Genomic Screen for Vacuolar Protein Sorting Genes in Saccharomyces cerevisiae (original) (raw)
Abstract
The biosynthetic sorting of hydrolases to the yeast vacuole involves transport along two distinct routes referred to as the carboxypeptidase Y and alkaline phosphatase pathways. To identify genes involved in sorting to the vacuole, we conducted a genome-wide screen of 4653 homozygous diploid gene deletion strains of Saccharomyces cerevisiae for missorting of carboxypeptidase Y. We identified 146 mutant strains that secreted strong-to-moderate levels of carboxypeptidase Y. Of these, only 53 of the corresponding genes had been previously implicated in vacuolar protein sorting, whereas the remaining 93 had either been identified in screens for other cellular processes or were only known as hypothetical open reading frames. Among these 93 were genes encoding: 1) the Ras-like GTP-binding proteins Arl1p and Arl3p, 2) actin-related proteins such as Arp5p and Arp6p, 3) the monensin and brefeldin A hypersensitivity proteins Mon1p and Mon2p, and 4) 15 novel proteins designated Vps61p-Vps75p. Most of the novel gene products were involved only in the carboxypeptidase Y pathway, whereas a few, including Mon1p, Mon2p, Vps61p, and Vps67p, appeared to be involved in both the carboxypeptidase Y and alkaline phosphatase pathways. Mutants lacking some of the novel gene products, including Arp5p, Arp6p, Vps64p, and Vps67p, were severely defective in secretion of mature α-factor. Others, such as Vps61p, Vps64p, and Vps67p, displayed defects in the actin cytoskeleton at 30°C. The identification and phenotypic characterization of these novel mutants provide new insights into the mechanisms of vacuolar protein sorting, most notably the probable involvement of the actin cytoskeleton in this process.
INTRODUCTION
Lysosomes have long been recognized as the major site for the degradation of both exogenous and endogenous macromolecules within mammalian cells (De Duve and Wattiaux, 1966; Kornfeld and Mellman, 1989). The biogenesis of lysosomes, however, remains poorly understood. Helpful insights have been gained from the study of the yeast vacuole, which is functionally analogous to the mammalian lysosome. To the extent that they have been compared, the biosynthetic pathways and molecular machineries involved in the formation of mammalian lysosomes and the yeast vacuole appear remarkably conserved (reviewed by Lemmon and Traub, 2000; Mullins and Bonifacino, 2001a). Thus, further studies of the yeast vacuole are likely to provide new insights into the mechanisms of lysosome biogenesis.
The main pathway for the delivery of newly synthesized proteins to the vacuole is referred to as the carboxypeptidase Y (CPY) pathway based on the utilization of this pathway by CPY (reviewed by Burd et al., 1998; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001a). CPY is a soluble hydrolase that normally resides within the vacuole lumen. Another soluble hydrolase, proteinase A (PrA), as well as the membrane-bound hydrolase carboxypeptidase S are also transported to the vacuole via the CPY pathway. This pathway involves transport from the late-Golgi complex (equivalent to the mammalian _trans_-Golgi network) to a prevacuolar compartment (PVC). From the PVC, some proteins are subsequently transferred to the vacuole, whereas others return to the late-Golgi complex for further rounds of transport. Membrane-bound enzymes involved in the processing of pro-α-factor, such as the Kex2p dibasic endopeptidase, are also transported along parts of the CPY pathway as they cycle between the late-Golgi complex and the PVC. A second pathway, referred to as the alkaline phosphatase (ALP) pathway, is followed by the membrane-bound hydrolase ALP and the t-soluble _N_-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) Vam3p and Nyv1p (Burd et al., 1998; Conibear and Stevens, 1998; Reggiori et al., 2000; Mullins and Bonifacino, 2001a). The ALP pathway differs from the CPY pathway in that it uses a different type of Golgi-derived carrier vesicle (Rehling et al., 1999) and bypasses the PVC (Cowles et al., 1997b; Piper et al., 1997) en route to the vacuole.
Genetic screens based on the detection of reduced proteolytic activity of the vacuole (Jones, 1977) or missorting of vacuolar hydrolases to the periplasmic space (Robinson et al., 1988; Rothman et al., 1989) have identified >40 vacuolar protein sorting (VPS) genes. Other screens devised for different purposes uncovered additional genes that, although not termed VPS, also participate in vacuolar protein sorting. The products of most vacuolar protein sorting genes identified to date control transport exclusively along the CPY pathway (reviewed by Burd et al., 1998; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001a). These include proteins involved in transport from the late-Golgi complex to the PVC (e.g., Vps1p, Vps8p, Vps9p, Vps10p, Vps15p, Vps19p, Vps21p, Vps34p, Vps45p, clathrin, the AP-1 complex, Gga1p/Gga2p, Drs2p, and Mvp1p); PVC maturation, including multivesicular body formation (e.g., Vps2p, Vps4p, Vps20p, Vps22p, Vps23p, Vps24p, Vps27p, Vps28p, Vps32p, and Vps36p); and retrieval from the PVC to the Golgi complex (e.g., Vps5p, Vps17p, Vps26p, Vps29p, Vps30p, Vps35p, Vps51p, Vps52p, Vps53p, Vps54p, and Grd19p). The ALP pathway is controlled by a surprisingly simpler machinery, of which the heterotetrameric adaptor protein complex AP-3 is the only specific component known to date (reviewed by Burd et al., 1998; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001a). AP-3 participates, together with Vps39p and Vps41p, in the formation of ALP carriers at the late-Golgi complex (Rehling et al., 1999). The CPY and ALP pathways converge at the step of fusion with the vacuole, for which reason they share a number of gene products involved in vacuole targeting and fusion (e.g., Vam3p, Vam7p, Vps11p, Vps16p, Vps18p, Vps33p, Vps39p, Vps41p, and Ypt7p).
As numerous as the vacuolar protein sorting genes may seem at present, the total number of genes involved in this process may be even higher. Indeed, studies over the past 2 yr alone have shown that mutations in several novel genes, including Mrl1p (Whyte and Munro, 2001b), Ric1p (Siniossoglou et al., 2000; Bensen et al., 2001), Rgp1p (Siniossoglou et al., 2000; Bensen et al., 2001), Did2p (Amerik et al., 2000), Did4p (Amerik et al., 2000), Ccz1p (Kucharczyk et al., 2000), Mos10p (Kranz et al., 2001), and Swa2p (Gall et al., 2000) impair CPY sorting to the vacuole. The steady pace of discovery of new vacuolar protein sorting genes suggests that previous screens were not saturating and that additional genes involved in this process might still remain to be identified.
To obtain a global view of all the genes involved in vacuolar protein sorting, we decided to perform a genome-wide screen for VPS genes in Saccharomyces cerevisiae. The approach used in our study consisted of screening a collection of 4653 homozygous diploid gene deletion strains (Winzeler et al., 1999) for secretion of CPY into the medium. This approach benefited from the fact that most vacuolar protein sorting genes are nonessential in yeast. We identified 146 mutant strains that secreted strong-to-moderate levels of CPY. Of these, only 53 of the corresponding genes had been previously implicated in vacuolar protein sorting. The identity of the remaining 93 genes and the phenotypic characterization of the mutant strains reported herein provide surprising new insights into vacuole biogenesis.
MATERIALS AND METHODS
Media, Strains, and Molecular Biology Procedures
S. cerevisiae cells were grown in yeast extract-peptone-dextrose (YEPD) medium. DNA manipulations and transformation into Escherichia coli DH5α cells were performed by standard protocols. Homozygous diploid deletion strains and Mat α haploid deletion strains were obtained from Research Genetics (Invitrogen, Huntsville, AL) and compared with the corresponding parental strains, BY4743 (diploid), BY4739 (haploid), and BY4742 (haploid). For genotypes see the Research Genetics Web site at http://www.resgen.com/products/YEASTD.php3.
CPY Colony Blots
The CPY colony blot assay was adapted from Roberts et al. (1991). Briefly, yeast strains were transferred from the 96-well plates to 15-cm2 YEPD plates with a pinning tool and placed at 30°C for 2 to 3 d. Once strains had grown in the round patches, they were replica-plated to a fresh YEPD plate and overlaid with a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany), taking care to remove any trapped bubbles. The plates were then incubated at 30°C for 18–22 h. The nitrocellulose membranes were washed several times with double-distilled H2O and then with standard phosphate-buffered saline. The membranes were then subjected to immunoblotting with mouse anti-CPY (Molecular Probes, Eugene, OR) performed as described previously (Mullins and Bonifacino, 2001b) with slight modifications. Briefly, the mouse anti-CPY was used at 1:1000 dilution. Donkey anti-mouse IgG-horseradish peroxidase (Amersham Biosciences, Piscataway, NJ) at 1:5000 dilution was used as the secondary antibody, followed by visualization with enhanced chemiluminescence reagents (PerkinElmer Life Sciences, Boston, MA) according to the manufacturer's protocols.
Metabolic Labeling and Immunoprecipitation
Metabolic labeling of yeast cells with 35S Express label (PerkinElmer Life Sciences), pulse-chase analyses, and immunoprecipitations were performed at 30°C as described by Bonifacino and Dell'Angelica (1998). A 10-min pulse, followed by a 10-min chase were used in all the pulse-chase assays. Immunoprecipitations were performed overnight at 4°C and the immunoprecipitates analyzed by SDS-PAGE and autoradiography. Mouse anti-CPY antibodies were purchased from Molecular Probes. Anti-PrA and rabbit anti-ALP antibodies were the generous gifts of Carol Woolford (Carnegie Mellon University, Pittsburgh, PA) and Tom Stevens (University of Oregon, Eugene, OR), respectively. For α-factor experiments, cells were pulse-labeled at 25°C for 7.5 min to visualize α-factor–processing intermediates (Graham and Emr, 1991) and immunoprecipitated with anti-α-factor (generously provided by Todd Graham, Vanderbilt University, Nashville, TN).
