recombinant proteins roughly similar to those found in extracts of MMR-proficient cells (Fig. 6A, cf., lanes HeLa and HCT15 + MutSα). Under these conditions, the hMSH6Δ77/hMSH2 hMutSα variant (20 ng) was unable to complement the MMR-deficient HCT15 extracts. However, this factor is proficient in mismatch binding, and the resulting protein–DNA complex is sensitive to ATP (P. Dufner and J. Jiricny, unpubl.), which suggests that both subunits are correctly folded and functional. We decided to test whether increasing the concentration of the hMSH6Δ77/hMSH2 factor altered the outcome of the experiment. As shown in Figure 6B, addition of a 10-fold higher amount (200 ng) of the truncated heterodimer to the repair assay restored the efficiency of MMR in the complemented extracts to wild-type levels, which suggested that the truncated factor is indeed functional, but that it fails to interact with the repair complex at low concentrations because of its lack of the N-terminal targeting sequence. However, at higher concentrations, the need for this targeting signal is no longer essential, as the protein can find its cognate partners without it. This is reminiscent of the PCNA interaction with FEN1, in which interaction of the two proteins at low concentration could be shown to be attenuated in the absence of the conserved interaction motif, but this effect could be overcome at higher FEN1 concentrations (Gomes and Burgers 2000).
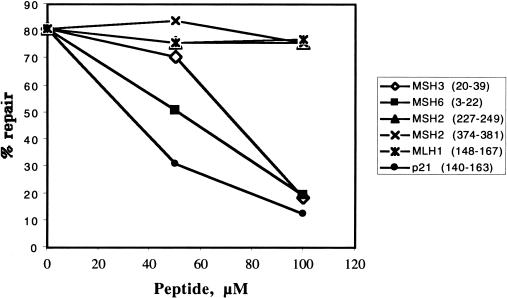
Peptides containing the consensus PCNA-binding motif of p21Cip1/WAF1, hMSH3, and hMSH6 inhibit MMR in vitro
Kunkel and colleagues showed that addition to the in vitro MMR assay of a 20-mer synthetic peptide KRRQTSMTDFYHSKRRLIFS, which contains the PCNA-binding motif of p21Cip1/WAF1, inhibited repair efficiency (Umar et al. 1996). To show that this consensus sequence was involved also in the binding of PCNA to hMSH6 and hMSH3, we challenged our MMR assays with the peptides RQSTLYSFFPKSPALSDANK and RQAVLSRFFQSTGSLKSTSS, containing the consensus PCNA-binding motifs of these two MMR proteins, respectively (residues thought to be essential for protein–protein interactions are set in boldface). As shown in Figure 7, both peptides reduced MMR efficiency, albeit somewhat less efficiently than the control p21Cip1/WAF1 peptide. As peptides with related sequences from hMSH2 and hMLH1, used as negative controls, failed to affect repair efficiency in this assay, our results imply that the putative consensus PCNA interaction motifs of hMSH6 and hMSH3 are indeed functional, and that the MMR defect associated with the hMSH6Δ77/hMSH2 factor is linked with the loss of this motif. The observed inhibitory effect of the hMSH3 and hMSH6 peptides was apparently specific, as repair efficiency could be largely restored by the addition of excess recombinant PCNA (data not shown).
Figure 7.
Inhibition of G/T mismatch repair with peptides carrying the consensus PCNA-binding sites of hMSH3 (amino acids 20–39), hMSH6 (amino acids 3–22), and p21Cip1/WAF1 (amino acids 140–163). Similar peptide sequences from hMSH2 (amino acids 227–249 and 373–392) and hMLH1 (amino acids 148–167) were used as controls (see Materials and Methods).
