PIAS Proteins Modulate Transcription Factors by Functioning as SUMO-1 Ligases (original) (raw)
Abstract
PIAS (protein inhibitor of activated STAT) proteins interact with and modulate the activities of various transcription factors. In this work, we demonstrate that PIAS proteins xα, xβ, 1, and 3 interact with the small ubiquitin-related modifier SUMO-1 and its E2 conjugase, Ubc9, and that PIAS proteins themselves are covalently modified by SUMO-1 (sumoylated). PIAS proteins also tether other sumoylated proteins in a noncovalent fashion. Furthermore, recombinant PIASxα enhances Ubc9-mediated sumoylation of the androgen receptor and c-Jun in vitro. Importantly, PIAS proteins differ in their abilities to promote sumoylation in intact cells. The ability to stimulate protein sumoylation and the interaction with sumoylated proteins are dependent on the conserved PIAS RING finger-like domain. These functions are linked to the activity of PIASxα on androgen receptor-dependent transcription. Collectively, our results imply that PIAS proteins function as SUMO-1-tethering proteins and zinc finger-dependent E3 SUMO protein ligases, and these properties are likely to explain their ability to modulate the activities of various transcription factors.
Members of the recently identified PIAS (protein inhibitor of activated STAT) protein family have been found to interact with several distinct nuclear proteins. PIAS1 and PIAS3 bind to STAT1 and STAT3, respectively, and inhibit their action (4, 28). PIASxα/ARIP3 (androgen receptor [AR]-interacting protein 3) was first characterized as an AR-interacting protein, and it modulates the transcriptional activity of the receptor (34). Other PIAS proteins have recently been demonstrated to function as coregulators for AR and other steroid receptors (25, 57). PIASxβ/Miz1 (Msx-interacting zinc finger) associates with a homeodomain-containing Msx2 protein (61), and GBP (Gu/RNA helicase II-binding protein), which is nearly identical to PIAS1, interacts with Gu/RNA II-helicase (59). More recently, PIAS proteins have been identified in many yeast two-hybrid screens, including PIASxα/ARIP3 from its interaction with mouse disabled 2 (mDab2) (3) and DJ-1 protein (54), PIAS1 from its interaction with p53 (9), and PIAS3 from its interaction with high-mobility-group protein HMGI-C (63) and zinc finger protein Gfi-1 (43). In addition to PIASx (PIASxα/ARIP3 and PIASxβ/Miz1), SUMO-1 (small ubiquitin-related modifier 1) was identified as a p73α-interacting protein by Minty et al. (32), who suggested that PIASx was isolated via the interaction with Smt3p (yeast SUMO) covalently linked to p73α. We have also detected SUMO-1 as a major PIASxα/ARIP3-interacting protein in yeast (unpublished results).
Members of the SUMO protein family, also known as Sentrin, GMP1, PIC1, and Ubl1 (31, 37, 62), are present in protozoa, metazoa, plants, and fungi. SUMO proteins from metazoa can be divided into two families: the SUMO-1 family and the SUMO-2 and -3 family (31, 37, 62). SUMO-2 and SUMO-3 are very similar at the amino acid level (97% identity for the human proteins), but they are only ∼50% identical to SUMO-1. SUMO-1 is only 18% identical to ubiquitin, but it has a ubiquitin fold common to ubiquitin-like proteins (2). The SUMO modification (sumoylation) pathway is mechanistically similar to that of ubiquitination, but the enzymes of the two processes are distinct. The genes encoding the enzymes of the SUMO modification process are essential in yeast, and the conjugation machinery is relatively well conserved (18, 27, 49). SUMO is activated by the specific activity of E1, which is a heterodimer comprising SAE1/Aos1 and SAE2/Uba2 proteins (6, 21). Subsequently, SUMO is transferred to E2-conjugating enzyme Ubc9 (7, 10, 18, 48). There are several E2 ubiquitin ligases, but only a single SUMO E2 has been identified thus far. In ubiquitination, the E3 ubiquitin protein ligases promote the transfer of ubiquitin from the E2 enzyme to the ɛ-amino group of the target lysine residue in the substrate. The E3 ubiquitin protein ligases are largely responsible for the substrate specificity of ubiquitination (60). Two distinct classes of ubiquitin ligases have been established: HECT (homologous to E6-AP carboxyl terminus) domain E3s and RING finger E3s (16). The HECT domain E3s form thiol-ester intermediates with ubiquitin as a part of the process, whereas the RING finger E3s are thought to facilitate direct transfer of ubiquitin from E2 to the substrate, probably without formation of thiol-ester intermediates (reference 29 and references therein).
The most conserved part of a PIAS protein is its central region, which is likely to form a zinc-binding domain termed the Miz zinc finger (61). A similar domain is found in many proteins from invertebrates, such as the Drosophila melanogaster PIAS homologue Zimp (zinc finger containing, Miz1, PIAS3-like), a predicted Caenorhabditis elegans protein, and Saccharomyces cerevisiae septin-interacting protein Nfi1p, as well as putative proteins from Schizosaccharomyces pombe, Vicia faba, and Arabidopsis thaliana.
Many interaction partners of PIAS or PIAS-like proteins, such as AR, p53, septins, and the DJ-1 protein (12, 19, 42, 45, 54), are modified by SUMO-1. Because of the ability of PIASxα/ARIP3 to interact with both sumoylated proteins and SUMO-1 (32), we examined the role of PIAS proteins in protein sumoylation. We demonstrate here that, in addition to associating with SUMO-1 and Ubc9, PIASxα/ARIP3 itself is sumoylated even though it does not contain recognizable consensus sites for SUMO attachment. More importantly, PIASxα/ARIP3 tethers other sumoylated proteins and enhances sumoylation of AR and c-Jun. We also show that PIAS proteins differ in their abilities to promote sumoylation in intact cells. The RING finger-like structure of PIASxα/ARIP3 is mandatory for all these functions. Likewise, the effects of PIASxα/ARIP3 and PIAS1 on AR-dependent transcription are dependent on their RING-like domain. Taken together, PIASxα/ARIP3 and other PIAS proteins not only interact with SUMO-1 and other sumoylated proteins but also function as E3-type SUMO protein ligases in a fashion that resembles the action of the RING-type E3 ubiquitin protein ligases.
MATERIALS AND METHODS
Materials.
Mouse monoclonal anti-FLAG antibody M2 was purchased from Sigma, and mouse monoclonal anti-GMP-1 was from Zymed. Rabbit polyclonal anti-AR antibody K333 and the anti-ARIP3 antibody have been described (24, 34). The rabbit polyclonal antibody against green fluorescent protein (GFP) was purchased from Santa Cruz Biotechnology. The anti-GRIP1 monoclonal antibody was a kind gift from M. Brown. Horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) and anti-mouse IgG were from Zymed. Rhodamine red X-conjugated goat anti-mouse IgG was from Jackson ImmunoResearch. The mouse monoclonal antibody against the five-His sequence was purchased from Qiagen. Testosterone was from Makor Chemicals.
Plasmids and their construction.
pFLAG-ARIP3, pFLAG-ARIP3Δ102-207, pFLAG-ARIP3Δ347-418, pFLAG-ARIP3Δ346-475, and pFLAG-ARIP3Δ467-547 were constructed by PCR as described previously (26). pFLAG-ARIP3Δ467-487 was also constructed by PCR; the ARIP3 (1-466) coding sequence was amplified by using a 5′ primer containing a _Hin_dIII site and a 3′ primer containing a _Kpn_I site. The ARIP3 (488-572) coding sequence was amplified by using 5′ and 3′ primers both containing _Kpn_I sites. PCR fragments were first ligated into pCMV-Tag2A (Stratagene) and then the ARIP3Δ467-487 coding sequence was transferred into pFLAG-CMV2 with _Eco_RI. In pFLAG-ARIP3(W383A), the Trp 383 codon was mutated to an Ala codon by using a site-directed mutagenesis system according to the manufacturer's instructions (Stratagene). pFLAG-PIAS3 and pFLAG-PIAS1 were gifts from K. Shuai, and Miz1 cDNA was a gift from R. Maxon. Full-length pFLAG-Miz1 was constructed as previously described (25). pFLAG-PIAS1Δ310-407 was constructed by amplifying the coding sequence for the mPIAS1 C terminus (amino acids 408 to 651) by PCR using primers containing _Xba_I and _Hin_dIII sites. The PCR fragment was ligated upstream of the _Xba_I site in the mPIAS1 coding sequence. pFLAG-PIAS1(W372A) was generated by using a site-directed mutagenesis system (Stratagene). The construction of pSG5-rAR, pcDNA-Flag-hAR, and pcDNA-Flag-hAR-K386R/K520R [AR(K→R)] have been described (39, 42). The ARIP3, ARIP3Δ347-418, ARIP3Δ467-487, Miz1, PIAS1, and PIAS3 coding sequences were cloned into pCMV-Tag2 for translation in vitro. GFP-ARIP3 (341-418) and GFP-ARIP3 (341-490) were generated by amplifying codons for amino acids 341 to 418 and 341 to 490 by PCR using primers containing _Eco_RI and _Bam_HI sites. PCR products were transferred into pEGFP-C1 (Clontech) digested with the same enzymes. Glutathione _S_-transferase (GST)-ARIP3 (pGEX4T3-ARIP3) and Miz1 (pGEX5X1-Miz1) have been described (25). pGEX6P1-ARIP3Δ347-418 and pGEX6P1-ARIP3Δ467-487 were generated by digesting coding sequences for ARIP3 with the deletions from pVP16-ARIP3Δ347-418 and pVP16-ARIP3Δ467-487, respectively, with _Eco_RI and ligating them into _Eco_RI-digested pGEX-6P1 vector. pGEX4T3-ARIP3(W383A) was cloned by transferring the ARIP3(W383A) coding sequence from pFLAG-ARIP3(W383A) to the pGEX-4T3 vector with _Eco_RI. pGEX5X1-SUMO-1 and pGEX5X1-Ubc9 have been described (42). GST-SUMOGG-1, corresponding to a mature SUMO-1 that is missing the last four C-terminal amino acids of the precursor (6), was constructed by PCR. pGEX-SAE1 and pGEX-SAE2 were gifts from R Hay. pSG5-His-SUMO-1 was provided by A. Dejean, and pSG5-GRIP1 was from M. Stallcup.