Labeling Yeast Vacuoles with FM4-64
Yeast vacuoles were visualized in vivo by labeling log-phase cells with 80 μM _N_-(3-triethylammoniumpropyl)-4-(_p_-diethyl-aminophenylhexatrienyl) pyridinium dibromide (FM4-64) (Vida and Emr, 1995; Bonangelino et al., 1997). Cells were viewed with a 100× objective lens on an Olympus IX-70 fluorescence microscope (excitation, 560 nm; dichroic mirror at 595 nm; emission, 630 nm) combined with a low level of transmitted light to reveal cell outlines. Images were captured digitally with an IMAGO charge-coupled device camera controlled by TILLvisION software (TILL Photonics, Eugene, OR). Images were processed using Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA).
Labeling Yeast Actin with Oregon-Green Phalloidin
Cells were grown in 5 ml of YEPD at 30°C to midlog phase (OD600 ∼0.5) and subsequently fixed by the addition of 1/9 volumes of 37.5% formaldehyde (final concentration ∼3.7%). This fixation was carried out at room temperature for 1 h with gentle agitation. The fixed cells were harvested and washed twice with phosphate-buffered saline. The cells were then resuspended in 100 μl of phosphate-buffered saline, added Oregon green-phalloidin (Molecular Probes) to 1.1 μM (20 μl of 6.6 μM stock), and incubated at 30°C with agitation in the dark overnight (∼16–20 h.). Labeled cells were viewed with a 100× objective lens on an IX-70 fluorescence microscope (excitation, 485 nm; dichroic mirror at 520 nm; emission, 540 nm; Olympus, Tokyo, Japan). Images were captured and processed as described above.
Analysis of α-Factor Secretion
Halo assays for α-factor secretion were performed essentially as described previously (Mullins and Bonifacino, 2001b). Briefly, the sst1-3 mutant strain RC634 (Mat a) was grown at 30°C to stationary phase in YEPD. Cells were harvested, washed with YEPD and resuspended in fresh medium to a final concentration of ∼9.0 OD600/ml. Then, a 120-μl aliquot of the sst1-3 mutant strain was spread evenly on a YEPD agar plate and used immediately in the assay. Strains to be tested for α-factor secretion were grown to 1–1.5 OD600/ml at 30°C, concentrated to 1 OD600/ml, and serially diluted in YEPD to the concentrations indicated in the figure. Three microliters of each dilution was spotted on plates containing RC634 cells and incubated at 30°C for ∼72 h to visualize the α-factor–induced growth inhibition (i.e., growth halo). Strains that produced no visible halos when spotted on the sst1-3 lawn were retested in the same way by using Xbhb-2C, a Mat α sst2-1 mutant strain, to ensure that these strains were not secreting a-factor.
RESULTS AND DISCUSSION
Screening of a Collection of Gene Deletion Strains for CPY Secretion
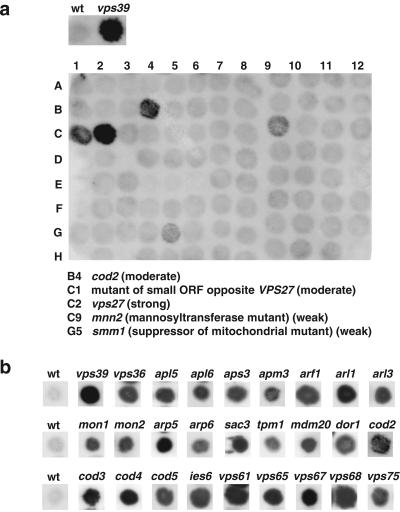
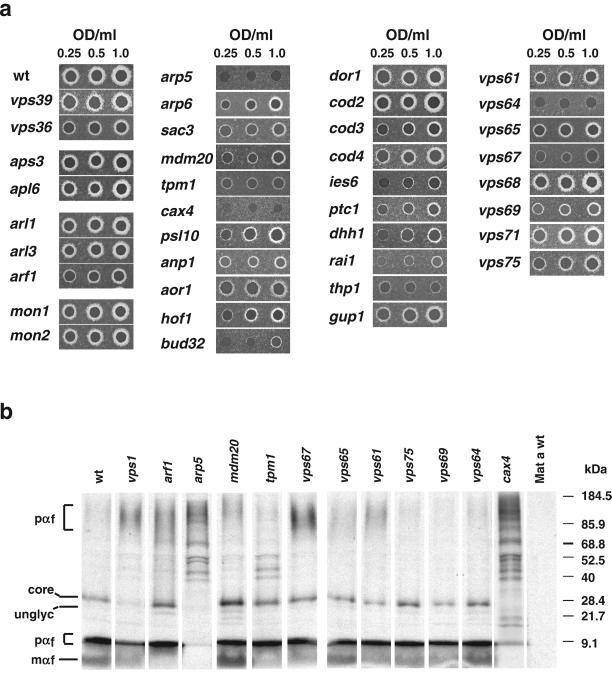
To identify vacuolar protein sorting mutants, we screened a collection of 4653 homozygous diploid deletion strains developed by the Saccharomyces Genome Deletion Project (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html) (Winzeler et al., 1999) and available through Research Genetics (http://www.resgen.com/products/YEASTD.php3). The collection was made by polymerase chain reaction-based disruption of all open reading frames (ORFs) larger than 100 codons in the BY4743 wild-type strain. Because the diploid strains are homozygous for the deletions, only nonessential genes (∼82% of the total) are represented in the collection. The primary screening consisted of analyzing each deletion strain for secretion of CPY by using a colony blotting assay (Roberts et al., 1991). In wild-type cells, CPY is efficiently sorted to the vacuole and therefore is not secreted. In vps mutants, on the other hand, CPY sorting to the vacuole is impaired and different levels of the protein are secreted. Figure 1a, top, shows examples of the CPY colony blotting assay for wild-type (negative control) and vps39, a vps mutant known to secrete high levels of CPY (positive control).
Figure 1.
Identification of CPY-secreting strains. The collection of 4653 homozygous diploid deletion strains developed by the Saccharomyces Genome Deletion Project was screened using a CPY colony blot assay (Roberts et al., 1991). The membranes were subjected to immunoblotting with mouse anti-CPY at 1:1000 dilution. Results for either one 96-well plate (a) or examples of several mutants taken from different blots (b) are shown. Wild-type (wt) and vps39 strains are included for comparison.
Screening of the complete collection resulted in the identification of 362 mutant strains (7.8% of the total) that secreted various amounts of CPY. The strains were categorized as strong (46 strains), moderate (100 strains), and weak (216 strains) based on a visual evaluation of the signal intensity, as exemplified by the colony blots shown in Figure 1, a and b. The majority of known VPS genes and other genes previously shown to be involved in vacuolar protein sorting were found distributed among the strong and moderate categories. For this reason, genes whose deletion gave strong and moderate phenotypes were treated as a single class in all subsequent analyses. Tables 1 and 2 list the genes with strong-moderate and weak phenotypes, respectively, that were identified in the screen. Supplemental Tables 1 and 2 are available online and contain additional information on these genes, their products and the phenotypes of the corresponding mutant strains. Genes in most tables are grouped according to the known or presumed functions of their products.
Table 1.
Genes identified in CPY secretion screen (strong or moderate phenotypes)
| Known VPS Genes (40) |
|---|
| VPS1, VPS3, VPS4, VPS5, VPS6, VPS8, VPS9, VPS10, VPS13, VPS16, VPS17, VPS18, VPS19, VPS20, VPS21, VPS22, VPS23, VPS24, VPS25, VPS26, VPS27, VPS28, VPS29, VPS30, VPS32, VPS33, VPS34, VPS35, VPS36, VPS37, VPS38, VPS39, VPS41, VPS44, VPS45, VPS52, VPS53, VPS54, VPS55, VPS60 |
| Other Genes Implicated in Vacuole Biogenesis or Function (13) |
| CCZ1, DID4, GOS1, MRL1, MVP1, PTC1, RGP1, RIC1, SYS1, TLG2, VAM7, YPT6, YPT7 |
| Vacuolar ATPase (12) |
| VMA1, VMA2, VMA4, VMA6, VMA8, VMA10, VMA11, VMA12, VMA13, VMA21, VMA22, VPH1 |
| Glycosylation (9) |
| FKS1, KRE6, MNN2, MNN9, MNN11, OCH1, OST3, PMT1, VAN1 |
| AP-3 Subunits (4) |
| APL5, APL6, APM3, APS3 |
| ARF1 and ARF-related (3) |
| ARF1, ARL1, ARL3 |
| Monensin and Brefeldin A Hypersensitive (2) |
| MON1, MON2 |
| Actin-related (10) |
| ANP1, AOR1, ARP5, ARP6, CAX4, HOF1, MDM20, PSL10, SAC3, TPM1 |
| Ribosomal Proteins (5) |
| RPL21A, RPL27A, RPL35A, RPP1A, RPS6A |
| Miscellaneous (35) |
| ADH1, ARC1, BDF1, BRO1, BUD14, BUD32, CKB1, COD2, COD3, COD4, COD5, DHH1, DOR1, GSG1, GUP1, GYP1, IES6, IRS4, KRE25, MAK3, MAK10, NPL6, OAR1, PER1, PKR1, PMR1, RAI1, SNA2, RSC2, SNF3, SPF1, STP4, THP1, TOM37, YAF9 |
| Hypothetical ORFs (15) |
| VPS61 (YDR136C), VPS62 (YGR141W), VPS63 (YLR261C), VPS64 (YDR200C), VPS65 (YLR322W), VPS66 (YPR139C), VPS67 (YKR020W), VPS68 (YOL129W), VPS69 (YPR087W), VPS70 (YJR126C), VPS71 (YML041C), VPS72 (YDR485C), VPS73 (YGL104C), VPS74 (YDR372C), VPS75 (YNL246W) |
Table 2.