Discussion
In the present study, we have confirmed previous findings that implicated PCNA in MMR (Johnson et al. 1996; Umar et al. 1996; Eissenberg et al. 1997; Gu et al. 1998). However, contrary to the published results (Umar et al. 1996), we have found no biochemical evidence of interaction between PCNA and MLH1 or MSH2 (Fig. 2; data not shown). Instead, we could show that the PCNA sliding clamp directly interacts with the hMSH6 and hMSH3 subunits of the human mismatch binding factors hMutSα and hMutSβ via their very N termini, which carry the consensus PCNA-binding motifs Qxx[LI]xxFF (Fig. 1). As deletion of the N-terminal 77 amino acid residues of hMSH6, which contain this motif, abrogated the interaction of hMutSα with PCNA in coimmunoprecipitation assays as well as in mismatch repair in vitro, we conclude that this interaction is essential for MMR.
The requirement for PCNA as the processivity factor of polymerase-δ during the late, gap-filling step of MMR is clear. But why should it be necessary for PCNA to interact with the mismatch binding factor(s), during the early stages of the process (Umar et al. 1996; Gu et al. 1998)? The most likely answer to this question lies in its propensity to be loaded and to remain bound at strand interruptions (Jonsson and Hubscher 1997). As noted earlier, the eukaryotic MMR systems appear to lack endonuclease activities such as MutH, which might be capable of specifically incising the newly synthesized strand, and we therefore must assume that the repair process can initiate solely at pre-existing strand interruptions. In the lagging strand of newly replicated DNA, gaps or nicks between Okazaki fragments might represent suitable (and numerous) MMR initiation sites. In the leading strand, which is synthesized continuously, the only available point at which exonucleolytic degradation of the newly synthesized strand could conceivably commence is the 3′ terminus of the primer strand. However, the 3′ terminus of the leading strand is blocked by the extending DNA polymerase-δ, whereas the gaps or nicks between Okazaki fragments are processed rapidly by RNaseHI, FEN1, and DNA ligase I (Levin et al. 1997). Thus, if the MMR system is to make use of these initiation points, it first must gain access to them. We postulate that this access is mediated by the interaction of the MMR proteins with PCNA, which is known to be present at these DNA termini, both in the leading and in the lagging strand, during replication. In the former, it acts as the processivity factor for polymerase-δ, whereas in the latter, it is thought to guide first FEN1 (Tom et al. 2000), then DNA ligase I (Montecucco et al. 1998), to their respective substrates (Jonsson and Hubscher 1997). It is therefore conceivable that in the leading strand, mismatch binding will induce a conformational change in hMutSα, which will then translocate along the DNA until it reaches the DNA polymerase complex. By interacting with PCNA, hMutSα might displace the replicating enzyme, thus liberating the 3′ terminus of the primer strand and allowing the assembly of the repairosome. In the lagging strand, hMutSα might translocate along the DNA to reach the terminus of the mismatch-containing Okazaki fragment, where it could either prevent the binding of FEN1 or DNA ligase I to PCNA or, in the case in which these complexes already have formed, bring about their dissociation. The MMR repairosome then could assemble at such sites, such that the helicase(s) and exonucleases that are part of the MMR complex could initiate the degradation of the error-containing strand.
Although plausible, this hypothesis might appear to have one major flaw: if the replicative polymerase synthesizes ∼100 nucleotides per second, how does the MMR system catch up after the detection of a mismatch? One possibility concerns the ability of hMutSα to translocate along the DNA. All MutS homologs contain a highly conserved ATP-binding region near their respective C termini, and several studies have shown that the integrity of this site is essential for the biological function of the mismatch binding factors (Wu and Marinus 1994; Iaccarino et al. 1998, 2000; Studamire et al. 1998). Recent reports showed that, after mismatch recognition, the bound heterodimeric factor undergoes a conformational change, which transforms it to a sliding clamp that can leave the mismatch site and translocate along the DNA helix (Allen et al. 1997; Blackwell et al. 1998; Gradia et al. 1999; Iaccarino et al. 2000). It may be able to reach the replication fork in this manner. The second scenario is based on the existence of the replication foci (Hozak et al. 1994; Leonhardt et al. 2000). It seems probable that the replication complex does not travel along the DNA, but rather that the DNA is being translocated through matrix-bound replication factories (Hozak et al. 1993; Leonhardt et al. 2000). Our present evidence shows that hMSH3 and hMSH6, and thus most likely also the other members of the MMR machinery, colocalize with PCNA, as well as with many other replication-associated polypeptides such as RF-C, DNA methyltransferase I, FEN1, DNA ligase I, polymerases α, δ, and ɛ to these factories (see Kelman 1997; Montecucco et al. 1998 and references therein). It is possible that hMutSα is scanning the newly replicated DNA for mismatches while situated in a close geographical proximity to the replicating polymerases. Its ability to displace polymerase-δ from its complex with PCNA after mismatch detection might then be substantially facilitated. This physical proximity of the MMR proteins to the replication complexes would, in a similar fashion, facilitate the interaction between the mismatch binding factor and the PCNA–DNA ligase I complex. Clearly, because of its similar biochemical properties, an analogous role in the repair of IDLs can be ascribed to hMutSβ.