Cell culture and transfections.
COS-1 cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle medium (DMEM) containing penicillin (25 U/ml), streptomycin (25 U/ml), and 10% (vol/vol) fetal bovine serum (FBS). HeLa (American Type Culture Collection) cells were maintained in DMEM containing penicillin, streptomycin, 10% FBS, and nonessential amino acids. For immunoprecipitation, 3 × 105 COS-1 cells were seeded on 6-cm-diameter dishes 24 h before transfection. Cells were transfected by the FuGene transfection method (Roche Molecular Biochemicals); the total concentration of DNA was 2.5 μg. Forty-eight hours after transfection cells were collected for immunoprecipitation. When testosterone-activated AR was studied, cells received a fresh medium containing 10% charcoal-stripped FBS 4 h before transfection and testosterone (100 nM) was added 24 h after transfection. For reporter gene assays, COS-1 or HeLa cells were seeded onto 12-well plates and transfected 24 h later by the FuGene reagent. In brief, each well received 200 ng of luciferase reporter pARE2TATA-LUC (35), 20 ng of pCMVβ (Clontech), 5 ng of AR expression plasmid, and various amounts of PIAS expression vectors as specified in the figure legends. The total amount of DNA was balanced by adding empty pFLAG-CMV2 when appropriate. Four hours before transfection, the medium was changed to one containing 10% charcoal-stripped FBS. Twenty hours after transfection, the cells received fresh medium containing 2% charcoal-stripped FBS with or without testosterone (100 nM). Forty-eight hours after transfection, the cells were harvested and lysed in reporter lysis buffer (Promega), and the cleared supernatants were used for luciferase measurements with reagents from Promega and a Luminoskan Ascent reader (ThermoLabsystems) and for β-galactosidase assays as described previously (42).
Immunoprecipitation and immunoblotting.
COS-1 cells were collected in phosphate-buffered saline containing 20 mM _N_-ethylmaleimide (NEM), and cell extracts were prepared in modified radioimmunoprecipitation assay 1 (RIPA-1) or RIPA-2 buffer (RIPA-1: 50 mM Tris-HCl [pH 7.8], 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 1% Nonidet P-40, 0.75% sodium deoxycholate, 1 mM dithiothreitol, 1:200-diluted protease inhibitor cocktail Σ, and 10 mM NEM; RIPA-2: 50 mM Tris-HCl [pH 7.8], 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 0.5% Nonidet P-40, 0.3% Triton X-100, 1 mM dithiothreitol, 1:200-diluted protease inhibitor cocktail, and 10 mM NEM). Immunoprecipitation with a mouse monoclonal anti-FLAG antibody was performed as described previously (35). Bound proteins were released in 2× sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred onto a Hybond enhanced chemiluminescence nitrocellulose membrane, and visualized by using the enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Purification of GST proteins and GST pull-down assays.
GST fusion proteins were produced in Escherichia coli BL21-CodonPlus bacteria (Stratagene) and purified with glutathione-Sepharose 4B (Amersham Pharmacia Biotech) as previously described (23). Lysis buffer containing 50 mM Tris-HCl (pH 7.8), 150 mM KCl, 0.1% Nonidet P-40, 0.1% Triton X-100, 0.5 mM EDTA, 10% glycerol, 5 mM MgCl2, and a 1:200-diluted protease inhibitor cocktail was used. ARIP3, ARIP3Δ347-418, ARIP3Δ467-487, PIAS3, PIAS1, and Miz1 were translated in vitro using the TNT-coupled transcription-translation system (Promega) in the presence of [35S]methionine. Protein-protein affinity chromatography with purified GST fusion proteins bound to glutathione-Sepharose and 10 μl of [35S]methionine-labeled in vitro-translated protein was carried out at 4°C for 2 h in a binding buffer containing 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 50 μM ZnCl2, 0.4% Nonidet P-40, 0.1% Triton X-100, and a 1:200-diluted protease inhibitor cocktail in a total volume of 500 μl. The resin was washed four times with 1 ml of binding buffer. Bound proteins were released by boiling the resin in SDS-PAGE sample buffer. After electrophoresis, the gels were fixed in methanol (45%)-acetic acid (10%), treated with Amplify (Amersham Pharmacia Biotech), and dried, and radioactive proteins were visualized by fluorography (40).
SUMO-1 conjugation in vitro.
GST-SUMOGG-1, GST-Ubc9, GST-SAE1, GST-SAE2, GST-ARIP3, GST-ARIP3Δ347-418, GST-ARIP3(W383A), and GST-ARIP3Δ467-487 were produced as described above. Purified proteins were eluted in a buffer containing 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 10% glycerol, and 20 mM glutathione. The c-Jun-His6 protein was purified as previously described (23). In vitro translations were performed by using the TNT-coupled transcription-translation system in the presence of [35S]methionine. For the conjugation assay, 1 μl of the in vitro translation product or 1 μg of His-tagged protein was incubated with 2 μl of GST-SUMOGG-1 (2 μg), 2 μl of GST-Ubc9 (2 μg), and 2.5 μl of the mixture of GST-SAE1 and GST-SAE2 (0.2 μg) at 30°C for 1 h in the presence of 1 mM dithiothreitol, 4 mM MgCl2, and 2 mM ATP. The amount of GST-PIAS proteins used in reactions was 0.2 μg. The reaction was terminated by adding 15 μl of 2× SDS sample buffer. The samples were heated at 95°C for 5 min, resolved by SDS-PAGE, and visualized by fluorography or immunoblotting.
Immunofluorescence.
COS-1 cells seeded on glass coverslips on six-well plates were transfected by using FuGene reagent with 0.75 μg of an expression plasmid encoding FLAG-tagged PIAS proteins and 0.75 μg of a plasmid encoding GFP-SUMO-1. Cells were fixed in 4% (wt/vol) paraformaldehyde in phosphate-buffered saline and permeabilized with Triton X-100, and PIAS proteins were detected with an anti-FLAG M2 monoclonal antibody (1:50 dilution; Sigma) and a rhodamine red X-conjugated goat anti-mouse antibody (1:200 dilution; Jackson ImmunoResearch Laboratories). Confocal imaging was performed by using a Bio-Rad MRC-1024 confocal laser system connected to a Zeiss Axiovert 135 M microscope with a 63×, 1.4-numerical-aperture oil immersion objective (33).
RESULTS
PIAS proteins interact with SUMO-1 and Ubc9 in vitro.
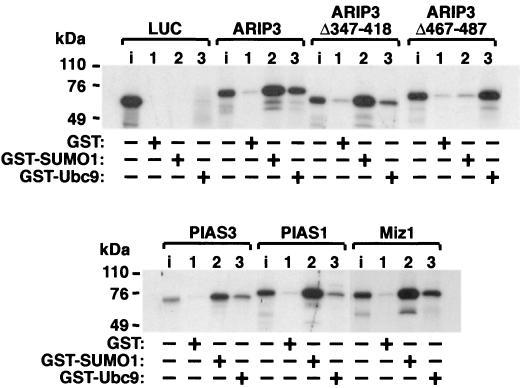
The ability of PIASxα/ARIP3 to interact with SUMO-1 and SUMO-1-conjugating enzyme Ubc9 was studied by GST pull-down assays. ARIP3 and two ARIP3 deletion mutants were translated in vitro in the presence of [35S]methionine, and the proteins were incubated with GST, GST-SUMO-1, or GST-Ubc9 bound to glutathione-Sepharose. As shown in Fig. 1, ARIP3 interacted efficiently with both SUMO-1 and Ubc9, whereas control protein luciferase did not bind to GST-SUMO-1 or GST-Ubc9. ARIP3 failed to bind GST alone. ARIP3Δ347-418, which lacks the putative zinc-binding structure containing the conserved cysteines and a histidine, bound to SUMO-1 as avidly as wild-type ARIP3, whereas the interaction with GST-Ubc9 was considerably weakened. Deletion of amino acids 467 to 487 abolished the interaction of ARIP3 with SUMO-1 completely. This region encompasses a motif similar to one in the PM-Scl75 protein shown to be sufficient for the interaction with SUMO-1 in yeast and proposed as a SUMO-1 interaction motif (32). The latter deletion did not influence Ubc9 binding. In sum, ARIP3 is capable of interacting via separate conserved domains with both SUMO-1 and the E2 SUMO-1 conjugase, Ubc9, in vitro; the short acidic region between amino acids 467 to 487 is critical for SUMO-1 binding, and the putative zinc-binding region is involved in the interaction with Ubc9. In agreement with the high conservation of the two domains among mammalian PIAS proteins, Miz1, PIAS1, and PIAS3 displayed similar abilities to interact with SUMO-1 and Ubc9 (Fig. 1).
FIG. 1.
PIAS proteins interact with SUMO-1 and Ubc9 in vitro. ARIP3, ARIP3Δ347-418, ARIP3Δ467-487, Miz1, PIAS1, and PIAS3 were labeled with [35S]methionine by translation in vitro and incubated with glutathione-Sepharose-bound GST, GST-SUMO-1, or GST-Ubc9. After extensive washings, bound proteins were eluted with SDS sample buffer, resolved by SDS-10% PAGE, and visualized by fluorography. Lanes i, input samples representing 10% of the amount of labeled proteins incubated with the matrices.