Genes identified in CPY secretion screen (weak phenotype)
| Vacuole Biogenesis or Function (11) |
|---|
| APG17, AUT4, BPH1, GRD19, IMH1, SNX4, SWA2, VAC14, VAM3, VID21, VID22 |
| Vacuolar ATPase (5) |
| RAV1, RAV2, VMA3, VMA7, VMA5 |
| Glycosylation or Cell Wall Assembly (11) |
| ALG5, CWH36, HOC1, KRE1, KRE11, KRE6, PMT2, RHK1, SMI1, YER083C, YUR1 |
| ARF GAPs (3) |
| AGE2, GCS1, GLO3 |
| Actin-related (3) |
| BNI1, SAC1, SAC7 |
| Brefeldin A Sensitive (3) |
| BRE5, BFR1, ERG4 |
| Protein Trafficking (13) |
| AKL1, AKRI, BST1, CNE1, ERD1, ERV41, LHS1, RUD3, SEC13, SEC22, SEC66, SSH1, YPT31 |
| Ribosomal Proteins (16) |
| RPL11B, RPL12A, RPL13B, RPL16B, RPL17B, RPL2A, RPL36A, RPL37A, RPL39, RPL40A, RPL6A, RPL7A, RPP1B, RPS21B, RPS29A, RPS9B |
| Miscellaneous (102) |
| ADE3, ADOI, APN1, ARO2, ATP11, BAS1, BIM1, BSD2, BTS1, BUD31, CDC10, CHL1, CTF19, CTF4, CTK1, CTK3, CTP1, CYK3, DAL1, DBP7, DOT5, DPP1, EFD1, ELM1, ERG24, ERG28, ESC2, GAS1, GCN5, GDA1, GEF1, GIF1, HCM1, HMT1, HNT3, HPR1, HTA1, HTL1, HTZI, IOC2, ISC1, ISWI, KIM3, KRE20, KRE28, LAS21, LOC1, LPP1, LSM6, LYP1, MAE1, MAL11, MIHI, MND2, MRT4, MSC1, MSL1, MSS18, MUD2, NEM1, NGL2, NIP100, NUP120, NUP84, OSM1, PFD1, PIN2, PLC1, PMC1, POS5, PRO1, RAD61, RAM1, REF2, RLR1, RSC1, SBP1, SGS1, SHP1, SICI, SKN7, SMM1, SNF1, SNF5, SPO7, STO1, SUM1, SWR1, SXM1, TCI1, TDH1, TIF3, UME6, UPF3, URM1, YAK1, YDL033C, YNL080C, YNL099C, YTA7, YVH1, ZMS1 |
| Hypothetical ORFs (47) |
| YAL014C, YAL011W, YAL004W, YLR111W, YML013C-A, YMR299C, YOR292C, YPL184C, YBR266C, YBR246W, YBR267W, YDR105C, YDR426C, YDR425W, YEL043W, YGR206W, YHL029C, YHR111W, YHR199C, YLR181C, YLR225C, YOR135C, YLR374C, YLR381W, YDR186C, YDR203W, YGL198W, YGL072C, YKR035C, YHR009C, YLR435W, YLR426W, YJR079W, YJR100C, YCR050C, YDL096C, YJR018W, YGR064W, YIL067C, YGL020C, YIL110W, YNL136W, YNL127W, YCR033W, YJL075C, YLR350W, YDL119C |
Genes whose deletions gave strong-moderate phenotypes encoded: most known Vps proteins, other proteins previously implicated in vacuolar protein sorting, vacuolar ATPase subunits, components of the glycosylation machinery, AP-3 subunits, Arf1 and Arf-related proteins, actin-related proteins, monensin and brefeldin A hypersensitivity proteins, ribosomal proteins, miscellaneous proteins, and the products of a number of hypothetical ORFs (Table 1 and Supplemental Table 1). Genes whose deletions gave weak phenotypes included members of some of the same groups discussed above, plus genes involved in protein trafficking to compartments other than the vacuole, and a large number of miscellaneous genes and hypothetical ORFs (Table 2 and Supplemental Table 2). Because of the greater likelihood that mutations in genes whose deletions have a weak phenotype could affect vacuolar protein sorting indirectly, we focused our subsequent analyses on deletion strains with strong-moderate CPY secretion.
The CPY-secretion phenotypes of 146 of the strong-moderate strains were confirmed at least twice by colony blotting of the homozygous diploid mutants and the corresponding haploid mutants. All diploid, as well as all but two haploid strains (adh1 and bud14), retested positive in these assays. Seventy-four mutant strains were selected for further analyses. Additional properties analyzed included growth at 30° or 37°C (Table 3); biosynthetic processing of CPY, PrA, and ALP (Figure 2); vacuole morphology (Figure 3); integrity of the actin cytoskeleton (Figure 4); and secretion of processed α-factor (Figure 5). Representative results for some of the mutants are presented in Figures 2–5, and a summary of these results is presented in Tables 3 and 4. The characteristics of selected genes and their products are discussed below.
Table 3.
Phenotypic characteristics of selected CPY-secreting strains
| Strain | CPY secretion | CPY processing | PrA processing | ALP processing | α-Factor halo | Growth at 30°C | Growth at 37°C |
|---|---|---|---|---|---|---|---|
| adh1 | +++ | + | ++ | +++ | mat a | +++ | +++ |
| aor1 | ++ | + | ++ | +++ | +++ | +++ | +++ |
| apl5 | ++ | ++ | ++ | − | +++ | +++ | +++ |
| apl6 | ++ | ++ | ++ | − | +++ | +++ | +++ |
| apm3 | ++ | ++ | ++ | − | +++ | +++ | +++ |
| aps3 | ++ | ++ | ++ | − | +++ | +++ | +++ |
| arf1 | ++ | + | + | + | + | +++ | +++ |
| arl1 | +++ | ++ | +++ | +++ | +++ | +++ | +++ |
| arl3 | ++ | ++ | +++ | +++ | +++ | +++ | +++ |
| arp5 | +++ | + | ++ | +++ | − | ++ | +/− |
| arp6 | ++ | ++ | ++ | +++ | + | +++ | +++ |
| bro1 | ++ | +/− | ++ | +++ | ++ | ++ | + |
| bud14 | ++ | +++ | ++ | +++ | + | +++ | +++ |
| bud32 | ++ | + | + | ++ | +/− | + | − |
| cax4 | +++ | ++ | ++ | ++ | − | + | + |
| ckb1 | ++ | ++ | ++ | +++ | +++ | ++ | + |
| cod2 | +++ | + | ++ | +++ | ++ | +++ | +++ |
| cod3 | +++ | +/− | + | + | +/− | + | + |
| cod4 | +++ | + | ++ | +++ | − | +++ | +++ |
| cod5 | ++ | ++ | ++ | +++ | ++ | + | + |
| dhh1 | ++ | + | ++ | +++ | ++ | +++ | +++ |
| did4 | ++ | + | ++ | +++ | +++ | ++ | ++ |
| dor1 | ++ | + | ++ | +++ | + | +++ | +++ |
| gos1 | +++ | + | + | ++ | +++ | +++ | +++ |
| gup1 | ++ | ND | ND | ++ | ++ | +++ | +++ |
| gyp1 | ++ | + | ++ | +++ | +++ | +++ | +++ |
| hex3 | + | ++ | ++ | +++ | +++ | ++ | ++ |
| hof1 | ++ | ++ | +++ | +++ | ++ | +++ | ++ |
| ies6 | ++ | + | ++ | +++ | − | + | +/− |
| irs4 | +++ | + | ++ | +++ | ++ | +++ | +++ |
| kre25 | ++ | + | + | +++ | +++ | ++ | − |
| mak3 | ++ | ++ | +++ | +++ | +++ | +++ | +++ |
| mak10 | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| mdm20 | ++ | ++ | + | ++ | +/− | + | − |
| mon1 | ++ | − | + | + | +++ | +++ | +++ |
| mon2 | ++ | + | ++ | − | +++ | +++ | + |
| mrl1 | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| mvp1 | ++ | + | ++ | +++ | + | + | + |
| pkr1 | ++ | + | + | +++ | + | +++ | +++ |
| psl10 | ++ | + | ++ | +++ | ++ | +++ | +++ |
| ptc1 | ++ | ++ | ++ | +++ | ++ | ++ | +/− |
| rai1 | ++ | ++ | ++ | +++ | + | ++ | ++ |
| ric1 | ++ | + | ++ | ++ | ++ | +++ | +/− |
| rgp1 | ++ | + | ++ | +++ | ++ | +++ | + |
| rpl27a | ++ | ++ | ++ | +++ | no haploid | ++ | ++ |
| sac3 | ++ | ++ | ++ | +++ | + | +++ | +++ |
| sna2 | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| snf3 | ++ | +++ | ++ | +++ | +++ | +++ | +++ |
| sys1 | ++ | + | ++ | +++ | +++ | +++ | +++ |
| thp1 | ++ | ++ | ++ | +++ | +/− | +++ | +++ |
| tpm1 | + | ++ | ++ | +++ | + | ++ | ++ |
| vps22 | ++ | + | + | +++ | + | +++ | +++ |
| vps36 | ++ | + | + | +++ | ++ | +++ | +++ |
| vps37 | +++ | +/− | + | +++ | ++ | +++ | +++ |
| vps39 | +++ | − | − | − | +++ | +++ | +++ |
| vps44 | +++ | + | + | +++ | + | +++ | +++ |
| vps55 | +++ | + | ++ | +++ | ++ | +++ | +++ |
| vma4 | +++ | +/− | + | +++ | + | + | + |
| vps61 | +++ | + | + | + | ++ | +++ | +/− |
| vps62 | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| vps64 | ++ | ++ | ++ | ++ | − | +++ | +++ |
| vps65 | ++ | + | ++ | +++ | ++ | + | +/− |
| vps66 | ++ | ++ | ++ | +++ | ++ | +++ | +++ |
| vps67 | +++ | +/− | ++ | + | − | + | +/− |
| vps68 | ++ | + | ++ | +++ | ++ | +++ | +++ |
| vps69 | ++ | ++ | ++ | ++ | ++ | ++ | + |
| vps70 | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| vps71 | ++ | + | ++ | +++ | + | +++ | +++ |
| vps72 | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| vps73 | ++ | ++ | +++ | +++ | ++ | +++ | +++ |
| vps74 | ++ | +++ | ++ | +++ | +++ | +++ | +++ |
| vps75 | ++ | ++ | + | ++ | +++ | ++ | ++ |
| yaf9 | ++ | ++ | ++ | +++ | +++ | +++ | ++ |
| WT | − | +++ | +++ | +++ | +++ | +++ | +++ |
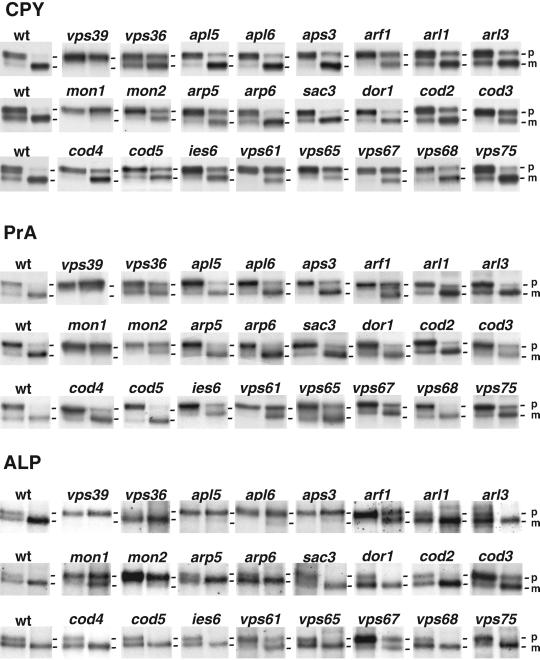
Figure 2.