hMSH3 and hMSH6 join the long line of proteins capable of interacting with PCNA. Although this factor is homotrimeric and thus potentially is capable of interacting with three partners, this was predicted to be unlikely due to steric reasons. However, the crystal structure of bacteriophage RB69 sliding clamp–DNA polymerase complex (Shamoo and Steitz 1999; Hingorani and O'Donnell 2000) may have revealed how the structurally highly conserved polymerase processivity factors might interact with more than one protein. The RB69 polymerase interacts with its clamp through the last 10 or so C-terminal amino acids, which hook the polymerase to the clamp. The final five residues have a stable helical structure, but the adjoining five act as a flexible tether, which may enable the polymerase to move rapidly on or off the 3′ terminus of the primer without completely dissociating from the template DNA. Thus, extrapolating to the eukaryotic systems, it is plausible that the vacant interdomain connector loops (IDCLs) of PCNA, which are the sites of protein–protein interactions (Gulbis et al. 1996), remain accessible even while the sliding clamp is in a complex with another protein. Transient dissociation of the polymerase would permit access to another protein containing a PCNA interaction motif. The striking structural similarities between the C-terminal peptide of the RB69 polymerase and the C terminus of p21Cip1/WAF1 (Shamoo and Steitz 1999; Hingorani and O'Donnell 2000) suggest that both these proteins interact with their respective sliding clamps in a similar manner. The PCNA-binding motifs of all the proteins listed in Figure 1A are located close to the protein termini. Their high degree of sequence conservation implies that they will form short, flexible connector domains analogous to that of RB69 polymerase. The hypothesis that polypeptides not only compete for binding to PCNA ab initio, but that they may be able to displace other proteins from pre-existing complexes with the sliding clamp thus appears highly plausible. Should this be the case, however, the competition for PCNA would have to be regulated. One way of achieving this goal is through a pecking order of protein–PCNA interactions, in which proteins would be able to displace polypeptides with weaker PCNA-binding motifs, but not those with stronger ones. Moreover, highly abundant proteins might displace less abundant ones. This would explain why the p21Cip1/WAF1 protein, which has a high affinity PCNA-binding motif and which is transcriptionally induced by p53 after DNA damage, apparently can abrogate all transactions at the replication fork. The system might have an even greater flexibility if we consider that the affinity for PCNA and thus the position of a given protein in the pecking order might be modulated by interactions with other proteins, conformational changes or post-translational modifications (Chen et al. 1996; Warbrick et al. 1997; Scott et al. 2000). Reversible phosphorylation of Thr 145 or Ser 146 of p21Cip1/WAF1, for example, has been shown to modulate its PCNA-binding (Scott et al. 2000). Assuming that the interaction of PCNA and the mismatch repair machinery is indeed required for initiation of the repair process and thus also for strand discrimination as suggested above, then hMSH3 and hMSH6 would have to be higher in the pecking order than DNA polymerase-δ and DNA ligase I. Experiments aimed at testing this hypothesis are currently in progress in our laboratory.