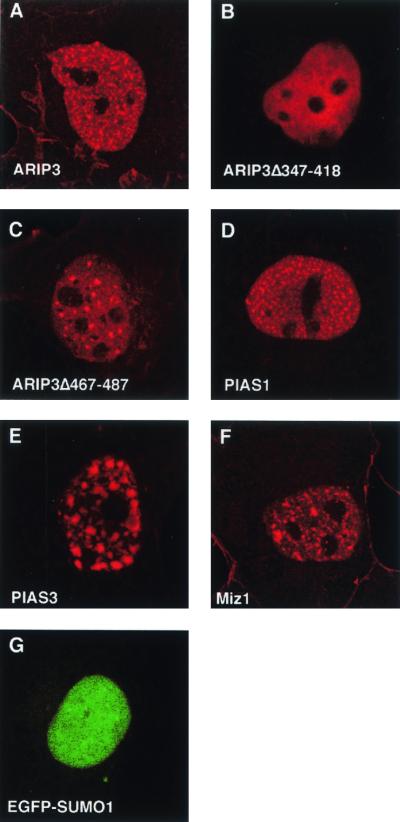
PIAS proteins and SUMO-1 colocalize in nuclear granules.
To determine whether ARIP3 colocalizes with SUMO-1 in mammalian cells, COS-1 cells were transfected with expression plasmids encoding FLAG-tagged ARIP3 (FLAG-ARIP3) and SUMO-1 fused to enhanced GFP (EGFP). ARIP3 was visualized by indirect immunofluorescence using a monoclonal antibody against the FLAG epitope and a rhodamine-conjugated secondary antibody. In the absence of ectopically expressed SUMO-1, ARIP3, Miz1, PIAS1, and PIAS3 displayed speckled patterns of nuclear distribution (Fig. 2A, D, E, and F). However, the number of nuclear speckles varied with PIAS protein. SUMO-1 transfected alone was typically diffusely distributed in the nucleoplasm (Fig. 2G). Interestingly, the deletion of amino acids 347 to 418 of ARIP3 rendered the protein evenly dispersed throughout the nucleoplasm, and ARIP3 devoid of amino acids 467 to 487 showed fewer nuclear granules than full-length ARIP3 (Fig. 2B and C).
FIG. 2.
Localization of PIAS proteins and SUMO-1 in COS-1 cells. Cells were transfected with expression plasmids encoding FLAG-ARIP3, FLAG-ARIP3Δ347-418, FLAG-ARIP3Δ467-487, FLAG-PIAS1, FLAG-PIAS3, FLAG-Miz1, or EGFP-SUMO-1 as described in Materials and Methods. Immunofluorescence labeling was performed with the M2 antibody against the FLAG epitope and a rhodamine red X-conjugated secondary antibody (A to F). Cells were analyzed by using a Bio-Rad MRC-1024 confocal laser scanning system connected to a Zeiss Axiovert 135 M microscope. EGFP was excited at 488 nm, and rhodamine red X was excited at 568 nm.
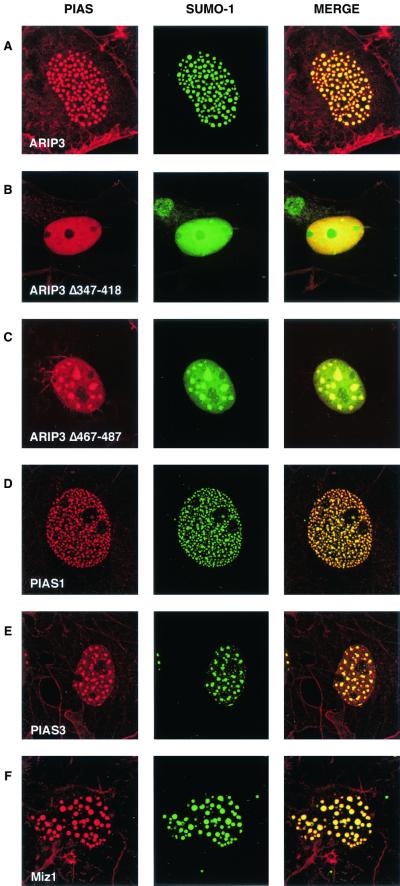
Coexpression of SUMO-1 with ARIP3 resulted in complete colocalization of SUMO-1 into nuclear granules with ARIP3 (Fig. 3A). The targeting was selective for SUMO-1, as the subcellular localization of EGFP was not influenced by ARIP3 (data not shown). ARIP3, Miz1, and PIAS1 were somewhat better concentrated to nuclear granules, i.e., they showed less staining that was dispersed throughout the nucleoplasm when expressed together with SUMO-1. Deletion of the SUMO-1-binding motif of ARIP3 (467-487) disrupted the colocalization partially, leading to fewer, but bigger granules and to a considerable amount of diffuse nuclear SUMO-1 (Fig. 3C). The putative zinc-binding region that was crucial for the formation of the ARIP3 granules was also needed for targeting SUMO-1 to the nuclear granules, since only diffuse nuclear SUMO-1 distribution was seen with ARIP3Δ347-418 (Fig. 3B). Thus, the regions important for binding to SUMO-1 and Ubc9 in vitro also influence intranuclear targeting of ARIP3 and its association with SUMO-1 in nuclei. In addition to PIASxα/ARIP3, its splicing isoform, PIASxβ/Miz1, and PIAS1 and PIAS3 were colocalized with SUMO-1 in COS-1 nuclei and displayed similar patterns of granule formation (Fig. 3D to F).
FIG. 3.
PIAS proteins and SUMO-1 colocalize in nuclear granules. COS-1 cells were cotransfected with expression plasmids encoding EGFP-SUMO-1 and FLAG-ARIP3, FLAG-ARIP3Δ347-418, FLAG-ARIP3Δ467-487, FLAG-PIAS1, FLAG-PIAS3, or FLAG-Miz1. Immunofluorescence labeling was performed with the M2 antibody against the FLAG epitope and a rhodamine-conjugated secondary antibody, and cells were analyzed as described for Fig. 2. Images were collected separately (EGFP was excited at 488 nm, and lissamine-rhodamine-conjugated anti-mouse IgG for M2 was excited at 568 nm) and merged as depicted.
PIAS proteins are modified by SUMO-1.
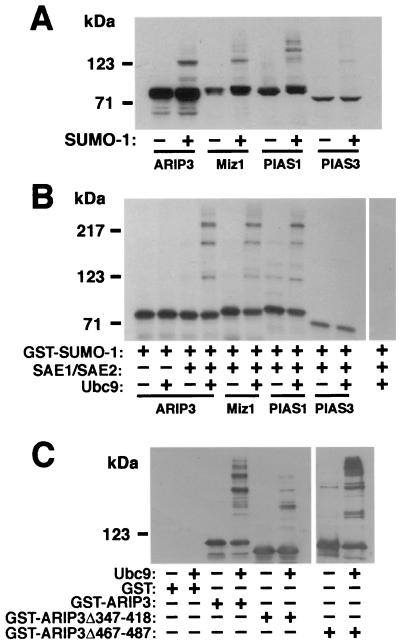
Since PIASxα/ARIP3 interacts with SUMO-1 and Ubc9 and colocalizes with SUMO-1, it was pertinent to test whether ARIP3 itself is a target for sumoylation. To this end, FLAG-tagged ARIP3 was transfected to COS-1 cells in the presence and absence of the SUMO-1 expression vector and ARIP3 was detected from the cell lysates by immunoblotting with an anti-FLAG antibody. Ectopic expression of SUMO-1 was needed, since COS-1 cells contain very small amounts of unconjugated endogenous SUMO-1 (31). NEM, a cysteine protease inhibitor that blocks the action of SUMO-1-deconjugating enzymes (11, 27, 53), was added to harvesting and lysis buffers to stabilize sumoylated proteins. Without coexpressed SUMO-1, only a doublet ARIP3 band migrating at ∼80 kDa was detected, whereas in the presence of ectopically expressed SUMO-1, higher-_M_r bands, most probably representing sumoylated forms of ARIP3, were also detected (Fig. 4A). The identities of these ARIP3 bands were confirmed with an anti-SUMO-1 antibody (see Fig. 5B). Miz1 and PIAS1 were also sumoylated in COS-1 cells. The sumoylation pattern of Miz1 was similar to that of ARIP3, whereas sumoylated PIAS1 forms were of somewhat higher _M_r. The migration of high-_M_r PIAS forms on SDS-PAGE gel suggests that up to three SUMO-1 moieties can be attached to these proteins and that the PIAS form containing two SUMO-1 moieties is the major sumoylated product under these conditions. PIAS3 was expressed at a level considerably lower than those at which other PIAS proteins were expressed, and therefore its sumoylation could not be accurately compared to that of other PIAS proteins (Fig. 4A).
FIG. 4.
Modification of ARIP3 and other PIAS proteins by SUMO-1 in mammalian cells and in vitro. (A) COS-1 cells grown on 6-cm-diameter dishes were transfected with 1 μg of pFLAG-ARIP3, pFLAG-Miz1, pFLAG-PIAS1, or pFLAG-PIAS3 and 1.5 μg of empty pSG5 vector or pSG5-His-SUMO-1. Forty-eight hours after transfection, the cells were lysed in RIPA-1 buffer containing 10 mM NEM, and the cell lysates were immunoblotted with the M2 FLAG antibody. (B) Sumoylation of PIAS proteins in vitro. In vitro-translated, 35S-labeled PIAS proteins were incubated with purified GST-SUMOGG-1 in the presence (+) or absence (−) of GST-SAE1 plus GST-SAE2 and GST-Ubc9 as indicated. Reactions were stopped by adding SDS-PAGE sample buffer, and the samples were resolved by SDS-PAGE and subjected to fluorography. (C) Sumoylation of purified GST-ARIP3, GST-ARIP3Δ347-418, and GST-ARIP3Δ467-487 (1 μg each) under conditions described for panel B. The reaction mixtures were immunoblotted with an anti-ARIP3 antibody.
FIG. 5.