Analysis of CPY, PrA, and ALP processing. Wild-type (wt) and deletion strains were pulse-labeled with [35S]methionine for 10 min (first lane of each pair) and chased for 10 min (second lane of each pair). Sequential immunoprecipitations were performed from the lysates by using antibodies against CPY (a), PrA (b), or ALP (c) as described in MATERIALS AND METHODS. Results are shown for wild-type (wt) and 24 mutant strains. The positions of precursor (p) and mature (m) forms of the hydrolases examined are indicated. The p1 and p2 precursor forms of CPY were not well resolved for all strains, in some cases due to aberrant Golgi glycosylation that resulted in comigration of both precursor species. A summary of results for these and other strains not shown in this figure is presented in Table 3.
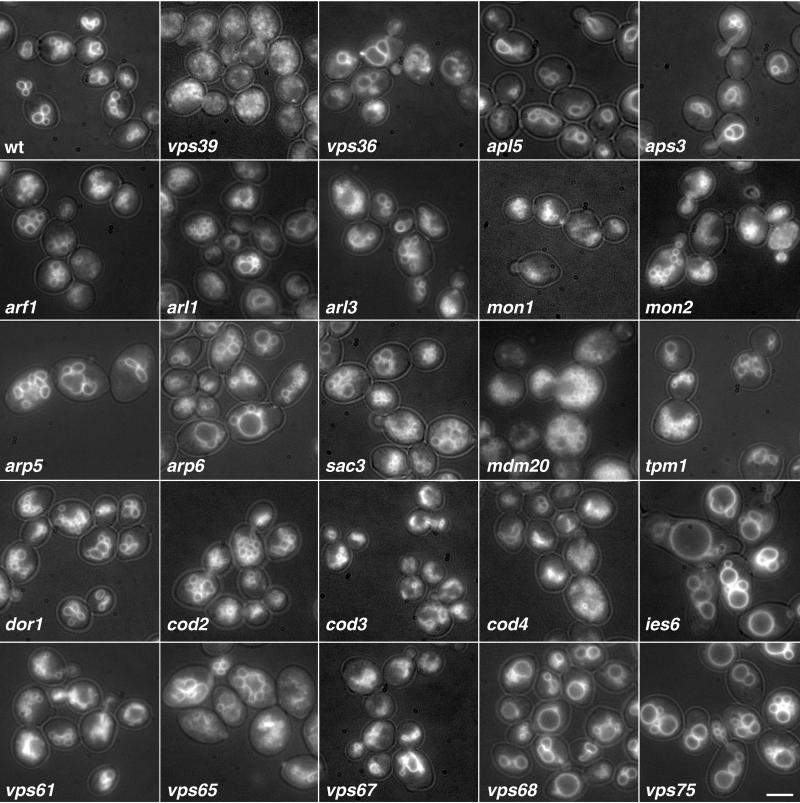
Figure 3.
Visualization of vacuole morphology in wild-type and mutant strains by light microscopy. Vacuole morphology was examined by labeling with FM4-64 as described in MATERIALS AND METHODS. The photographs were taken with both fluorescence and a low level of transmitted light. Results are shown for wild-type (wt), vps39 (as a control for aberrant vacuole morphology), and 23 selected vacuolar sorting mutants identified in our screen. Many of the deletions resulted in fragmented vacuole morphology, which was intermediate, and not as severe as the class B vps mutants (vps39). These include arf1, arl1, arl3, mon1, mon2, mdm20, tpm1, dor1, cod2, cod3, vps61, and vps67. Bar, 9 μm.
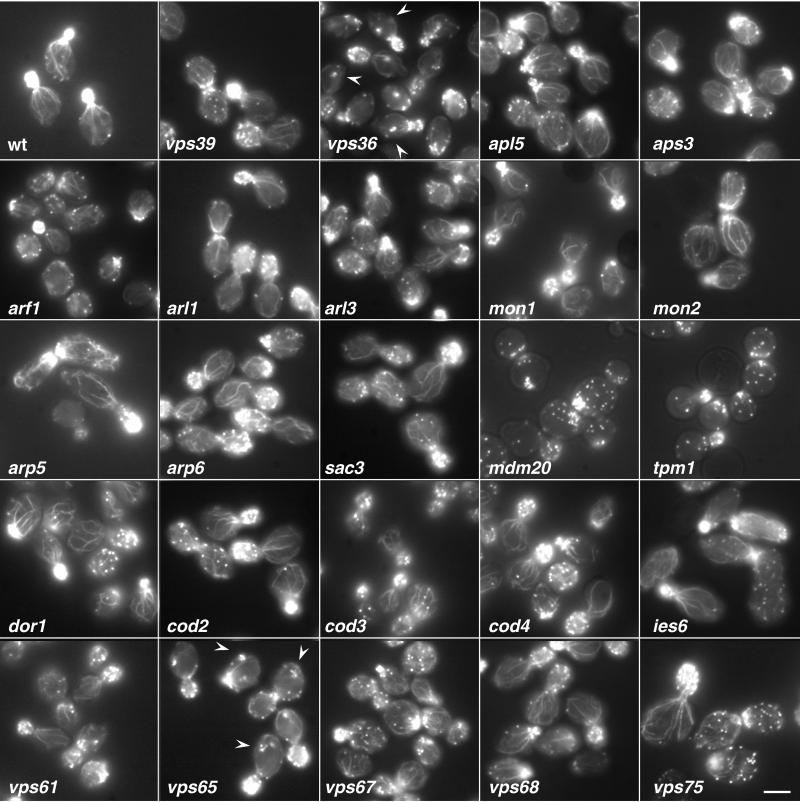
Figure 4.
Visualization of the actin cytoskeleton in wild-type and mutant strains by light microscopy. The actin cytoskeleton was examined by labeling fixed log-phase cells with 1.1 μM Oregon green-phalloidin at 30°C for ∼16–20 h as described in MATERIALS AND METHODS. Results are shown for wild-type (wt), vps39, and 23 selected vacuolar sorting mutants. The most common phenotype observed was the decrease in number or an absence of polarized actin cable in the mother cells, as is seen in arf1, mdm20, tpm1, and vps61. Other phenotypes included reduced polarized actin cables with the appearance of a large actin aggregate in the mother cell, seen in vps36 and vps65 (arrowheads), and slightly disorganized actin filaments, observed in arp5 and arp6. Bar, 9 μm.
Figure 5.
Analysis of α-factor secretion and processing. (a) To test for α-factor secretion, wild-type and mutant strains were grown to log phase and serially diluted to the concentrations indicated, and equal amounts were spotted on YEPD plates containing a freshly spread lawn of the α-factor–sensitive strain RC634 (MAT a, sst1-3), as described in MATERIALS AND METHODS. The plates were then incubated at 30°C for 48 h to assess α-factor secretion as indicated by the relative growth inhibition of the sst1-3 mutant strain (halo). (b) To test for processing of α-factor, strains were metabolically labeled with [35S]methionine at 25°C for 7.5 min as described in MATERIALS AND METHODS. Mature α-factor (mαf), α-factor–processing intermediates and pro-α-factor were immunoprecipitated with anti-α-factor antibodies, and resolved by SDS-PAGE (4–20% acrylamide gradient gels). Various forms of α-factor were detected, including pro-α-factor (pαf) that includes a range of high molecular weight intermediates containing the proregion and various degrees Golgi-derived glycosylation (∼ 90– 150 kDa), unglycosylated core-α-factor (unglyc) (∼20 kDa), ER core-glycosylated α-factor (core) (∼26 kDa), low molecular weight intermediates in α-factor processing (interαf) that most likely represent the tetrapeptide in various states of processing with the proregion removed (∼6–8 kDa), and the mature α-factor (mαf), the 13-amino acid terminally processed secreted form (∼2 kDa).