While this article was under review, Clark et al. (2000) reported the existence of a direct interaction between PCNA and MSH3 and MSH6 from both S. cerevisiae and human. They could show that substitution of the conserved residues within the respective PCNA-binding motifs of these proteins with alanine abolished their interactions with the yeast PCNA. Their data thus provide further evidence in support of our prediction that the interactions of the human MMR proteins with PCNA indeed are mediated by the conserved N-terminal motif Qxx[LI]xx[FF].
Materials and methods
Antibodies and reagents
Dulbecco's modified Eagle medium (DMEM), RPMI-1640 medium, fetal bovine serum (FBS), and Geneticin were from GIBCO BRL Life Technology. Hygromycin B and complete protease inhibitor cocktail were from Boehringer Mannheim GmbH.
The rabbit anti-MSH6 polyclonal serum was described previously (Palombo et al. 1995). The mouse anti-hMSH6 mAbs 21F10 and 66H6 were purchased from Serotec. Polyclonal anti-MSH3 rabbit antiserum (NTH3) was generated at Eurogentec by immunization with a His 6–tagged N-terminal polypeptide of hMSH3 (amino acids 1–200), according to standard protocols.
The mouse monoclonal antibodies against MSH2 (Ab-2) and PMS2 (Ab-1) were from Calbiochem-Novabiochem, the anti-MLH1 antibody (13271A) was from PharMingen, and the antibodies against PCNA (19F4) and BrdU (5-bromo-2‘-deoxyuridine-5′-monophosphate) conjugated to fluorescein were from Boehringer Mannheim. The mouse monoclonal antibody against β-tubulin (N357), the HRP-linked anti-mouse and anti-rabbit antibodies, polyvinylidene difluoride (PVDF) transfer membrane (Hybond-P, 0.45 μm), and Western blotting detection reagent enhanced chemiluminescence (ECL) were purchased from Amersham Pharmacia Biotech. The mouse monoclonal antibody against PCNA (PC10), the anti-mouse and the anti-rabbit IgGs conjugated to FITC or tetramethylrhodamine (TRITC), and DAPI were from Sigma-Aldrich. Other chemicals were purchased from Sigma-Aldrich unless indicated otherwise.
Peptides
The following peptides were synthesized by Chiron Technologies Pty: hMSH6 (3–22), RQSTLYSFFPKSPALSDANK; hMSH3 (20–39), RQAVLSRFFQSTGSLKSTSS; hMSH2 (227–249), RKKAD FSTKDIYQDLNRLLKGKK; hMSH2 (373–392), RQTLQEDLLR RFPDLNRLAK; hMLH1 (148–167), TQITVEDLFYNIATRRKALK; hCDN1 (p21Cip1/WAF1, 140–163), RKRRQTSMTDFYHSKRR LIFSKRK. The peptides derived from hMSH2 (227–249) and hCDN1 (p21Cip1/WAF1) were kindly provided by Felix R. Althaus (Institute of Pharmacology and Toxicology, University of Zürich, Switzerland). All peptides were dissolved in milli-Q water at a concentration of 1 mg/mL and stored at −80°C.
Cell lines and extract preparation
The HeLa and HCT15 (MSH6−/−) cells were grown in DMEM, supplemented with 10% FBS. TK6 cells were maintained in RPMI-1640 medium, supplemented with 10% FBS. The HCT15 cell line and M13 mp2 phage DNA were kindly provided by Thomas Kunkel (National Institute of Environmental Health Sciences, Research Triangle Park, NC). Transfections of HCT15 cells with hMSH6 (Nicolaides et al. 1996) and hMSH6Δ77 (Palombo et al. 1995) cDNA expression vectors were performed as described (Lettieri et al. 1999). Selected clones HCT15/MSH6 and HCT15/MSH6Δ77 were maintained in RPMI-1640 medium, supplemented with 20% FBS and Geneticin (900 μg/mL) or hygromycin B (200 μg/mL), respectively.
