SSRP1 functions as a co-activator of the transcriptional activator p63 (original) (raw)
Abstract
The p53 homolog p63 is a transcriptional activator. Here, we describe the identification of an HMG1-like protein SSRP1 as a co-activator of p63. Over expression of wild-type, but not deletion mutant, SSRP1 remarkably enhanced p63γ-dependent luciferase activity, G1 arrest, apoptosis and expression of endogenous PIG3, p21Waf1/cip1 and MDM2 in human p53-deficient lung carcinoma H1299 cells and mouse embryonic fibroblasts. Also, SSRP1 interacted to p63γ in vitro and in cells, and resided with p63γ at the p53-responsive DNA element sites of the cellular endogenous MDM2 and p21Waf1/cip1 promoters. Moreover, N-terminus-deleted p63 (ΔN-p63) bound to neither SSRP1 nor its central domain in vitro. Accordingly, SSRP1 was unable to stimulate ΔN-p63-mediated residual luciferase activity and apoptosis in cells. Finally, the ectopic expression of the central p63-binding domain of SSRP1 inhibited p63-dependent transcription in cells. Thus, these results suggest that SSRP1 stimulates p63 activity by associating with this activator at the promoter.
Keywords: apoptosis and cell cycle/co-activator/p63/SSRP1/transcription
Introduction
The p63 gene, also known as p51, encodes a family of alternatively spliced proteins that were identified based upon its homology with the tumor suppressor p53 (Osada et al., 1998; Yang et al., 1998). The N-terminally intact α, β and γ isoforms of p63 (referred to as p63 herein), like p53, possess trans-activation activity on most of the p53-responsive genes and are able to bind to the p53-responsive DNA elements (p53REs) of these genes in a sequence-specific manner (Osada et al., 1998; Yang et al., 1998). In contrast, the N-terminally truncated isoforms of p63 (referred to as ΔN-p63) lack these p53-like functions and instead act as dominant-negative inhibitors of p63 as well as of the other p53 family members (Osada et al., 1998; Yang et al., 1998). This suggests that the N-terminal domain of p63, similar to that of p53 and of p73 (Ko and Prives, 1996; Levrero et al., 2000; Irwin and Kaelin, 2001; Yang and McKeon, 2000), is crucial for executing its transcriptional activity. However, it remains unclear which nuclear components interact with this domain of p63 to mediate its transcriptional function.
In spite of sharing transcriptional activity with p53, p63 displays a physiologically distinct role. p63 is essential for ectodermal differentiation and development during embryogenesis (Mills et al., 1999; Yang et al., 1999). Knocking out the p63 gene led to death of mice within a week after birth (Mills et al., 1999; Yang et al., 1999), whereas p53 knock-out mice developed normally with an early onset of tumorigenesis in different tissues (Donehower et al., 1992). Human patients harboring point mutations at the central DNA-binding domain of p63 suffer from EEC (ectrodactyly, ectodermal dysplasia and cleft lip) syndromes (Celli et al., 1999; Wessagowit et al., 2000) and split hand/split foot malformation (SHFM) syndrome (Ianakiev et al., 2000) with certain craniofacial and limb defects and changes in tissues similar to those of p63 null baby mice, which show short or no limbs and defects in a variety of tissues (Mills et al., 1999; Yang et al., 1999). The same point mutations of p53, however, are closely linked to human cancers (Hollstein et al., 1994). These differences between p63 and p53 suggest that the two similar transcriptional activators must be regulated through distinct mechanisms. Thus, deciphering the mechanisms for regulating p63-dependent transcriptional activation is also critical for a better understanding of the biological role of p63.
Transcriptional activation is a complex process involving communication between the RNA polymerase II-containing machinery and transcriptional activators, such as p53 or p63, on a nuclear chromatin template. This communication requires intermediary proteins called co-activators (Lewin, 1990). Some of the co-activators, such as TATA box protein-associated factors (TAFs) (Green, 2000) or p300/CBP (Goodman and Smolik, 2000), have been shown to mediate transcriptional activation by many transcriptional activators including the p53 family members (Avantaggiati et al., 1997; Gu et al., 1997; Lill et al., 1997; Zeng et al., 1999, 2000, 2001a; Costanzo et al., 2002). In addition to these co-activators, the high mobility group (HMG) protein family has also been suggested to regulate transcription in cells (Grosschedl et al., 1994; Baxevanis and Landsman, 1995).
HMG proteins were identified originally as a group of non-histone chromatin-associated proteins with a high mobility property on SDS–polyacrylamide gels (Grosschedl et al., 1994; Baxevanis and Landsman, 1995). They all possess a conserved DNA-binding domain of ∼80 amino acids called the HMG box. However, other HMG box-containing proteins with a variety of molecular masses have also been identified (Grosschedl et al., 1994; Baxevanis and Landsman, 1995). Based upon their DNA-binding activity and roles in transcription regulation, HMG proteins are classified into at least two subgroups. One group is able to bind to DNA in a sequence-specific manner, thus acting as transcriptional activators themselves, such as LEF-1 (Giese et al., 1991; Travis et al., 1991), hUBF (McStay et al., 1991) or SRY (Ferrari et al., 1992). The other group (HMG-1/HMG-2) lacks intrinsic transcriptional activity, but is able to influence the activity of other transcriptional activators such as Oct-1, Oct-2, Oct-6, HOXD9 (Dailey et al., 1994), MLTF (Watt and Molloy, 1988) or p53 (Jayaraman et al., 1998). This subgroup is also able to bind to DNA in a structure-specific manner. However, one unique protein is the structure-specific recognition protein (SSRP1), which was identified originally as a cisplatin-modified DNA-binding protein (Bruhn et al., 1992). Although it possesses one HMG-1-like domain and binds to a specific DNA structure (Bruhn et al., 1992), SSRP1 can act as either a transcriptional activator or a co-activator. For instance, SSRP1 was shown to stimulate the transcriptional activity of the serum-responsive transcription factor SRF (Spencer et al., 1999). Also, SSRP1, though lacking an intrinsic activation domain, was reported to bind to the specific DNA element of the human embryonic β-like globin gene and to activate its expression (Dyer et al., 1998). More recently, SSRP1 was found to form a heterodimer with Spt16, facilitating transcriptional elongation on chromatin templates in vitro (Orphanides et al., 1999). These studies suggest that SSRP1 plays multiple roles in transcription regulation.