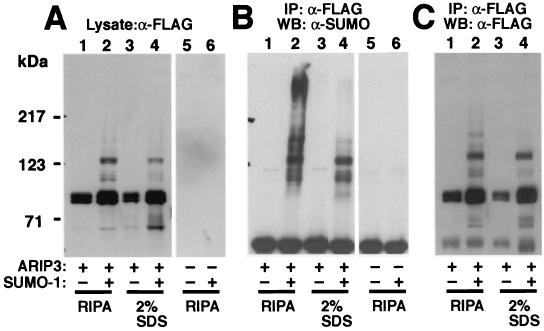
ARIP3 interacts with other sumoylated proteins. COS-1 cells were transfected with 1 μg of empty pFLAG-CMV2 or pFLAG-ARIP3 and 1.5 μg of empty pSG5 or pSG5-His-SUMO-1. Forty-eight hours after transfection, cells were collected and lysed either in RIPA-1 buffer containing 10 mM NEM or in a denaturing buffer containing 2% SDS in 10 mM Tris-HCl (pH 8.0)-150 mM NaCl. Samples lysed in the latter buffer were heated at 95°C for 10 min. Before immunoprecipitation (IP), these samples were diluted (1:10) with a mixture of 10 mM Tris-HCl (pH 8.0) and 150 mM NaCl containing 1% Triton X-100. Five percent of the cell extracts were immunoblotted with the anti-FLAG antibody (A), and the rest of the samples were immunoprecipitated with the anti-FLAG antibody followed by immunoblotting with the anti-SUMO-1 antibody (B) or the anti-FLAG antibody (C). WB, Western blotting.
To investigate PIAS protein sumoylation under cell-free conditions, 35S-labeled PIAS proteins produced by in vitro translation were incubated in the presence of ATP and purified fusions of GST with SAE1, SAE2, Ubc9, and SUMO-1. The in vitro sumoylation reactions produced at least three additional higher-molecular-mass ARIP3 bands (Fig. 4B) that, due to the fusion of GST with SUMO-1, migrated considerably more slowly than the sumoylated forms in panel A. The formation of these bands was dependent on the presence of both SUMO-1 E1 (SAE1 plus SAE2) and E2 (Ubc9) conjugase activity in the reaction. Incubation of purified GST-ARIP3 in this system yielded a similar pattern of sumoylated forms (Fig. 4C). Deletion of amino acids 467 to 487 from ARIP3 did not reduce the extent of sumoylation, whereas GST-ARIP3Δ347-418 was very weakly sumoylated. Under these in vitro conditions, ARIP3 served as a SUMO-1 acceptor that was at least as good as promyelocytic leukemia protein (PML) (data not shown). Sumoylation of Miz1 and PIAS1 was similar to that of ARIP3 in vitro, whereas sumoylation of PIAS3 was undetectable under these conditions (Fig. 4B), which agrees with the COS-1 cell data. Together these results show that ARIP3, Miz1, and PIAS1 can be efficiently sumoylated on more than one site.
PIASxα/ARIP3 interacts with other SUMO-1-modified proteins.
To verify that the high-molecular-mass ARIP3 bands (Fig. 4A) correspond to sumoylated ARIP3 forms, COS-1 cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-SUMO-1 antibody or an anti-FLAG antibody. The anti-SUMO-1 antibody detected the same slowly migrating bands (between ∼100 and 140 kDa) as those detected both in cell extracts and in immunoprecipitates with the anti-FLAG antibody (Fig. 5), indicating that they indeed represent SUMO-1-modified ARIP3. In addition, the anti-SUMO-1 antibody revealed the presence of other SUMO-1-containing proteins, especially a heavy smear of immunoreactivity migrating at >230 kDa (Fig. 5B, lane 2). The slowly migrating bands were assumed to represent other sumoylated proteins that were coimmunoprecipitated with ARIP3. To examine this possibility, the cells cotransfected with ARIP3 and SUMO-1 were lysed in a buffer containing 2% SDS instead of a standard immunoprecipitation buffer and proteins were denatured by heating at 95°C for 10 min, which should break noncovalent protein-protein interactions. This treatment abolished anti-SUMO-1 immunoreactive bands with molecular masses >140 kDa, and only the same ∼100- to 140-kDa bands as those detected with the anti-FLAG antibody were seen with the anti-SUMO-1 antibody (cf. lanes 4 in Fig. 5A and B). Thus, in addition to being covalently modified by SUMO-1, PIASxα/ARIP3 binds other sumoylated proteins in a noncovalent fashion. The pattern of sumoylated proteins interacting with PIASxβ/Miz1 was similar to that for ARIP3, whereas proteins recruited by PIAS1 were shifted to a lower-molecular-mass range (data not shown).
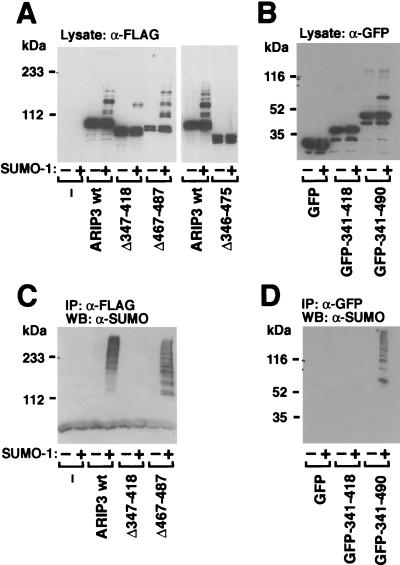
We next sought to identify the regions of ARIP3 involved in sumoylation and needed for the interaction with other sumoylated proteins. In view of the efficient sumoylation of ARIP3, it was surprising that the ARIP3 sequence does not contain consensus SUMO-1 attachment sites with sequences of the form ΨKXE, where Ψ is a large hydrophobic residue and X represents any amino acid (5, 8, 19, 44, 51). To address this issue, versions of ARIP3 with different deletions were expressed in COS-1 cells and their sumoylation was examined (Fig. 6A). Removal of the SUMO-1-binding motif (amino acids 467 to 487) alone or a longer fragment encompassing this region (amino acids 467 to 547) altered the pattern of sumoylated bands without markedly decreasing the extent of modification (Fig. 6 and data not shown). By contrast, when amino acids 347 to 418 were deleted, sumoylation of ARIP3 was dramatically weakened and deletion of the ARIP3 region comprising amino acids 346 to 475 completely abolished the sumoylation (Fig. 6A). Even though the latter region does not contain consensus sumoylation sites, lysine pairs 324 and 326, 379 and 380, 390 and 391, and 430 and 431 were mutated to arginines and sumoylation of the double mutants in COS-1 cells was studied. As these lysines are not embedded in a consensus sequence for sumoylation, it was not unexpected that the mutations failed to alter the pattern of ARIP3 sumoylation (data not shown). To confirm that the central region of ARIP3 indeed harbors a site(s) for SUMO-1 attachment and that the lack of sumoylation of the deletion mutants was not due to the improper folding of the other ARIP3 regions, we performed a converse experiment in which ARIP3 regions comprising amino acids 341 to 418 and 341 to 490 were fused to EGFP and the sumoylation of the fusion proteins in COS-1 cells was examined. Immunoblotting extracts derived from cells transfected with GFP-ARIP3 (341-490) with the anti-GFP antibody indeed revealed an additional slower-migrating band in the presence of coexpressed SUMO-1, but no sumoylation was detectable with GFP-ARIP3 (341-418) (Fig. 6B). Thus the central ARIP3 region harbors at least one SUMO-1 attachment site.
FIG. 6.
Zinc finger region of ARIP3 is required for the recruitment of other sumoylated proteins. (A) FLAG-tagged wild-type ARIP3, ARIP3Δ347-418, ARIP3Δ467-487, and ARIP3Δ346-475 were coexpressed in COS-1 cells without or with pSG5-His-SUMO-1 as described for Fig. 4. Cells were lysed in RIPA-1 buffer, and samples from the lysate were immunoblotted with the anti-FLAG antibody. (B) Expression vectors encoding EGFP, EGFP-ARIP3 (341-418), or EGFP-ARIP3 (341-490) were cotransfected into COS-1 cells without or with pSG5-His-SUMO-1. Cells were lysed in RIPA-1 buffer, and lysates were immunoblotted with an antibody against GFP. (C) The rest of the lysates from panel A were immunoprecipitated (IP) with the anti-FLAG antibody, and immunoprecipitates were immunoblotted with the anti-SUMO-1 antibody. (D) The rest of the lysates from panel B were immunoprecipitated with the anti-GFP antibody, and immunoprecipitates were immunoblotted with the anti-SUMO-1 antibody. WB, Western blotting.
Immunoprecipitation of the COS-1 cell lysates containing ARIP3 with deletions with the FLAG antibody and subsequent immunoblotting with the SUMO-1 antibody revealed that the potential zinc-binding structure of ARIP3 (amino acids 347 to 418) is required for the recruitment of other SUMO-1-conjugated proteins, since no sumoylated proteins were tethered to this mutant (Fig. 6C). An ARIP3 deletion affecting the free SUMO-1-binding motif (amino acids 467 to 487) did not abolish the recruitment but rather shifted the pattern of the recruited proteins to a lower _M_r range. Even though the putative zinc finger region of ARIP3 is mandatory for the tethering of sumoylated proteins, the region in isolation is not sufficient for the recruitment, as no sumoylated proteins were immunoprecipitated with EGFP-ARIP3 (341-418) (Fig. 6D). However, EGFP-ARIP3 (341-490) was capable of tethering other sumoylated proteins. Thus, in addition to being covalently modified by SUMO-1 at nonclassical attachment sites, PIASxα/ARIP3 (and PIASxβ/Miz1 and PIAS1; data not shown) also interacts with other sumoylated proteins; the predicted zinc-binding region and the SUMO-1-binding motif are both involved in tethering other sumoylated proteins.
PIASxα/ARIP3 stimulates protein sumoylation in intact cells.