Protein extracts were prepared from HeLa, TK6, HCT15, HCT15/MSH6, and HCT15/MSH6Δ77 cell lines by using 5 × 108 cells harvested in the logarithmic growth phase, as described (Marra et al. 1998). Briefly, after cell resuspension in ice-cold hypotonic buffer to yield a cell density of 1 × 108 cells/mL, the cells were allowed to swell in a glass Dounce homogenizer (Kontes) for 20 min on ice, and then lysed mechanically by applying four or more strokes with a tight (type B) pestle. When >80% cells were lysed, the nuclei were pelleted, and the supernatant was centrifuged at 12,000_g_ for 10 min at 2°C, frozen in liquid nitrogen in aliquots, and stored at −80°C. The nuclei (prepared as above) were suspended in extraction buffer containing 25 mM HEPES/KOH (pH 7.5), 155 mM KCl, 10% sucrose, 0.5 mM dithiothreitol (DTT), 1 μg/mL leupeptin, 1 mM PMSF, and the mixture was agitated on a rotating wheel for 1 h at 4°C. After pelleting the nuclear debris at 14,500_g_ for 20 min at 4°C, the supernatants were extensively dialyzed against buffer containing 25 mM HEPES/KOH (pH 7.6), 50 mM KCl, 0.1 mM EDTA, 10% sucrose, 0.5 mM DTT, 1 μg/mL leupeptin, 1 mM PMSF. The extracts then were clarified at 20,000_g_, 15 min at 4°C, frozen in liquid nitrogen in aliquots, and stored at −80°C. The protein concentrations were estimated by the Bradford method (BioRad) using BSA as standard.
Expression and purification of the recombinant proteins
The baculovirus vectors carrying cDNA inserts encoding hMSH6, hMSH2, and hMSH3 (Palombo et al. 1996) as well as hMSH6Δ77 proteins (Palombo et al. 1995) were used to infect cultures of Sf9 cells (GIBCO). Coinfection with hMSH2 and hMSH6, hMSH2 and hMSH6Δ77, or hMSH2 and hMSH3 viral vectors resulted, respectively, in the expression of hMutSα, hMSH6Δ77/MSH2, and hMutSβ in high yields. The procedure for protein recovery and purification was described previously (Palombo et al. 1995). The hMSH3 (1–200) polypeptide was expressed in E. coli and purified as described previously (Marra et al. 1998). Recombinant PCNA(a kind gift of Zophonias Jonsson and Ulrich Hübscher, Institute of Veterinary Biochemistry, University of Zürich, Switzerland) and FEN1 were kindly provided by Zophonias Jonsson and Ulrich Hübscher.
Western blots
Aliquots of cytoplasmic extracts (50 μg), nuclear extracts (20 μg), or recombinant proteins (5 pmoles) were loaded on 7.5% or 10% SDS–polyacrylamide gels. After electrophoresis, the proteins were transferred to Hybond-P PVDF membranes by electroblotting at 30 V, 4°C overnight. The membranes were blocked with 5% low-fat milk in TBST (100 mM Tris-HCl at pH 8, 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature. They then were incubated for 1 h at room temperature with the primary mouse monoclonal IgGs: anti-hMSH6 (21F10, 1.3 ng/mL); anti-hMSH2 (Ab-2, 0.1 μg/mL); anti-PCNA (19F4, 0.5 μg/mL); anti–β-tubulin (N357, 7.5 ng/mL) or polyclonal rabbit anti-MSH3 (NTH3, diluted 1:5000). After three washes with TBST containing 5% low-fat milk, the membranes were incubated for 1 h with HRP-conjugated anti-mouse IgG (1:5000) or anti-rabbit IgG (1:5000). The immunoreactive proteins were visualized by ECL.