Table 4.
Summary of phenotypes
| Aberrant Intracellular Trafficking (73 Analyzed): |
|---|
| Affect CPY and PrA Sorting (53): adh1, aor1, arl1, arl3, arp5, arp6, bro1, bud14, ckb1, cod2, cod4, cod5, dhh1, did4, dor1, gyp1, hex3, hof1, ies6, irs4, kre25, mak3, mak10, mrl1, mvp1, pkr1, psl10, ptc1, rai1, rgp1, rpla27, sac3, sna2, snf3, sys1, thp1, tpm1, vma4, vps22, vps36, vps37, vps44, vps55, vps62, vps65, vps66, vps68, vps70, vps71, vps72, vps73, vps74, yaf9 |
| Affect CPY, PrA and ALP Sorting (20): apl5, apl6, apm3, aps3, arf1, bud32, cax4, cod3, gos1, gup1, mdm20, mon1, mon2, ric1, vps39, vps61, vps64, vps67, vps69, vps75 |
| Affect α-factor secretion (41): arf1, arp5, arp6, bro1, bud14, bud32, cax4, cod2, cod3, cod4, cod5, dhh1, dor1, gup1, hof1, ies6, irs4, mdm20, mvp1, pkr1, psl10, ptc1, rai1, rgp1, sac3, thp1, tpm1, vma4, vps22, vps36, vps37, vps44, vps55, vps61, vps64, vps66, vps67, vps68, vps69, vps71, vps73 |
| Aberrant vacuole morphology (71 Analyzed): |
| Class B (35): aor1, arf1, arl1, arl3, bud14, ckb1, cod2, cod4, cod5, dhh1, dor1, gos1, hof1, ies6, irs4, mdm20, mon1, mon2, pkr1, ptc1, ric1, rgp1, snf3, sys1, thp1, tpm1, vps61, vps64, vps66, vps67, vps69, vps71, vps72, vps73, vps75 |
| Class C (2): cax4, gup1 |
| Class D (1): bud32 |
| Class E (4): vps22, vps36, vps37, vps44 |
| Class F (16): arp5, arp6, bro1, hex3, mak10, mvp1, rai1, sac3, thp1, vma4, vps55, vps62, vps65, vps68, vps71, yaf9 |
| Aberrant Actin Cytoskeleton at 30°C (71 Analyzed): |
| Disorganized actin cables (2): arp5, arp6 |
| Reduced number of polarized actin cables (9): anp1, aor1, arf1, cax4, hof1, mdm20, psl10, tpm1, and vps52 |
| Reduced actin cables and aberrant actin patches (4): vps36, vps61, vps65, vps67 |
Known VPS Genes and Other Genes Previously Implicated in Vacuolar Protein Sorting
Forty genes identified in our screen had been previously reported in the literature or listed in the Saccharomyces Genome Database as VPS genes (Table 1 and Supplemental Table 1). Of the VPS genes characterized to date (reviewed by Burd et al., 1998; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001a), only VPS11 and_VPS15_ were not identified in our screen, but this was because these genes were not represented in the deletion collection. The recovery of virtually all known VPS genes attests to the accuracy of the collection and the reliability of the CPY secretion assay. From this, we infer that the majority of the other genes have been accurately identified. Thirteen other genes (CCZ1, DID4, GOS1, MRL1, MVP1, PTC1, RGP1, RIC1, SYS1, TLG2, VAM7, YPT6, YPT7) had also been previously implicated directly or indirectly in vacuolar protein sorting (Table 1 and Supplemental Table 1). These known genes will not be further discussed herein, except for their relationship to other genes of interest.
Vacuolar ATPase
Twelve genes whose deletions resulted in strong-moderate phenotypes (Table 1 and Supplemental Table 1) and six genes whose deletions gave weak phenotypes (Table 2 and Supplemental Table 2) encoded either subunits of the vacuolar proton-translocating ATPase or proteins involved in its assembly or regulation (reviewed by Graham et al., 2000). The subunits belonged to both the catalytic V1 subcomplex (Vma1p, Vma2p, Vma4p, Vma5p, Vma7p, Vma8p, Vma10p, and Vma13p) and the proton-translocating V0 subcomplex (Vma3p, Vma6p, Vma11p, and Vph1p). The only remaining subunit of the V0 subcomplex, Vma16p, was not represented in the deletion collection used for the screen. The assembly factors included Vma12p, Vma21p, and Vma22p, which facilitate assembly but are not stable components of either subcomplex. Thus, the full integrity of the vacuolar ATPase appears to be required for efficient CPY sorting to the vacuole. The identification of vacuolar ATPase genes in our screen is in line with previous observations that mutations in the VMA2 gene (Yamashiro et al., 1990) or treatment with the vacuolar ATPase inhibitor, bafilomycin A1 (Klionsky and Emr, 1989), cause partial missorting and secretion of CPY. This points to a possible requirement for acidification of the vacuole and perhaps the PVC for efficient CPY sorting. However, the V0 subcomplex has also been shown to participate directly in vacuole fusion, in a role independent of acidification (Peters et al., 2001); thus, it is possible that this subcomplex may also play a more direct role in CPY sorting.
Glycosylation
Nine genes whose deletions resulted in strong-moderate phenotypes (Table 1 and Supplemental Table 1) encoded proteins involved in glycosylation or modification of carbohydrate chains. Some of these proteins (e.g., Van1p, Mnn9p, Mnn11p, and Anp1, the latter of which is listed under actin-related proteins) are known to form multiprotein complexes with mannosyl-transferase activity in the Golgi complex (Jungmann and Munro, 1998; Kojima et al., 1999). At present it is unclear how defects in glycosylation could cause missorting of CPY, although it has been reported that unglycosylated CPY is transported more slowly through the secretory pathway (Winther et al., 1991). Thus, it is possible that abnormal glycosylation of CPY or transmembrane components of the vacuolar protein sorting machinery (e.g., Vps10p, Mrl1p) could decrease the efficiency of sorting to the vacuole. Given that glycosylating enzymes are also involved in the biosynthesis of cell wall materials, it is also possible that their deficiency renders the cell wall fragile and causes lytic release of vacuolar CPY.
AP-3
We were surprised to find that strains with deletions of the genes encoding the four subunits of the AP-3 complex (Apl5p, Apl6p, Apm3p, and Aps3p) (Cowles et al., 1997a; Panek et al., 1997; Stepp et al., 1997) secreted moderate levels of CPY (Figure 1b and Table 1 and Supplemental Table 1). AP-3 had been shown to be involved in the ALP pathway but not the CPY pathway (Cowles et al., 1997a; Stepp et al., 1997). Pulse-chase analyses revealed delayed processing of the Golgi precursor forms of pro-CPY and pro-PrA in the AP-3 mutant strains, although this was much less severe than that of the strongest vps mutants (Figure 2). Processing of ALP was completely blocked in the AP-3 mutants (Figure 2), as reported previously (Cowles et al., 1997a; Stepp et al., 1997). The detection of a slight CPY sorting defect in our study could be due to the greater sensitivity of the assays or to the different genetic background of the yeast strains used herein. We suspect that this CPY sorting defect could be an indirect consequence of the drastic inhibition of the ALP pathway. The vacuolar t-SNAREs Vam3p and Nyv1p, for example, normally traffic via the ALP pathway, but function in vesicle fusion in the CPY pathway (Darsow et al., 1997; Reggiori et al., 2000). Although, Vam3p and Nyv1p are rerouted through the CPY pathway in AP-3 mutants (Darsow et al., 1998; Reggiori et al., 2000), it is possible that this rerouting does not completely compensate for the loss of the AP-3 pathway.
The apl5, apl6, apm3, and aps3 mutant strains have similar vacuole morphology, with slightly fewer lobes than the wild-type strain and lobes that seem somewhat unfolded (Figure 3). This phenotype, although subtle, may indicate a decrease in vacuole fission, or an increase in vacuole fusion. This may also be an indirect consequence of reduced Vam3p at the vacuole, as factors required for normal vacuole morphology may have difficulty reaching the vacuole under these conditions.
ARF1 and ARF-related Proteins
Three strong-moderate genes encoded the small GTP-binding proteins of the Ras superfamily, Arf1p (ADP-ribosylation factor 1 product), Arl1p (Arf-like 1 product), and Arl3p (Arf-like 3 product) (Table 1 and Supplemental Table 1). In addition, three weak genes encoded the Arf GTPase-activating proteins Age2p, Gcs1p, and Glo3p (Table 2 and Supplemental Table 2). Arf proteins had been previously implicated in the regulation of protein trafficking along the secretory pathway by virtue of their interactions with multiple effectors, including several coat proteins (Donaldson and Jackson, 2000). S. cerevisiae expresses two Arf proteins, Arf1p and Arf2p, of which Arf1p is the most abundant (Stearns et al., 1990). Disruption of both genes is lethal, whereas disruption of only ARF1 is not (Stearns et al., 1990). Although viable, arf1 mutant strains display various phenotypes, including slower kinetics of CPY transport through the secretory pathway (Chen and Graham, 1998; Huang et al., 1999; Yahara et al., 2001).