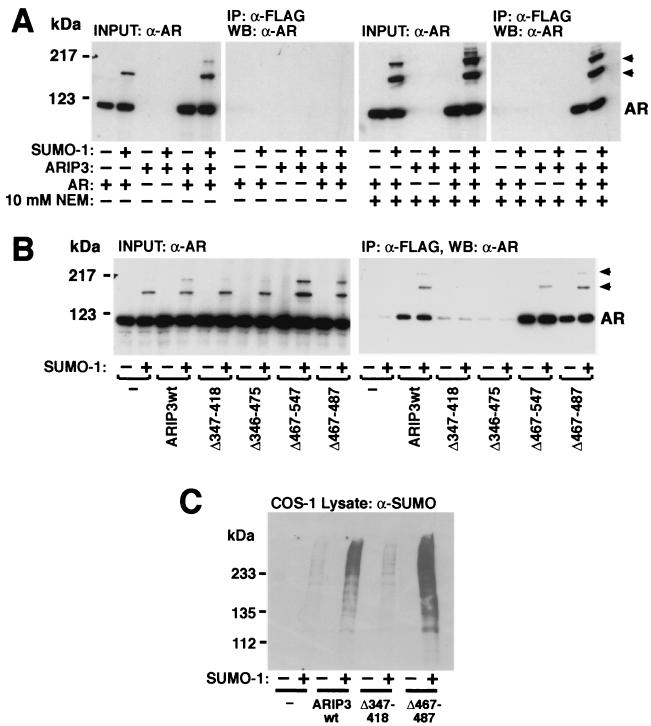
ARIP3 was originally characterized as an AR-interacting protein that functions as a coregulator in AR-dependent transcription (34). We have recently shown that AR is sumoylated at two sites and that this modification modulates the transcriptional activity of AR (42). Since ARIP3 interacts with sumoylated proteins (Fig. 6), it was of interest to test whether protein sumoylation influences the interaction between ARIP3 and AR. To this end, COS-1 cell lysates derived from cells transfected with FLAG-ARIP3 and AR in the presence and absence of SUMO-1 were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-AR antibody. AR was coimmunoprecipitated with ARIP3 from cell lysates only in the presence of NEM in the immunoprecipitation buffer (Fig. 7A). However, both unmodified and sumoylated AR proteins were coimmunoprecipitated with ARIP3, with the latter AR forms showing somewhat better recovery than the unmodified receptor. Moreover, the sumoylation-deficient AR mutant (K386R/K520R [42]) was also coimmunoprecipitated with ARIP3 in a NEM-sensitive fashion (data not shown). These results indicate that covalent attachment of SUMO-1 to AR is not mandatory for the AR-ARIP3 interaction. However, the stabilizing effect of NEM suggests that thiol-ester bonds are involved in the interaction. Again, coimmunoprecipitation experiments indicated that the zinc-binding region of ARIP3 is crucial for the interaction with AR, since AR failed to bind to ARIP3Δ347-418 or ARIP3Δ346-475 (Fig. 7B).
FIG. 7.
Role of ARIP3 domains in the interaction with AR and effect of ARIP3 on protein sumoylation. (A) COS-1 cells grown on 6-cm-diameter plates were transfected with 0.7 μg of empty pFLAG-CMV2 vector or pFLAG-ARIP3, 0.9 μg of pSG5 or pSG5-rAR, and 0.9 μg of pSG5 or pSG5-His-SUMO-1. The cells were supplied with 100 nM testosterone 24 h after transfection and were collected 48 h after transfection and lysed in RIPA-2 buffer in the presence (+) or absence (−) of NEM. Five percent of the lysate was immunoblotted with the K333 antibody against AR, and the rest of the sample was subjected to immunoprecipitation (IP) with the anti-FLAG antibody. Immunoprecipitates were blotted with anti-AR antibody. WB, Western blotting. (B) The zinc finger region of ARIP3 is required for the NEM-dependent interaction with AR. Versions of FLAG-tagged ARIP3 with different deletions were coexpressed with AR in COS-1 cells. Cells were lysed in RIPA-2 buffer containing 10 mM NEM, and immunoblotting of cell extracts and immunoprecipitation were performed as described for panel A. Arrowheads indicate sumoylated forms of AR. (C) Overexpression of ARIP3 enhances the sumoylation of endogenous COS-1 cell proteins. ARIP3, ARIP3Δ347-418, or ARIP3Δ467-487 were coexpressed in COS-1 cells without or with SUMO-1. Cells were lysed in RIPA-1 buffer, and lysates were immunoblotted with the anti-SUMO-1 antibody. wt, wild type.
The expression of ARIP3 with AR and SUMO-1 enhanced especially the intensity of the most slowly migrating AR band (Fig. 7), corresponding to a receptor form with two SUMO-1 moieties (42). This suggests that ARIP3 is capable of promoting protein sumoylation in intact cells. To address whether ARIP3 also influences sumoylation of endogenous COS-1 cell proteins, lysates derived from the cells transfected with or without ARIP3 and SUMO-1 were immunoblotted with the anti-SUMO-1 antibody. As shown in Fig. 7C, full-length ARIP3, but not ARIP3Δ347-418, markedly increased the amount of SUMO-1 attached to endogenous COS-1 proteins, indicating that PIASxα/ARIP3 indeed possesses a general ability to facilitate protein sumoylation.
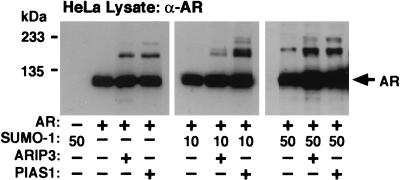
The influence of ARIP3 on AR sumoylation in COS-1 cells with ectopically expressed SUMO-1 was rather modest. Since COS-1 cells contain very small amounts of unconjugated SUMO-1, we studied the effects of ARIP3 and PIAS1 on the modification of AR in another cell line. In HeLa cells, both ARIP3 and PIAS1 promoted sumoylation of AR even in the presence of endogenous SUMO-1 only but no sumoylated AR forms were detectable without ectopic expression of PIAS proteins (Fig. 8).
FIG. 8.
ARIP3 and PIAS1 enhance the sumoylation of AR in HeLa cells. HeLa cells seeded onto six-well plates were cotransfected with 100 ng of pcDNA-Flag-hAR; 100 ng of pFLAG-CMV2, pFLAG-ARIP3, or pFLAG-PIAS1; and 0, 10, or 50 ng of pSG5-His-SUMO-1. Cells were lysed in RIPA-1 buffer, and the lysates were immunoblotted with the anti-AR antibody. The difference between the migration of AR forms sumoylated with endogenous SUMO-1 and that of ectopically expressed SUMO-1 is due to the His tag present in the SUMO-1 expression vector. The identities of sumoylated AR forms were verified by immunoblotting the anti-AR antibody-immunoprecipitated AR with the anti-SUMO-1 antibody (not shown).
PIASxα/ARIP3 enhances in vitro sumoylation of AR and c-Jun.
To examine whether ARIP3 also facilitates sumoylation of AR in vitro, GST-ARIP3 was expressed in E. coli and purified. It was subsequently used in an in vitro sumoylation system containing recombinant SUMO-1 E1 and E2 enzymes, GST-SUMO-1, and 35S-labeled AR, produced by translation in vitro, as a substrate. The formation of the two bands corresponding to sumoylated AR was dependent on the presence of Ubc9 (Fig. 9A). GST-ARIP3 in the reaction mixture increased significantly the extent of AR sumoylation, whereas GST-ARIP3Δ347-418 was inactive in this respect. When lower concentrations of Ubc9 were employed, the effect of ARIP3 was even more pronounced (data not shown). Recombinant GST-Miz1 was also capable of facilitating SUMO-1 modification of AR in vitro (data not shown).
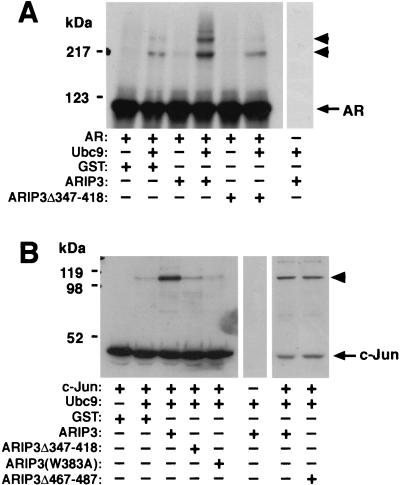
FIG. 9.
ARIP3 enhances the sumoylation of AR and c-Jun in vitro. (A) Effect of ARIP3 on sumoylation of in vitro-translated AR. 35S-labeled, in vitro-translated AR was incubated with purified GST-SUMOGG-1 (in all reactions), GST-SAE1 plus GST-SAE2 (in all reactions), and GST-Ubc9 in the presence or absence of GST, GST-ARIP3, or GST-ARIP3Δ347-418 as indicated. Samples were resolved by SDS-PAGE and subjected to fluorography. (B) ARIP3 enhances the sumoylation of purified recombinant c-Jun. His-tagged c-Jun was incubated with GST-SUMOGG-1, GST-SAE1 plus GST-SAE2, and GST-Ubc9 in the presence of GST alone, GST-ARIP3, GST-ARIP3Δ347-418, GST-ARIP3(W383A), or GST-ARIP3Δ467-487 as indicated. Samples were resolved by SDS-PAGE and immunoblotted with the anti-His antibody. Arrowheads indicate sumoylated forms of proteins.
To study whether sumoylation of other specific proteins is promoted by ARIP3, purified recombinant c-Jun was used as the substrate in an in vitro sumoylation reaction. c-Jun is sumoylated in vivo at a single lysine residue (36). ARIP3 clearly increased the extent of sumoylation of purified recombinant c-Jun, and the effect of ARIP3 in this system, containing entirely recombinant proteins, was at least as strong as that seen with reticulocyte lysate-produced AR as the substrate (Fig. 9B). Again, the deletion of the zinc-binding structure of ARIP3 (ARIP3Δ347-418) abolished the ability of ARIP3 to promote sumoylation, whereas the deletion of amino acids 467 to 487 did not weaken the activity of ARIP3 toward c-Jun (Fig. 9B).