Far-Western
Recombinant hMutSα, hMSH2/hMSH6Δ77, hMutSβ, hMSH2 (Iaccarino et al. 1998), and hMSH3 N-terminal 200 amino acids (Marra et al. 1998) and FEN1 (0.2–6 pmoles each) were blotted onto Protran nitrocellulose (Schleicher and Schuell GmbH), using a Bio-Rad manifold. Alternatively, 5 pmoles of hMutSα, hMSH2/hMSH6Δ77, and hMutSβ was separated on a 7.5% SDS–polyacrylamide gel and transferred onto a Hybond-P PVDF membrane by electroblotting at 30 V, 4°C overnight. The membranes were blocked with 5% low-fat milk in TBST for 1 h and then incubated with 3 μg/mL of recombinant PCNA at 4°C overnight. After extensive washing, the membranes were incubated for 1 h at room temperature with anti-PCNA mAb (19F4), followed by secondary HRP-conjugated anti-mouse IgG. The antibodies and PCNA were diluted in TBST containing 5% low-fat milk. The signals were visualized using the ECL system.
Immunoprecipitations
All the steps were performed at 4°C. Monoclonal antibodies against MSH6 (66H6) or PCNA (PC10; Sigma) were coupled to Pan Mouse IgG Dynabeads (Dynal), and affinity-purified polyclonal antibody against MSH3 (NTH3) was coupled to Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology) in 200 μL PBS containing 0.1% BSA for 1 h with rotation. Beads were then washed extensively with PBS/BSA buffer and incubated with 6 pmoles of PCNA mixed with either 2 pmoles of hMutSα, hMSH2/hMSH6Δ77, or MutSβ complexes in 80 μL of binding buffer (20 mM Tris-HCl at pH 7.5, 40 mM KCl, 5 mM MgCl2, 1 mM glutathione, 0.05% BSA), 1 × complete protease inhibitor cocktail, 1 mM PMSF, 10% sucrose for 2 h with rotation. After incubation, the beads were washed with 4 × 160 μL of binding buffer containing 150 mM NaCl and 0.1% Tween 20, resuspended in 30 μL of loading buffer (0.2 M DTT, 2% SDS, 1% bromophenol blue, 20% glycerol), and denatured for 5 min at 100°C, and 15 μL was loaded on 10% SDS-PAGE. The absence of nonspecific binding was shown by mixing PCNA with the beads precoupled with anti-MSH6 antibody. Alternatively, 200 μg of TK6 or HCT15 nuclear extracts was incubated with 30 μL of beads precoupled with anti-MSH6 antibody in 80 μL of binding/washing buffer (50 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.05% BSA, 0.1% Triton X-100), 1 × complete protease inhibitor cocktail, 2 mM PMSF, 10% glycerol, for 2 h with rotation. Subsequently, the beads were washed with 4 × 160 μL of the same buffer, resuspended in 30 μL of loading buffer, and denatured for 5 min at 100°C, and 15 μL was loaded on 10% SDS-PAGE.
After gel electrophoresis, Western blotting was performed as described above.
Indirect immunofluorescence
Subconfluent HeLa, HCT15, HCT15/MSH6, or HCT15/MSH6Δ77 cells grown on coverslips were rinsed with PBS at pH 7.4 and fixed in ice-cold methanol for 10 min at −20°C. After 30 min incubation in 10% FCS to block nonspecific protein binding, the fixed cells were incubated with the following primary antibodies: affinity-purified rabbit anti-MSH6 pAb (1:5), affinity-purified rabbit anti-MSH3 pAb (1:5), or mouse anti-PCNA mAb (PC10, 1:500) diluted in PBS containing 3% low-fat milk at 37°C for 1 h or at 4°C overnight. After washing with PBS containing 3% low-fat milk, the fixed cells were incubated with FITC-conjugated anti-rabbit IgG antibody (1:750) or TRITC-conjugated anti-mouse IgG antibody (1:50) diluted in PBS containing 3% low-fat milk, at 37°C for 1 h. After washing, the fixed cells were stained with DAPI at 0.1 μg/mL to confirm nuclear localization and washed again, and the coverslips were mounted on slides by using MOWIOL 4–88 (Calbiochem).