In agreement with these reports, we found that deletion of the ARF1 gene results in delayed processing of CPY and PrA (Figure 2). Some of this delay could be due to a reduced rate of transport from the endoplasmic reticulum (ER) to the Golgi complex. However, our finding that the _ARF1_-deletion strain secretes CPY (Figure 1) indicates that Golgi-to-vacuole sorting is also compromised in this strain. This interpretation agrees with the observation that ARF1 displays genetic interactions with clathrin and auxilin (Swa2p, also identified in our screen; Table 2 and Supplemental Table 2), two proteins involved in late-Golgi transport events (Chen and Graham, 1998). Moreover, Arfs are involved in the recruitment of the GGA adaptor proteins to the late-Golgi complex (Zhdankina et al., 2001). We also observed delayed processing of ALP in the _ARF1_-deletion strain (Figure 2), which could be due to inhibition of either ER-to-Golgi or Golgi-to-vacuole transport. In mammalian cells, Arf1 and Arf3 regulate the association of AP-3 with membranes (Ooi et al., 1998; Drake et al., 2000), and it is possible that Arf1p could play a similar role in S. cerevisiae. In addition to defects in protein trafficking, the arf1 strain exhibited defects in vacuole morphology, actin cytosketelon, and α-factor secretion. The vacuole morphology of arf1 was fragmented, exhibiting a larger number of vacuole lobes than the wild-type (Figure 3). However, the degree of fragmentation in arf1 was less severe than that observed for vps39, in which protein traffic is more impaired. This suggests that inefficient trafficking of factors required for vacuole fusion or biogenesis may account for the vacuole morphology observed in arf1. We also observed a decrease in the number and thickness of polarized actin filaments in arf1 mutants (Figure 4). Finally, we found that the arf1 mutant strain secreted less mature α-factor, as determined by a halo assay for growth arrest (Figure 5A). This reduced secretion of mature α-factor is likely due to a block in Kex2p recycling to the Golgi complex as glycosylated pro-α-factor accumulates in arf1 (Figure 5B), suggesting a deficiency in α-factor processing.
The three Arf GAPs, Age2p, Gcs1p, and Glo3p, that were identified among the weak secretors might play overlapping roles in the inactivation of yeast Arfs at the late-Golgi complex. In support of this notion, a recent study has shown that yeast mutants lacking both Age2p and Gcs1p function exhibit substantial defects in CPY and ALP sorting to the vacuole (Poon et al., 2001).
S. cerevisiae Arl1p and Arl3p are structurally related to Arf1p (55 and 37% amino acid sequence identity, respectively), but their functions are less well understood. An _ARL1_-deletion strain was previously reported to display normal biosynthetic processing of CPY (Lee et al., 1997). Another study showed that deletion of ARL3 also resulted in normal processing of CPY, but processing of ALP was delayed at the nonpermissive temperature of 15°C (Huang et al., 1999). We observed that _ARL1_- or _ARL3_-deletion strains exhibited secretion of CPY into the medium (Figure 1), delayed processing of both CPY and PrA (Figure 2), and normal processing of ALP (Figure 2) at 30°C. Both arl1Δ and arl3Δ have vacuole morphology defects similar to those of arf1Δ (Figure 3). Unlike Arf1p, however, Arl1p and Arl3p do not seem to be required for a normal polarized actin cytoskeleton (Figure 4) or in α-factor secretion (Figure 5).
These observations place Arl1p and Arl3p in the CPY pathway of biosynthetic sorting to the vacuole. Like Arf1p, Arl1p and Arl3p would be expected to exert their functions through GTP-dependent binding to effector proteins. Human Arl1 has been shown to bind to various putative effector proteins, including one termed SCOCO (for short coiled-coil) (Van Valkenburgh et al., 2001). This protein has two S. cerevisiae homologs, Vps30p and Imh1p, both of which were also identified in our screen (Tables 1 and 2 and Supplemental Tables 1 and 2).
Monensin and Brefeldin A Hypersensitivity Genes
Two genes whose deletions gave strong-moderate phenotypes, MON1 and MON2, and two genes whose deletions gave weak phenotypes, BRE5 and ERG4, identified in our screen had been previously found in a screen for mutant strains hypersensitive to the drugs monensin and/or brefeldin A (Muren et al., 2001). We observed that both _MON1_- and _MON2_-deletion strains secreted CPY (Figure 1). Proteolytic processing of CPY and PrA was completely blocked in mon1, as in the strongest of the vps mutants (Figure 2). Processing of ALP was partially blocked in mon1 (Figure 2). Processing of all three vacuolar enzymes was also impaired in the mon2 strain, but the pattern differed from that of the mon1 strain in that processing of CPY and PrA was less affected than that of ALP (Figure 2). Interestingly, the block in ALP processing in mon2 was similar to that observed in vps39 and the AP-3 mutants (Figure 2).
Consistent with defects in Golgi-to-vacuole traffic, both mon1 and mon2 have fragmented vacuoles (Figure 3). However, the vacuoles in mon1 are smaller and tend to remain clustered together (Figure 3), whereas those of mon2 are slightly larger but more dispersed (Figure 3). The distinct vacuolar phenotypes of these mutants may reflect their preferential function in the CPY and ALP pathways, respectively. Neither Mon1p nor Mon2p is required for secretion of α-factor (Figure 5).
A noteworthy feature of Mon2p is a 115-amino acid segment of homology (26% amino acid sequence identity) to Sec7p, a guanine nucleotide exchange factor (GEF) for Arf (Peyroche et al., 1996). The homologous segment, however, does not overlap with the so-called Sec7 domain, which has intrinsic Arf GEF activity (Chardin et al., 1996).
Actin-related Proteins
Another intriguing finding was the identification in our screen of 10 genes (ANP1, AOR1, ARP5, ARP6, CAX4, HOF1, MDM20, PSL10, SAC3, and TPM1) encoding proteins that are structurally or functionally related to actin (Table 1 and Supplementary Table 1). Arp5p and Arp6p are two of 10 actin-related proteins identified in S. cerevisiae (Schafer and Schroer, 1999). In addition to secreting CPY (Figure 1), arp5 and arp6 mutant strains displayed delayed processing of CPY and PrA (arp5 more than arp6), but not ALP (Figure 2), placing them in the CPY pathway. Both arp5 and arp6 have similar defects in vacuolar morphology, often displaying a large vacuole lobe surrounded by much smaller lobes (Figure 3). Furthermore, both mutant strains were defective in secretion of mature α-factor (Figure 5), defects that seem to stem from a delay in α-factor processing indicated by the accumulation of pro-α-factor and α-factor–processing intermediates in these strains (Figure 5B; our unpublished data). Neither Arp5 nor Arp6 has severe defects in the actin cytoskeleton structure at 30°C, although polarized actin cables in the mother cells may be slightly more disorganized than those observed in wild type (Figure 4). Other ARPs are components of stable complexes with other proteins (Schafer and Schroer, 1999). Arp2p and Arp3p, for example, are part of a complex that regulates the assembly of actin networks and also participates in endocytosis in S. cerevisiae (Moreau et al., 1997). In mammalian cells, the Arp2/3 complex mediates propulsion of endosomes in the cytoplasm (Taunton et al., 2000). It is thus tempting to speculate that Arp5p and Arp6p could play a role in transport to the vacuole analogous to that of Arp2p and Arp3p in endocytosis.
anp1, psl10, cax4, aor1, and hof1 displayed severe defects in the actin cytoskeleton with few or no polarized actin cables visible in the mother cells [Novick et al., 1989; Kamei et al., 1998; Sekiya-Kawasaki et al., 1998; our unpublished data), similar to those observed for tpm1 and mdm20 (Liu and Bretscher, 1989; Hermann et al., 1997; Figure 4). All of these mutants also exhibited defects in secretion of mature α-factor (Figure 5). cax4, which, like arp5, secretes very little mature α-factor, also accumulates a large amount of α-factor–processing intermediates (Figure 5B). In fact, all the actin mutants analyzed accumulate some of these intermediates, suggesting that the integrity of the actin cytoskeleton may be important for recycling of Kex2 from the prevacuolar compartment back to the Golgi.
The actin cytoskeleton is known to mediate vacuole movements in the process of mitotic vacuolar inheritance (Catlett and Weisman, 2000). Thus, it is possible that some of the actin-related proteins identified in our screen could participate in the translocation of vesicular intermediates on their way to the vacuole. This role could be analogous to the well-established function of the actin cytoskeleton in endocytosis in yeast (Munn, 2001). It is also possible that some of the genes in this group could be more directly involved in vesicle tethering or fusion events and only indirectly involved in association with the actin cytoskeleton. A case in point is the product of the VPS52 gene, a component of the Vps52p-Vps53p-Vps54p complex involved in tethering endosome-derived vesicles to the Golgi apparatus (Conibear and Stevens, 2000; Siniossoglou and Pelham, 2001), which was first identified as a suppressor of actin mutations, Sac2p (Novick et al., 1989). Interestingly, sac2 also displays similar defects in the actin cytoskeleton as the other mutants, exhibiting a decrease in the number of polarized actin cables visible in the mother cells (Novick et al., 1989; our unpublished data).
Ribosomal Proteins
Five strong-moderate genes (Table 1 and Supplemental Table 1) and 16 weak genes (Table 2 and Supplemental Table 2) encoded components of both the large and small ribosome subunits. One intriguing explanation for the secretion of CPY by these mutants would be the existence of a regulatory network that reduces vacuolar protein sorting when translation is impaired. In this regard, RIC1 was initially identified in a screen for mutations that decrease ribosome synthesis (Mizuta et al., 1997), but was later found to encode a protein that, together with the product of RGP1, functions as a GEF for Ypt6p (Siniossoglou et al., 2000; Bensen et al., 2001) (both RIC1 and RGP1 were identified in our screen; Table 1 and Supplemental Table 1). We cannot rule out, however, that decreased translation of one or more key components of the vacuolar protein sorting machinery might underlie the CPY sorting defects observed in ribosomal proteins mutants.