A number of active RING-type ubiquitin ligases, such as c-Cbl, contain a conserved Trp408 residue in their RING domains (17, 50), the mutation of which eliminates the E3 ligase activity of c-Cbl (17). Interestingly, PIASxα/ARIP3 and other PIAS proteins also harbor a conserved Trp in the middle of their potential zinc fingers. Mutation of this Trp to Ala (W383A) inhibited the sumoylation by ARIP3 of c-Jun (Fig. 9B) and AR (data not shown), indicating that this bulky amino acid residue plays an essential role in the domain structure, which may be similar to that of the conserved Trp in RING E3 ubiquitin ligases. Sumoylation is selectively influenced by the W383A mutation, as the mutant maintains its ability to interact with SUMO-1 and Ubc9 in GST pull-down assays (data not shown). Collectively, our data indicate that PIASxα/ARIP3 and other PIAS proteins with a Miz zinc domain participate in protein sumoylation and function as E3-type SUMO-1 protein ligases in a fashion that resembles the action of RING-type E3 ubiquitin protein ligases.
PIAS proteins differ in their abilities to promote sumoylation of GRIP1.
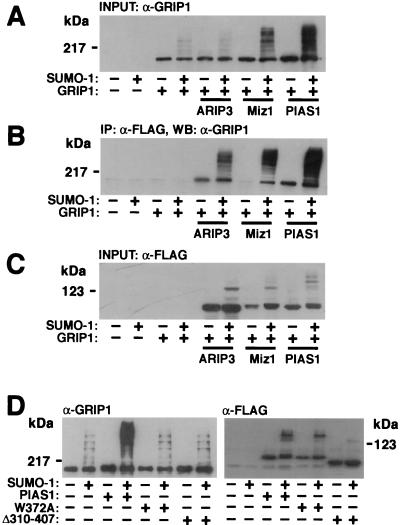
We have shown that PIASx and PIAS1, in addition to interacting with steroid receptors, interact with a p160 nuclear receptor coactivator, glucocorticoid receptor-interacting protein 1 (GRIP1) (26; see also reference 14). To investigate whether these interactions are linked to SUMO-1 modifications of GRIP1, various FLAG-tagged PIAS proteins were coexpressed with GRIP1 in the presence or absence of a SUMO-1-encoding vector. GRIP1 was detected by immunoblotting COS-1 cell lysates with a monoclonal anti-GRIP1 antibody, or the proteins immunoprecipitated with PIAS proteins (anti-FLAG antibody) were subjected to immunoblotting with the anti-GRIP1 antibody. GRIP1 is indeed a target for covalent SUMO-1 modifications but, similar to what was found for AR, both unmodified GRIP1 and sumoylated GRIP1 interact with PIASxα/ARIP3 and PIAS1 (Fig. 10). There are four potential SUMO acceptor sites in GRIP1 (lysines 239, 731, 788, and 1452), which explains, at least in part, the heterogeneity of sumoylated GRIP1 forms. It is currently unknown why unsumoylated GRIP1 interacted poorly with PIASxβ/Miz1 in the absence of coexpressed SUMO-1 (Fig. 10B). Interestingly, the ability of PIASxα/ARIP3 to influence SUMO-1 attachment to GRIP1 was clearly weaker than that of two other PIAS proteins examined, PIASxβ/Miz1 and PIAS1. Immunoblotting the samples with the anti-FLAG antibody ruled out the possibility that the dissimilar abilities of PIAS proteins to promote GRIP1 sumoylation derive from differences in their expression levels (Fig. 10C). PIAS1(W372A) (the conserved Trp residue in the zinc-binding domain converted to Ala) and PIAS1Δ310-407 (the whole zinc-binding domain deleted) mutants were unable to facilitate sumoylation of GRIP1 (Fig. 10D), verifying the importance of the PIAS RING-like region in the reaction. Again, these results were not caused by lower expression of the PIAS1 mutants. These results imply that, even though the PIAS proteins are sumoylated and able to bind other SUMO-1-conjugated proteins, their ability to facilitate protein sumoylation exhibits substrate selectivity.
FIG. 10.
PIAS proteins enhance the sumoylation of GRIP1. COS-1 cells grown on 6-cm-diameter plates were transfected with 0.7 μg of empty pFLAG-CMV2 vector or a vector encoding the FLAG-tagged PIAS protein, 0.9 μg of pSG5 or pSG5-GRIP1, and 0.9 μg of pSG5 or pSG5-His-SUMO-1. The cells were harvested 48 h after transfection and lysed in RIPA-2 buffer containing 10 mM NEM. Five percent of the lysate was immunoblotted with the anti-GRIP1 antibody (A), and the rest was subjected to immunoprecipitation (IP) with the anti-FLAG antibody followed by immunoblotting with the anti-GRIP1 antibody (B). WB, Western blotting. (C) The amounts of PIAS proteins were verified by immunoblotting the cell lysates with the anti-FLAG antibody. (D) The predicted zinc-binding region of PIAS1 is required for stimulation of the sumoylation of GRIP1. COS-1 cells were cotransfected with 0.9 μg of pSG5-GRIP1; 0.7 μg of pFLAG-CMV2, pFLAG-PIAS1, pFLAG-PIAS1(W372A), or pFLAG-PIAS1Δ310-407; and 0.9 μg of pSG5 or pSG5-His-SUMO-1. Cells were lysed in RIPA-2 buffer, and lysates were immunoblotted with either the anti-GRIP1 antibody or the anti-FLAG antibody.
The role of SUMO-1-tethering and ligase domains of PIAS proteins in the regulation of AR-dependent transcription.
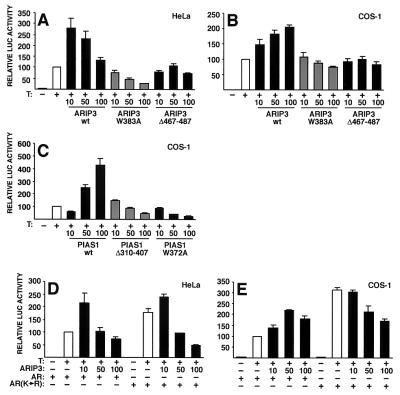
The preceding results suggest that the modulatory effects of PIAS proteins on steroid receptors, such as AR, could be exerted through their SUMO-1-binding and SUMO-1 ligase activities. To link these findings to transcriptional regulation, we examined the effects of ARIP3(W383A) and ARIP3Δ467-487 mutants on AR-dependent transcription using a luciferase reporter driven by two androgen response elements (ARE) of the C3 (1) gene in front of a TATA box (pARE2TATA-LUC). Ectopic expression of wild-type ARIP3 activated AR function in both COS-1 and HeLa cells, whereas ligase-deficient ARIP3(W383A) was repressive or inert and ARIP3Δ467-487, in which the SUMO-binding motif was deleted, failed to influence AR-dependent transcription (Fig. 11A and B). It is of note that, even though these ARIP3 mutants show altered nuclear localization patterns (Fig. 2), they are still capable of interacting with AR (Fig. 7) (our unpublished results). Therefore, the altered activity of these mutants in AR-dependent transcription cannot simply be due to their mislocalization. Similar to what was found for ARIP3, the RING-like region of PIAS1 was critical for the activation of AR-dependent transcription, as both the Trp372-to-Ala mutation and the deletion of amino acids 310 to 407 converted PIAS1 from a coactivator to a corepressor of AR function in COS-1 cells (Fig. 11C). Consistent with our recent results (42), the AR mutant with the two major sumoylation sites (K386 and K520) converted to arginines [AR(K→R)] was more active than the wild-type receptor on the simple ARE-driven reporter (Fig. 11D and E). In contrast to wild-type AR, AR(K→R) was not significantly activated by ARIP3 in HeLa or COS-1 cells. The reason for the repression by ARIP3 of the mutated AR is not currently known, but it may be due to ARIP3-enhanced sumoylation of an AR(K→R)-interacting protein that does not normally interact with wild-type AR. Taken together, both the SUMO-1-binding and ligase activities of PIAS proteins are critical for these proteins to regulate transcriptional functions. We cannot formally rule out the possibility that the effects of ARIP3 and PIAS1 also involve subnuclear targeting of AR which is brought about by interactions of the PIAS SUMO-1-binding domain with other nuclear proteins. However, we have not been able to detect PIAS-induced alterations in the subnuclear localization of AR.
FIG. 11.
The SUMO-1-tethering region and the ligase domain are critical for the ability of PIAS proteins to coregulate AR-dependent transcription. HeLa (A) and COS-1 cells (B) cultured on 12-well plates were cotransfected with 200 ng of pARE2TATA-LUC, 20 ng of pCMVβ, 5 ng of pcDNA-Flag-hAR, and 10, 50, or 100 ng of pFLAG-ARIP3, pFLAG-ARIP3 (W383A), or pFLAG-ARIP3Δ467-487 and treated or not treated with 100 nM testosterone (T). After normalization for transfection efficiency by using β-galactosidase activity, reporter gene activities were expressed relative to those of AR plus T without a coregulator (set at 100). wt, wild type. (C) The same experiment as in panel B, but instead of ARIP3 expression plasmids, COS-1 cells were cotransfected with pFLAG-PIAS1, pFLAG-PIAS1(W372A), or pFLAG-PIAS1Δ310-407. (D and E) Effect of cotransfected ARIP3 on the AR mutant with the SUMO-1 attachment lysines mutated to arginines [AR(K→R)] compared to that on wild-type AR. The experimental conditions were the same as those in panels A and B. The values represent means ± standard deviations from three to six independent experiments.