To simultaneously detect proteins and sites of DNA synthesis, we grew the cells on coverslips in 10 μM BrdU for 60 min immediately before methanol fixation. After sequential incubation with the MSH6 or MSH3 antibodies and anti-rabbit TRITC-conjugated secondary antibody (diluted 1:40), antigen–antibody complexes were fixed by immersion in ice-cold methanol for 10 min at −20°C. Subsequently, the fixed cells were treated with 2M HCl at 37°C for 60 min to denature the DNA and then neutralized with 0.1 M borate buffer (pH 8.5) with three changes at room temperature. In addition, the fixed cells were washed with PBS containing 3% low-fat milk and subsequently incubated for 60 min at 37°C with mouse monoclonal anti-BrdU fluorescein antibody diluted 1:5 in PBS containing 3% low-fat milk. After washing with PBS, the coverslips were mounted on slides by using MOWIOL 4–88 (Calbiochem). The HCT15 (MSH6−/−) cell line was used as control for nonspecific binding of MSH6 antibodies. Controls of HeLa cells with primary antibodies omitted were also included. When photographed under the same conditions as the samples, the controls did not show any fluorescence above background (see also Fig. 5A).
Fluorescence microscopy was performed using a computer-interfaced, laser-scanning Leica DM IRB E microscope (Leica Mikroscopie) equipped with Confocal Laser Scanning System TCSSP (Leica Lasertechnik), by using HCxPLAPO 100x/1.4 oil immersion objective. Immunolabeled slides (n = 3–4 representative fields per slide) were sectioned optically at 0.5-μm intervals through the cell monolayer to obtain the appropriate focal depth. A series of optical sections through the cells were collected and images assembled using Adobe PhotoShop 5.5 software.
In vitro mismatch repair assays
The efficiency of the cell extracts in repairing DNA mismatches was tested as described previously (Thomas et al. 1991). Briefly, a 13mp2 DNA heteroduplex, containing a G/T mispair in the coding sequence of the lacZ α complementation gene, was obtained by hybridizing a single-strand viral (+) DNA with the (−) strand of replicative form DNA as described (Thomas et al. 1991). One fmole of the heteroduplex was used in a repair reaction (25 μL) with 30 mM HEPES (pH 7.8), 7 mM MgCl2, 4 mM ATP, 200 μM each CTP, GTP, and UTP, 100 μM each dATP, dGTP, and dCTP, 40 mM creatine phosphate, 100 fmoles of creatine phosphokinase, 15 mM sodium phosphate (pH 7.5), 50 μg of extracted protein. In complementation studies, purified recombinant proteins were added to the extracts: 20 ng or 200 ng of wild-type or mutant hMutSα factors or 100–400 ng of PCNA. In peptide-inhibition studies, 50-μM or 100-μM concentrations of the polypeptide were used. The incubation was performed at 37°C for 20 min. The repair was directed to the (−) strand of M13mp2 by the presence of a nick. The DNA heteroduplex then was purified and introduced by electroporation into E. coli NR9162 (mutS strain) and plated on minimal medium in a soft agar layer containing 0.5 mL of a log phase culture of CSH50 (the α-complementation strain), 0.5 mg of isopropyl-β-D-thiogalactopyranoside, 2 mg of 5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside. After incubating for 20 h at 37°C, the plaques were assigned to one of the following phenotypes: blue, colorless, or mixed. If no repair occurred, mixed plaques were observed containing both blue and colorless progeny. Repair of the substrate in a cell extract reduced the percentage of mixed plaques and increased the percentage of pure-color plaques.
Acknowledgments
The authors thank Claudia Perrera, Patrick Dufner, Ingram Iaccarino, and Primo Schär for many helpful discussions, Thomas Bächi for fluorescence microscopy, and Christine Hemmerle for excellent technical assistance. We are also grateful to Ulrich Hübscher for critical reading of the manuscript and for the gift of PCNA and FEN1, and to Felix R. Althaus for providing the synthetic MSH2 and p21 peptides. The generous financial support of the Swiss National Science Foundation is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jiricny@imr.unizh.ch; FAX 41-1-634-8904.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.191201.