Miscellaneous Proteins
This group includes the products of 35 genes whose deletions resulted in strong-moderate phenotypes but that were previously identified in screens other than for vacuolar protein sorting (Table 1 and Supplemental Table 1). A subgroup of these genes has been implicated in Golgi function. These include GYP1, PMR1, BRO1, DOR1, COD2, COD3, COD4, and COD5. The products of five of these genes (DOR1, COD2, COD3, COD4, and COD5) have recently been shown to be components of an eight-subunit complex with Sec34p and Sec35p (Whyte and Munro, 2001a) referred to as the Sec34/35 complex (Kim et al., 1999; VanRheenen et al., 1999). This complex has been proposed to mediate vesicle tethering to the Golgi complex based on the weak homology of some of its subunits to subunits of the Vps52p-Vps53p-Vps54p (Conibear and Stevens, 2000; Siniossoglou and Pelham, 2001) and exocyst (TerBush et al., 1996) complexes (Whyte and Munro, 2001a). Deletion of genes encoding some of the subunits of the Sec34/35 complex has been shown to result in abnormal accumulation of intracellular membranes and glycosylation defects (Whyte and Munro, 2001a). Other observations suggest that the role of this complex may not be restricted to the Golgi complex. For example, the Golgi-plasma-membrane v-SNARE Snc1p became trapped in internal membranes in dor1 mutants (Whyte and Munro, 2001a). Moreover, a sec34 (also known as grd20) mutant strain exhibited mislocalization of Kex2p and secretion of CPY (Spelbrink and Nothwehr, 1999).
We found that the _DOR1_-, _COD2-, COD3_-, _COD4_-, and _COD5-_deletion strains displayed secretion of CPY (Figure 1) and delayed processing of CPY and PrA (Figure 2). These observations are consistent with the notion that the Sec34/35 complex could play a role in late-Golgi or post-Golgi sorting events. However, only cod3 had a significant delay in ALP processing (Figure 2). In addition, dor1, cod2, cod4, and cod5 had similar vacuolar morphologies, exhibiting fragmented vacuoles (Figure 3), whereas the vacuoles of cod3 were smaller and often exhibited unusual tubular structures (Figure 3). All of the mutants with the exception of cod2 displayed defects in α-factor secretion with cod3 having the most severe defect (Figure 5). The observations that these mutants share only some of the phenotypes and have some distinct phenotypes is consistent with previous reports, and may suggest that some of the proteins have additional roles besides their function in the sec34/35 complex.
Hypothetical ORFs
Fifteen hypothetical ORFs were among the strong-moderate genes (Table 1 and Supplemental Table 1). For convenience, we designated these ORFs VPS61-VPS75. Many of these are small open reading frames (under 300 amino acids) and may have been missed in previous screens that depended on random mutagenesis, as the smaller target size of the genes lowered the probability of them being hit. Of these new putative ORFs, the deletion of two, VPS61 (YDR136C) and VPS67 (YKR020W), resulted in secretion of high levels of CPY (Figure 1) and caused a significant block in the processing of CPY, PrA, and ALP (Figure 2). Interestingly, vps67 has similar vacuolar morphology to cod3, exhibiting small fragmented vacuoles as well as some tubular structures (Figure 3). VPS61 is a small ORF opposite of 5′-untranslated region of RGP1, which has previously been implicated in vacuole protein sorting. Thus, it is still unclear whether deletion of VPS61 is responsible for all or some of the phenotypes observed. However rgp1Δ did not display a defect in ALP processing (our unpublished data), suggesting that the lack of Vps61p may be responsible for this defect. Deletion of the other ORFs resulted in secretion of moderate levels of CPY (Figure 1; and Table 3) as well as defects in CPY and PrA processing that were less severe (Figure 2 and Table 3). In addition, many of them exhibited defects in α-factor secretion and/or processing (Figure 5). vps67 and vps64 (deletion of YDR200C) displayed the most severe phenotype (Figure 5), with little to no mature α-factor being secreted. Intriguingly, although vps67 accumulates α-factor–processing intermediates (Figure 5B), vps64, which has a more severe secretion defect, exhibits almost normal levels of mature α-factor and α-factor–processing intermediates compared with wild type. Although in many of the strains analyzed, aberrant Kex2p localization and recycling to the Golgi compartment may result in delayed processing of α-factor and thus indirectly cause an α-factor secretion defect, this is not always the case. Some of the proteins involved in vacuole protein sorting, such as Vps64p, may be directly involved in the secretion of mature α-factor as well.
Of the 15 new ORFs, seven displayed some homology to mammalian gene products. However, only Vps74p (product of YDR372C) exhibited homology to a known protein, having ∼40% amino acid sequence identity to a mammalian protein termed GMx33 (Wu et al., 2000) or GPP34/Golgi phosphoprotein 3 (Bell et al., 2001).
Because several actin-related proteins were found to be important for CPY sorting in our screen, it seemed possible that some of the new ORFs could encode proteins involved in the organization or stability of the actin cytoskeleton. Thus, we examined the actin cytoskeleton through staining F-actin with Oregon green-phalloidin in all of the new open reading frame deletions, as well as some additional control strains (Figure 4; our unpublished data). Of the new ORFs examined, only vps61, vps65 (deletion of YLR322W), and vps67 displayed defects in the actin cytoskeleton at 30°C. Interestingly, both vps36 and vps65 had a similar staining pattern, displaying a greatly reduced number of polarized actin cables as well as a large aggregate (indicated by arrowheads) in the mother cells (Figure 4). Although it is still unclear whether this aberrant structure is associated with any cellular organelles or whether it is simply a cytosolic actin aggregate, these observations further confirm that the need to establish a normal actin cytoskeleton may be important in intracellular Golgi-to-vacuole trafficking. Further characterization of these proteins and their interaction with the actin cytoskeleton will help to clarify the role of actin in trafficking to the vacuole.
Concluding Remarks
Many more genes had been previously implicated in the CPY pathway than in the ALP pathway (reviewed by Burd et al., 1998; Conibear and Stevens, 1998; Lemmon and Traub, 2000; Mullins and Bonifacino, 2001a). In fact, only the genes encoding the four subunits of AP-3 were known to be specifically involved in the ALP pathway (Cowles et al., 1997a; Stepp et al., 1997). Other gene products that controlled the ALP pathway were shared with the CPY pathway. These included Vps39p and Vps41p, which play dual roles in the budding of AP-3–coated intermediates from the late-Golgi complex (Rehling et al., 1999; Darsow et al., 2001) and tethering/fusion of both CPY and ALP carriers to the vacuole (Price et al., 2000; Wurmser et al., 2000). They also included other proteins involved in vesicle formation in the late-Golgi complex and tethering to the vacuole (reviewed by Burd et al., 1998; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001a). More than two-thirds of all known VPS genes, however, were specific to the CPY pathway.
The work reported herein resulted in the identification of many new genes involved in vacuolar protein sorting. The fact remains, however, that the majority of them controls trafficking along the CPY pathway (summarized in Table 4). This could reflect a bias due to the use of CPY secretion as the assay for identification of the mutants. More likely, however, is that the overabundance of CPY pathway genes reflects the complex roles of the PVC in this pathway. The interposition of the PVC between the Golgi complex and the vacuole enables the recycling of cargo receptors such as the CPY and PrA receptor, Vps10p. In addition, the PVC allows for certain membrane proteins to be transported into the vacuole lumen through invagination of its limiting membrane (Odorizzi et al., 1998; Katzmann et al., 2001). This process is important for the delivery of some vacuole resident proteins (e.g., carboxypeptidase S) into the lumen and for the regulated turnover of proteins whose primary function is in other compartments (e.g., Kex2p).
Our studies did nonetheless identify several novel proteins involved in the ALP pathway. Mon2p was particularly interesting because its absence affected the ALP pathway more than the CPY pathway. The absence of Mon1p, Cod3p, Vps61p, and Vps67p also impaired both pathways, with the CPY pathway being more affected than the ALP pathway. These proteins could thus mediate either formation of both CPY and ALP carriers at the late-Golgi complex or tethering/fusion of Golgi-derived ALP carriers or PVC-derived CPY carriers to the vacuole. It is likely that screens specifically designed for analysis of ALP, Vam3p, or Nyv1p missorting could result in the identification of novel ALP pathway genes. Given that even AP-3 subunit mutants secrete some CPY, however, it is plausible that some ALP pathway genes may be represented among the “weak” genes identified in our screen (Table 2 and Supplemental Table 2).
We do not expect the list of genes identified in our study to represent the complete repertoire of vacuolar protein sorting genes, for the following reasons: 1) Some VPS genes might be essential and therefore would not be represented in the collection. For example, Sec18p is the product of an essential gene that plays a role in vesicle fusion in various pathways, including transport to the vacuole (Graham and Emr, 1991). 2) Other VPS genes might be redundant, as is the case for those encoding Gga1p and Gga2p (Dell'Angelica et al., 2000; Hirst et al., 2000; Costaguta et al., 2001; Zhdankina et al., 2001). 3) Some knockout strains might cease secretion of CPY due to an adaptive or compensatory response, as is the case for clathrin mutants (Seeger and Payne, 1992). 4) Genes <100 codons were not targeted for deletion. 5) Manifestation of the CPY secretion phenotype might be dependent on the genetic background of the parental strains. Most of the deletion mutants used herein were made in the BY4743 background, whereas most previously characterized vps mutants were derived from other strains (e.g., SEY6210, SEY6211, and SF839–1D). 6) Some genes may not have been deleted due to technical problems. For example, vps11 and vps15 mutants are not present in the collection. 7) Some genes could have been erroneously identified, or the mutants mishandled. Given the high accuracy with which VPS genes were identified in our screen, however, we suspect that the cases of mistaken identity are few.