DISCUSSION
PIAS proteins can bind to distinct classes of nuclear proteins and enhance or repress transcriptional activities of structurally unrelated factors, such as steroid receptors and STATs (4, 28, 34, 57), and their biological functions are clearly not restricted to the inhibition of STAT signaling. Even though PIAS1 and Miz1 possess modest intrinsic transcription activation functions and ARIP3 and PIAS3 lack this activity, the pleiotropic effects of PIAS proteins on transcriptional regulation can hardly derive from these functions (25). The mechanism of PIAS protein action has remained elusive, and one study has suggested that it involves formation of complexes with other coregulatory proteins (25).
Using PIASxα/ARIP3 as the main representative of PIAS proteins, we demonstrate in this work that ARIP3 interacts via separate conserved structural domains with both SUMO-1 and its E2 conjugase, Ubc9. In addition, ARIP3, Miz1, and PIAS1 are heavily sumoylated even though their sequences do not harbor recognizable consensus SUMO-1 attachment motifs. Many distinct proteins have recently been shown to be modified by SUMO-1 (31), and the ability of a given protein to interact with Ubc9 and SUMO-1 generally reflects its capability of becoming sumoylated. The fact that ARIP3, Miz1, and PIAS1 interact with other sumoylated proteins and possess a peculiar sumoylation feature, i.e., they lack consensus SUMO-1 acceptor sites, suggests that these proteins participate in the sumoylation process itself.
It has been suggested that protein motifs that contain Ser-rich and Glu- and Asp-rich stretches preceded by hydrophobic residues mediate interactions with free SUMO-1 (32). In agreement with this notion, deletion of the region comprising amino acids 467 to 487, fulfilling the characteristics of a potential SUMO-1 binding motif, abolished the interaction of ARIP3 with unconjugated SUMO-1 in vitro. This deletion did not, however, reduce sumoylation of ARIP3 in vivo or in vitro, but it influenced the recruitment of other sumoylated proteins. By contrast, ARIP3Δ347-418, which is devoid of the zinc finger-like region, was both very weakly sumoylated and incapable of tethering other sumoylated proteins. Colocalization of SUMO-1 and PIAS proteins in the same subnuclear sites is likely to reflect both the covalent attachment of SUMO-1 to PIAS proteins and their ability to bind other sumoylated proteins at those subnuclear domains. These results are in line with the findings that deletion of the SUMO-1-binding motif weakened the formation of nuclear cogranules by ARIP3 and SUMO-1 and that removal of the zinc finger-like structure abolished the nuclear granules.
In addition to binding proteins in a zinc finger-like domain-dependent fashion, ARIP3, Miz1, and PIAS1 clearly promoted sumoylation of other proteins in a manner that was also dependent on this domain. Both ARIP3 and PIAS1 were capable of stimulating SUMO-1 attachment to AR in intact cells, whereas PIAS1, but not ARIP3, enhanced sumoylation of GRIP1. Furthermore, ARIP3 enhanced sumoylation of AR and c-Jun in a RING-like domain-dependent manner in a cell-free system utilizing purified recombinant proteins. The activity of many E3-type ubiquitin protein ligases relies on their RING finger structures. It is, therefore, intriguing that the central regions of mammalian PIAS proteins, except for PIAS3, harbor seven cysteine residues and a histidine which are mandatory for a C3HC4-type RING finger motif. Indeed, threading analysis of this PIAS region suggests that its three-dimensional structure is similar to a C3HC4 RING finger fold. However, the spacing between potential zinc-coordinating residues and the amino acid composition of the mammalian PIAS RING-like structure differ substantially from those of the RING motifs of E3 ubiquitin ligases. Interestingly, PIAS3, which is poorly sumoylated in cell-free conditions and in intact cells, does not have a cysteine residue at the fourth position of the putative C3HC4 motif.
The crystal structure of c-Cbl E3 ubiquitin ligase bound to a cognate E2 and a substrate peptide suggests that the RING finger domains not only recruit the substrate and the E2 enzyme but also serve as scaffolds that position and orient them optimally for ubiquitin transfer (64). A number of RING finger proteins which function as ubiquitin E3 ligases or which have been implicated in protein degradation possess a conserved Trp residue between cysteines at the fifth and sixth positions of their RING (17). This Trp in c-Cbl forms part of the RING interface for the interaction with E2 ligase, and Trp mutations eliminate the E3 activity. All PIAS proteins contain a Trp residue in their central Cys-rich regions. It is conserved in more distantly related proteins possessing a similar Cys-rich structure. Conversion of the Trp to Ala in PIASxα/ARIP3 (W383A) and PIAS1 (W372A) abolished their ability to promote protein sumoylation, indicating that this amino acid residue plays an important role in the sumoylation process. Thus, PIAS proteins harboring a Miz zinc finger structure seem to function as E3 SUMO-1 ligases, facilitating Ubc9-dependent sumoylation in a fashion that resembles the action of RING-type E3 ligases in ubiquitination.
Ubc9 interacts physically with almost all known SUMO acceptors, which may indicate that it is sufficient for substrate recognition in vivo. It has been suggested that sumoylation sites per se act as major determinants of Ubc9 binding and SUMO-1 modification, which argues against the need for E3 ligase activity in sumoylation (47). However, even though purified or recombinant E1 and E2 enzymes are sufficient to sumoylate target proteins in vitro, these reactions are slow and inefficient compared to reactions in intact cells (1, 5, 31, 58). The notion that Ubc9 alone is not likely to be able to discriminate between different SUMO proteins and, therefore, is not capable of conferring specificity also strongly points to the existence of additional regulatory factors in sumoylation (58). The high degree of homology between Ubc9 and ubiquitin E2 ligases suggests that the transfer of SUMO-1 to target proteins is very similar to that in ubiquitination and that substrate specificity in sumoylation requires additional E3-type ligases. Our results showing that PIAS proteins differ in their abilities to promote SUMO-1 attachment to GRIP1 indeed support the notion that different PIAS proteins act as E3 SUMO ligases with dissimilar substrate preferences for target proteins.
Enzymes of sumoylation are predominantly nuclear, and proteins involved in various nuclear functions (DNA replication and repair, mitosis, and transcriptional regulation) are substrates for SUMO attachment (31, 37). There is compelling evidence for a critical role of SUMO modifications in the regulation of protein-protein interactions and subcellular and subnuclear localization (31, 37, 62). Conjugation of SUMO-1 to RanGAP1 targets it to the nuclear pore complex (30), and modification of PML by SUMO-1 directs it to subnuclear domains termed PML nuclear bodies (38). The latter modification is also needed for PML-mediated recruitment of other proteins (15), which may also be the case with PIAS proteins. Sumoylation may also prevent degradation of the target protein, as exemplified by IκBα (5). In view of the role of SUMO-1 in protein complex formation, one can envision that sumoylation modulates transcription factor-coregulator interactions (12, 36, 41, 42, 45). Our present data on the PIAS1-promoted sumoylation of coactivator GRIP1 support this possibility. In addition to interacting with various sequence-specific transcription factors and influencing their sumoylation state, PIAS proteins may act as platforms to recruit other sumoylated proteins or proteins to be sumoylated, such as transcriptional coactivators and corepressors. Notably, the effects of ARIP3 and PIAS1 on AR-dependent transcription are dependent on both the SUMO-binding motif and the ligase domain. However, all effects of PIAS proteins on AR-dependent transcription cannot be explained through events based solely on the sumoylation of AR. It is important to note that PIAS proteins have multiple interaction partners in transcriptional regulation, including steroid receptor coactivator GRIP1, and their effects (positive or negative) on a given promoter are most likely combinatorial and reflect the overall effects of the interaction partners. In agreement with this notion, we have recently shown that PIAS proteins and GRIP1 exhibit synergistic effects on steroid receptor-dependent signaling and that the RING-like domain of PIAS proteins plays a crucial role also in this process (26).
The function of PIAS or PIAS-like proteins in the regulation of chromosome structure and function may also be linked to protein sumoylation. The Drosophila PIAS protein homologue Su(var)2-10 (Zimp) is required for proper chromosome structure and inheritance, and one study suggested that it controls multiple aspects of chromosome function by maintaining chromosome organization in interphase nuclei (13). The finding that defects in chromosome condensation in S. cerevisiae caused by the loss of the SMT4 gene are bypassed when the SIZ1 gene is overexpressed (52) connects a PIAS-like protein to sumoylation processes. Specifically, SMT encodes an evolutionarily conserved protease exhibiting SUMO-cleaving isopeptidase activity and Siz1 shows extensive similarity to animal PIAS proteins. Indeed, while this work was being finalized for publication, Siz1 and related protein Siz2 (Nfi1) were reported to promote sumoylation in yeast (20, 56) and Siz1 was shown to enhance sumoylation of septins in vitro (20, 55). Moreover, contemporaneous studies suggest that PIAS1 and PIASy function as SUMO ligases for p53 and LEF1, respectively (22, 46).
In summary, our results along with very recent data from other laboratories convincingly argue that proteins with the conserved Miz zinc finger function as E3 SUMO-1 ligases. Mammalian PIAS proteins themselves undergo sumoylation in a fashion that resembles automodification of RING finger E3 ubiquitin ligases (29), which may play a self-regulatory role. Furthermore, the domains responsible for SUMO-1 binding and SUMO-1 conjugation are segregated, and both of these structures are critical for nuclear targeting of and transcriptional regulation by the PIAS proteins. Importantly, mammalian PIAS proteins display target protein selectivity in intact cells. In view of these considerations, we suggest that many of the effects of the PIAS protein on various transcription factors are exerted through their SUMO-tethering and SUMO ligase activities and that these properties are linked to their ability to interact with and modulate the activities of distinct transcription factors.
Acknowledgments
We thank Kati Saastamoinen and Saija Kotola for technical assistance; Hetti Poukka, Anne Dejean, Ronald T. Hay, Rob Maxon, Ke Shuai, and Michael R. Stallcup for plasmids; and Christophe Roos for structure threading analysis.