References
- Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F, Marinus MG. The great GATC: DNA methylation in E. coli. Trends Genet. 1989;5:139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Martik D, Bjornson KP, Bjornson ES, Modrich P. Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J Biol Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- Chen U, Chen S, Saha P, Dutta A. p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc Natl Acad Sci. 1996;93:11597–11602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang WH, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2–hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- Gu L, Hong Y, McCulloch S, Watanabe H, Li GM. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Hingorani MM, O'Donnell M. Sliding clamps: A (tail)ored fit. Curr Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- Holmes JJ, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Hozak P, Jackson DA, Cook PR. Replication factories and nuclear bodies: The ultrastructural characterization of replication sites during the cell cycle. J Cell Sci. 1994;107:2191–2202. doi: 10.1242/jcs.107.8.2191. [DOI] [PubMed] [Google Scholar]
- Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino I, Marra G, Dufner P, Jiricny J. Mutation of the magnesium binding site of hMSH6 disables the hMutS-α sliding clamp from translocating along DNA. J Biol Chem. 2000;275:2080–2086. doi: 10.1074/jbc.275.3.2080. [DOI] [PubMed] [Google Scholar]
- Jiricny J. Mediating mismatch repair. Nat Genet. 2000;24:6–8. doi: 10.1038/71698. [DOI] [PubMed] [Google Scholar]
- Jiricny J, Nyström-Lahti M. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157–161. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Jiricny J, Su SS, Wood SG, Modrich P. Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res. 1988;16:7843–7853. doi: 10.1093/nar/16.16.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kovvali GK, Guzder SN, Amin NS, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Hubscher U. Proliferating cell nuclear antigen: More than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- Kelman Z. PCNA: Structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- Kolodner RD. Mismatch repair: Mechanisms and relationship to cancer susceptibility. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Lacks SA, Dunn JJ, Greenberg B. Identification of base mismatches recognized by the heteroduplex-DNArepair system of Streptococcus pneumoniae. Cell. 1982;31:327–336. doi: 10.1016/0092-8674(82)90126-x. [DOI] [PubMed] [Google Scholar]
- Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettieri T, Marra G, Aquilina G, Bignami M, Crompton NE, Palombo F, Jiricny J. Effect of hMSH6 cDNA expression on the phenotype of mismatch repair-deficient colon cancer cell line HCT15. Carcinogenesis. 1999;20:373–382. doi: 10.1093/carcin/20.3.373. [DOI] [PubMed] [Google Scholar]
- Levin DS, Bai W, Yao N, O'Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: Implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Shivji MK, Chen C, Kolodner R, Wood RD, Dutta A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem. 1998;273:1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989;264:6597–6600. [PubMed] [Google Scholar]
- ————— Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: Identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Robetorye RS, Pereira-Smith OM, Smith JR. The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem. 1995;270:17060–17063. doi: 10.1074/jbc.270.29.17060. [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Palombo F, Kinzler KW, Vogelstein B, Jiricny J. Molecular cloning of the N-terminus of GTBP. Genomics. 1996;31:395–397. doi: 10.1006/geno.1996.0067. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D'Arrigo A, Truong O, Hsuan JJ, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D'Arrigo A, Markowitz S, Willson JK, Kinzler KW, et al. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- Parker BO, Marinus MG. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Marra G, Nyström-Lahti M, Schar P, Jiricny J. Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Warbrick E, Fantes PA, MacNeill SA. Essential interaction between the fission yeast DNA polymerase delta subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21(Cip1)-like PCNA binding motif. EMBO J. 2000;19:1108–1118. doi: 10.1093/emboj/19.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MT, Morrice N, Ball KL. Reversible phosphorylation at the C-terminal regulatory domain of p21(Waf1/Cip1) modulates proliferating cell nuclear antigen binding. J Biol Chem. 2000;275:11529–11537. doi: 10.1074/jbc.275.15.11529. [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA. Building a replisome from interacting pieces: Sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol Cell Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SS, Lahue RS, Au KG, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro J. Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom S, Henricksen LA, Bambara RA. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J Biol Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: A potential mechanism to co-ordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- Wu TH, Marinus MG. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J Biol Chem. 1999;274:26647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]