Our studies support the conclusion that the vacuolar protein sorting machinery is highly conserved between yeast and higher eukaryotes. BLAST searches by using the strong and moderate genes as queries revealed that >50% of them have mammalian homologs. This percentage is considerably higher than the 31% of all yeast genes that have definite homologs among mammals (Botstein et al., 1997). This confirms the presumption that studies of vacuolar protein sorting in yeast are particularly relevant to the understanding of lysosome biogenesis in mammals. The biochemical and functional characterization of the novel gene products identified in this study should considerably further our understanding of the molecular mechanisms that underlie the biogenesis of the yeast vacuole and mammalian lysosomes.
Supplementary Material
MBC Supplementary Material
ACKNOWLEDGMENTS
We thank Carol Woolford, Tom Stevens, and Todd Graham for generous gifts of reagents; Peter Rubenstein, Kuo-Kuang Wen, and Runa Musib for discussion of actin related genes and protocols; and Cathy Jackson and Chris Mullins for helpful discussions and critical review of the manuscript. C. Bonangelino was supported by Pharmacology Research Associates Training Award from the NIGMS.
Footnotes
D⃞
Online versions of this article contain complete data sets.
REFERENCES
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AW, et al. Proteomics characterization of abundant Golgi membrane proteins. J Biol Chem. 2001;276:5152–65. doi: 10.1074/jbc.M006143200. [DOI] [PubMed] [Google Scholar]
- Bensen ES, Yeung BG, Payne GS. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol Biol Cell. 2001;12:13–26. doi: 10.1091/mbc.12.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Catlett NL, Weisman LS. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol Cell Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Dell'Angelica EC. Immunoprecipitation. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada K, editors. Current Protocols in Cell Biology. New York: John Wiley & Sons; 1998. [Google Scholar]
- Botstein D, Chervitz SA, Cherry JM. Yeast as a model organism. Science. 1997;277:1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Babst M, Emr SD. Novel pathways, membrane coats and PI kinase regulation in yeast lysosomal trafficking. Semin Cell Dev Biol. 1998;9:527–533. doi: 10.1006/scdb.1998.0255. [DOI] [PubMed] [Google Scholar]
- Catlett NL, Weisman LS. Divide and multiply: organelle partitioning in yeast. Curr Opin Cell Biol. 2000;12:509–516. doi: 10.1016/s0955-0674(00)00124-1. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Chen CY, Graham TR. An arf1Delta synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics. 1998;150:577–589. doi: 10.1093/genetics/150.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997a;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997b;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Burd CG, Emr SD. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J Cell Biol. 1998;142:913–922. doi: 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Katzmann D, Cowles CR, Emr SD. Vps41p function in the alkaline phosphatase pathway requires homo-oligomerization and interaction with AP-3 through two distinct domains. Mol Biol Cell. 2001;12:37–51. doi: 10.1091/mbc.12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- Drake MT, Zhu Y, Kornfeld S. The assembly of AP-3 adaptor complex-containing clathrin-coated vesicles on synthetic liposomes [In Process Citation] Mol Biol Cell. 2000;11:3723–3736. doi: 10.1091/mbc.11.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Higginbotham MA, Chen C, Ingram MF, Cyr DM, Graham TR. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]
- Graham LA, Powell B, Stevens TH. Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J Exp Biol. 2000;203:61–70. doi: 10.1242/jeb.203.1.61. [DOI] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–53. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Buu LM, Yu WL, Lee FJ. Characterization of a novel ADP-ribosylation factor-like protein (yARL3) in Saccharomyces cerevisiae. J Biol Chem. 1999;274:3819–3827. doi: 10.1074/jbc.274.6.3819. [DOI] [PubMed] [Google Scholar]
- Jones EW. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kim DW, Sacher M, Scarpa A, Quinn AM, Ferro-Novick S. High-copy suppressor analysis reveals a physical interaction between Sec34p and Sec35p, a protein implicated in vesicle docking. Mol Biol Cell. 1999;10:3317–3329. doi: 10.1091/mbc.10.10.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Hashimoto H, Yoda K. Interaction among the subunits of Golgi membrane mannosyltransferase complexes of the yeast Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1999;63:1970–1976. doi: 10.1271/bbb.63.1970. [DOI] [PubMed] [Google Scholar]
- Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Kranz A, Kinner A, Kolling R. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol Biol Cell. 2001;12:711–723. doi: 10.1091/mbc.12.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk R, Dupre S, Avaro S, Haguenauer-Tsapis R, Slonimski PP, Rytka J. The novel protein Ccz1p required for vacuolar assembly in Saccharomyces cerevisiae functions in the same transport pathway as Ypt7p. J Cell Sci. 2000;113:4301–4311. doi: 10.1242/jcs.113.23.4301. [DOI] [PubMed] [Google Scholar]
- Lee FJ, Huang CF, Yu WL, Buu LM, Lin CY, Huang MC, Moss J, Vaughan M. Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J Biol Chem. 1997;272,:30998–31005. doi: 10.1074/jbc.272.49.30998. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Mizuta K, Park JS, Sugiyama M, Nishiyama M, Warner JR. RIC1, a novel gene required for ribosome synthesis in Saccharomyces cerevisiae. Gene. 1997;187:171–178. doi: 10.1016/s0378-1119(96)00740-8. [DOI] [PubMed] [Google Scholar]
- Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS. The molecular machinery for lysosome biogenesis. Bioessays. 2001a;23:333–343. doi: 10.1002/bies.1048. [DOI] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS. Structural requirements for function of yeast GGAs in vacuolar protein sorting, alpha-factor maturation, and interactions with clathrin. Mol Cell Biol. 2001b;21:7981–7994. doi: 10.1128/MCB.21.23.7981-7994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL. Molecular requirements for the internalization step of endocytosis: insights from yeast. Biochim Biophys Acta. 2001;1535:236–257. doi: 10.1016/s0925-4439(01)00028-x. [DOI] [PubMed] [Google Scholar]
- Muren E, Oyen M, Barmark G, Ronne H. Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast. 2001;18:163–172. doi: 10.1002/1097-0061(20010130)18:2<163::AID-YEA659>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek HR, Stepp JD, Engle HM, Marks KM, Tan PK, Lemmon SK, Robinson LC. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Nothwehr SF, Singer RA, Johnston GC. The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J Cell Biol. 2001;17:17. doi: 10.1083/jcb.200108075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A, Wickner W, Ungermann C. Proteins needed for vesicle budding from the Golgi complex are also required for the docking step of homotypic vacuole fusion. J Cell Biol. 2000;148:1223–1229. doi: 10.1083/jcb.148.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in Class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Black MW, Pelham HR. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol Biol Cell. 2000;11:3737–3749. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Darsow T, Katzmann DJ, Emr SD. Formation of AP-3 transport intermediates requires Vps41 function. Nat Cell Biol. 1999;1:346–353. doi: 10.1038/14037. [DOI] [PubMed] [Google Scholar]
- Roberts, C.J., Raymond, C.K., Yamashiro, C.T., and Stevens, T.H. (1991). Methods for studying the yeast vacuole. Methods Enzymol. 194, [DOI] [PubMed]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Howald I, Stevens TH. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Schroer TA. Actin-related proteins. Annu Rev Cell Dev Biol. 1999;15:341–363. doi: 10.1146/annurev.cellbio.15.1.341. [DOI] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya-Kawasaki M, Botstein D, Ohya Y. Identification of functional connections between calmodulin and the yeast actin cytoskeleton. Genetics. 1998;150:43–58. doi: 10.1093/genetics/150.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyzes nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink RG, Nothwehr SF. The yeast GRD20 gene is required for protein sorting in the trans-Golgi network/endosomal system and for polarization of the actin cytoskeleton. Mol Biol Cell. 1999;10:4263–4281. doi: 10.1091/mbc.10.12.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast AP-3 complex is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J Cell Biol. 2000;148:519–530. doi: 10.1083/jcb.148.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Van Valkenburgh H, Shern JF, Sharer JD, Zhu X, Kahn RA. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J Biol Chem. 2001;276:22826–22837. doi: 10.1074/jbc.M102359200. [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Sapperstein SK, Chiang EC, Lupashin VV, Barlowe C, Waters MG. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J Cell Biol. 1999;147:729–742. doi: 10.1083/jcb.147.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001a;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S. A yeast homolog of the mammalian mannose 6-phosphate receptors contributes to the sorting of vacuolar hydrolases. Curr Biol. 2001b;11:1074–1078. doi: 10.1016/s0960-9822(01)00273-1. [DOI] [PubMed] [Google Scholar]
- Winther JR, Stevens TH, Kielland-Brandt MC. Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivo stability, or activity. Eur J Biochem. 1991;197:681–689. doi: 10.1111/j.1432-1033.1991.tb15959.x. [DOI] [PubMed] [Google Scholar]
- Winzeler, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wu CC, Taylor RS, Lane DR, Ladinsky MS, Weisz JA, Howell KE. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1:963–975. [PubMed] [Google Scholar]
- Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara N, Ueda T, Sato K, Nakano A. Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol Biol Cell. 2001;12:221–238. doi: 10.1091/mbc.12.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro CT, Kane PM, Wolczyk DF, Preston RA, Stevens TH. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol Cell Biol. 1990;10:3737–3749. doi: 10.1128/mcb.10.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdankina O, Strand NL, Redmond JM, Boman AL. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MBC Supplementary Material