This work was supported by grants from the Academy of Finland, the Finnish Foundation for Cancer Research, the Sigrid Jusélius Foundation, Biocentrum Helsinki, and the Helsinki University Central Hospital.
REFERENCES
- 1.Azuma, Y., S. H. Tan, M. M. Cavenagh, A. M. Ainsztein, H. Saitoh, and M. Dasso. 2001. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 10**:**1825-1827. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, P., A. Arndt, S. Metzger, R. Mahajan, F. Melchior, R. Jaenicke, and J. Becker. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280**:**275-286. [DOI] [PubMed] [Google Scholar]
- 3.Cho, S. Y., J. W. Jeon, S. H. Lee, and S. S. Park. 2000. p67 isoform of mouse disabled 2 protein acts as a transcriptional activator during the differentiation of F9 cells. Biochem. J. 352**:**645-650. [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278**:**1803-1805. [DOI] [PubMed] [Google Scholar]
- 5.Desterro, J. M. P., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2**:**233-239. [DOI] [PubMed] [Google Scholar]
- 6.Desterro, J. M. P., M. S. Rodriguez, G. D. Kemp, and R. T. Hay. 1999. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274**:**10618-10624. [DOI] [PubMed] [Google Scholar]
- 7.Desterro, J. M. P., J. Thompson, and R. T. Hay. 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417**:**297-300. [DOI] [PubMed] [Google Scholar]
- 8.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112**:**381-393. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher, W. M., M. Argentini, V. Sierra, L. Bracco, L. Debussche, and E. Conseiller. 1999. MBP1: a novel mutant p53-specific protein partner with oncogenic properties. Oncogene 18**:**3608-3616. [DOI] [PubMed] [Google Scholar]
- 10.Gong, L., T. Kamitani, K. Fujise, L. S. Caskey, and E. T. H. Yeh. 1997. Preferential interaction of Sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272**:**28198-28201. [DOI] [PubMed] [Google Scholar]
- 11.Gong, L., S. Millas, G. G. Maul, and E. T. H. Yeh. 2000. Differential regulation of sentrinized proteins by a novel Sentrin-specific protease. J. Biol. Chem. 275**:**3355-3359. [DOI] [PubMed] [Google Scholar]
- 12.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18**:**6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hari, K. L., K. R. Cook, and G. H. Karpen. 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15**:**1334-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong, H., K. Kohli, M. J. Garabedian, and M. R. Stallcup. 1997. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17**:**2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. H. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147**:**221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102**:**549-552. [DOI] [PubMed] [Google Scholar]
- 17.Joazeiro, C. A. P., S. S. Wing, H.-K. Huang, J. D. Leverson, T. Hunter, and Y.-C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286**:**309-312. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272**:**26799-26802. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, E. S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147**:**981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106**:**735-744. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16**:**5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8**:**713-718. [DOI] [PubMed] [Google Scholar]
- 23.Kallio, P. J., H. Poukka, A. Moilanen, O. A. Jänne, and J. J. Palvimo. 1995. Androgen receptor-mediated transcriptional regulation in the absence of direct interaction with a specific DNA element. Mol. Endocrinol. 9**:**1017-1028. [DOI] [PubMed] [Google Scholar]
- 24.Karvonen, U., P. J. Kallio, O. A. Jänne, and J. J. Palvimo. 1997. Interaction of androgen receptors with androgen response element in intact cells. J. Biol. Chem. 272**:**15973-15979. [DOI] [PubMed] [Google Scholar]
- 25.Kotaja, N., S. Aittomäki, O. Silvennoinen, J. J. Palvimo, and O. A. Jänne. 2000. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol. Endocrinol. 14**:**1986-2000. [DOI] [PubMed] [Google Scholar]
- 26.Kotaja, N., M. Vihinen, O. A. Jänne, and J. J. Palvimo. 2002. Androgen receptor-interacting protein 3 and other PIAS proteins cooperate with glucocorticoid receptor-interacting protein 1 in steroid receptor-dependent signaling. J. Biol. Chem. 277**:**17781-17788. [DOI] [PubMed]
- 27.Li, S.-J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398**:**246-251. [DOI] [PubMed] [Google Scholar]
- 28.Liu, B., J. Liao, R. Xiaoping, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95**:**10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96**:**11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88**:**97-107. [DOI] [PubMed] [Google Scholar]
- 31.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16**:**591-626. [DOI] [PubMed] [Google Scholar]
- 32.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2000. Covalent modification of p73α by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275**:**36316-36323. [DOI] [PubMed] [Google Scholar]
- 33.Moilanen, A.-M., U. Karvonen, H. Poukka, O. A. Jänne, and J. J. Palvimo. 1998. Activation of androgen receptor function by a novel nuclear protein kinase. Mol. Biol. Cell 9**:**2527-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moilanen, A.-M., U. Karvonen, H. Poukka, W. Yan, J. Toppari, O. A. Jänne, and J. J. Palvimo. 1999. A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J. Biol. Chem. 274**:**3700-3704. [DOI] [PubMed] [Google Scholar]
- 35.Moilanen, A.-M., H. Poukka, U. Karvonen, M. Häkli, O. A. Jänne, and J. J. Palvimo. 1998. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. 18**:**5128-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller, S., M. Berger, F. Lehembre, J.-S. Seeler, Y. Haupt, and A. Dejean. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275**:**13321-13329. [DOI] [PubMed] [Google Scholar]
- 37.Müller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell. Biol. 2**:**202-210. [DOI] [PubMed] [Google Scholar]
- 38.Müller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17**:**61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palvimo, J. J., P. J. Kallio, T. Ikonen, M. Mehto, and O. A. Jänne. 1993. Dominant negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol. Endocrinol. 7**:**1399-1407. [DOI] [PubMed] [Google Scholar]
- 40.Palvimo, J. J., P. Reinikainen, T. Ikonen, P. J. Kallio, A. Moilanen, and O. A. Jänne. 1996. Mutual transcriptional interference between RelA and androgen receptor. J. Biol. Chem. 271**:**24151-24156. [DOI] [PubMed] [Google Scholar]
- 41.Poukka, H., P. Aarnisalo, U. Karvonen, J. J. Palvimo, and O. A. Jänne. 1999. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 274**:**19441-19446. [DOI] [PubMed] [Google Scholar]
- 42.Poukka, H., U. Karvonen, O. A. Jänne, and J. J. Palvimo. 2000. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 97**:**14145-14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rödel, B., K. Tavassoli, H. Karsunky, T. Schmidt, M. Bachmann, F. Schaper, P. Heinrich, K. Shuai, H. P. Elsässer, and T. Möröy. 2000. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 19**:**5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276**:**12654-12659. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez, M. S., J. M. P. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response to p53. EMBO J. 18**:**6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15**:**3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampson, D., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276**:**21664-21669. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz, S. E., K. Matuschewski, D. Liakopoulos, M. Schefner, and S. Jentsch. 1998. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the Ubc9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95**:**560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seufert, W., B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373**:**78-81. [DOI] [PubMed] [Google Scholar]
- 50.Skowyra, D., D. M. Koepp, T. Kamura, N. M. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284**:**662-665. [DOI] [PubMed] [Google Scholar]
- 51.Sternsdorf, T., K. Jensen, B. Reich, and H. Will. 1999. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274**:**12555-12566. [DOI] [PubMed] [Google Scholar]
- 52.Strunnikov, A. V., L. Aravind, and E. V. Koonin. 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158**:**95-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki, T., A. Ichiyama, H. Saitoh, T. Kawakami, M. Omata, C. H. Chung, M. Kimura, N. Shimbara, and K. Tanaka. 1999. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J. Biol. Chem. 274**:**31131-31134. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi, K., T. Taira, T. Niki, C. Seino, S. M. M. Iguchi-Ariga, and H. Ariga. 2001. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASxα to the receptor. J. Biol. Chem. 276**:**37556-37563. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi, Y., T. Kahyo, A. Toh-e, H. Yasuda, and T. Kikuchi. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3-ligase for septin components and functions as an adaptor between conjugating enzymes and substrates. J. Biol. Chem. 276**:**48973-48977. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi, Y., A. Toh-e, and T. Kikuchi. 2001. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275**:**223-231. [DOI] [PubMed] [Google Scholar]
- 57.Tan, J.-A., S. H. Hall, G. Katherine, K. G. Hamil, G. Grossman, P. Petrusz, J. Liao, K. Shuai, and F. S. French. 2000. Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol. Endocrinol. 14**:**14-26. [DOI] [PubMed] [Google Scholar]
- 58.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276**:**35368-35374. [DOI] [PubMed] [Google Scholar]
- 59.Valdez, B. C., D. Henning, L. Perlaky, R. K. Busch, and H. Busch. 1997. Cloning and characterization of Gu/RNA II-binding protein. Biochem. Biophys. Res. Commun. 234**:**335-340. [DOI] [PubMed] [Google Scholar]
- 60.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2**:**169-178. [DOI] [PubMed] [Google Scholar]
- 61.Wu, L., H. Wu, L. Ma, F. Sangiorgi, N. Wu, J. R. Bell, G. E. Lyons, and R. Maxson. 1997. Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech. Dev. 65**:**3-17. [DOI] [PubMed] [Google Scholar]
- 62.Yeh, E. T. H., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248**:**1-14. [DOI] [PubMed] [Google Scholar]
- 63.Zentner, M. D., H. H. Lin, H.-T. Deng, K.-J. Kim, H.-M. Shih, and D. K. Ann. 2001. Requirement for high mobility group protein HMGI-C interaction with STAT3 inhibitor PIAS3 in repression of α-ENaC transcription by Ras activation in salivary epithelial cells. J. Biol. Chem. 276**:**29805-29814. [DOI] [PubMed] [Google Scholar]
- 64.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102**:**533-539. [DOI] [PubMed] [Google Scholar]