In our search for p63 regulators, we have observed that SSRP1, when overexpressed, markedly stimulated p63-dependent transcription in transient transfection– luciferase experiments using human p53 null small cell lung carcinoma H1299 cells and mouse p53–/– mouse embryonic fibroblasts (MEFs). Also, SSRP1 remarkably enhanced the p63γ-induced expression of endogenous p53 target genes, such as MDM2 (Wu et al., 1993), PIG3 (Polyak et al., 1997) and p21waf1/cip1 (el-Deiry et al., 1993; Harper et al., 1993) in H1299 cells. Consistently, SSRP1 augmented p63-induced G1 arrest and apoptosis of these cells. The stimulation of p63 activity by SSRP1 is probably mediated through a direct interaction between SSRP1 and p63γ, as they bound to each other both in vitro and in cells. As revealed in chromatin immunoprecipitation (ChIP) assays, SSRP1 co-resided with p63γ at the p53-responsive element of the endogenous MDM2 promoter. Also, SSRP1 associated with p63γ that bound to the p53RE motif in vitro. Domain mapping indicated that the central domain of SSRP1 interacted with the N-terminal domain of p63γ. In line with this, an SSRP1 deletion mutant failed to positively affect p63-dependent transcription and apoptosis in cells. Instead, the p63-binding fragment of SSRP1 inversely inhibited p63-dependent transcription. Also, SSRP1 apparently did not affect the residual activity of ΔN-p63γ. These results demonstrate that SSRP1 serves as a co-activator of the intact isoform of p63.
Results
SSRP1 stimulates the transcription activity of p63γ, but not ΔN-p63γ
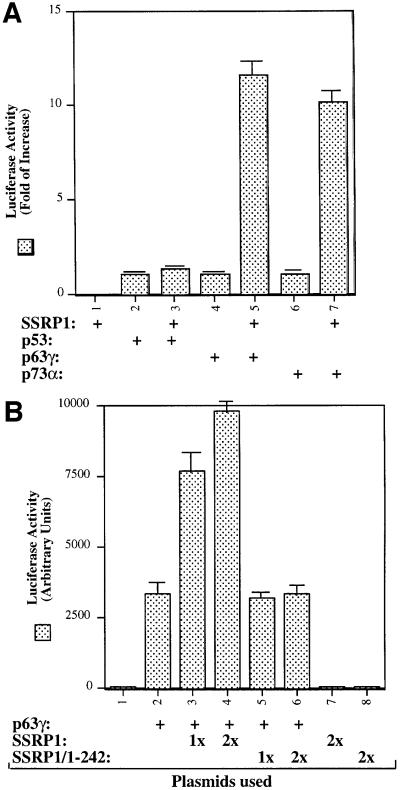
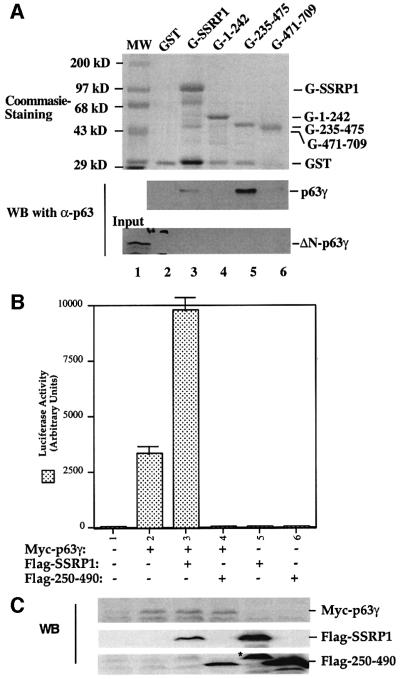
Our recent study reveals SSRP1 as one of the components purified along with the UV-responsive p53 kinase activity (Keller et al., 2001). In our initial attempt to determine whether SSRP1 alone influences the transcription activity of the p53 family members, we performed transient transfection–luciferase experiments and surprisingly found that SSRP1, when overexpressed, stimulated transcription mediated by p63 and by p73, but not by p53 (Figure 1A). Due to the space limitation, this paper primarily describes the regulation of p63 activity by SSRP1. Because our recently identified non-p53 p53RE-binding protein is p63γ (Zeng et al., 1998, 2001b), which is the most transcriptionally active isoform among the p63 family members (Yang et al., 1998), we decided to focus on this p63 isoform in this study. First, we transfected human p53 null small cell lung carcinoma H1299 cells, which contain a low level of p63, with mammalian expression vectors encoding Myc-tagged p63γ alone or together with Flag-tagged SSRP1 in the presence of a luciferase reporter plasmid driven by the p53RE derived from the MDM2 promoter (Wu et al., 1993), and then carried out luciferase assays. As shown in Figure 1B, ectopic expression of SSRP1 in H1299 cells markedly stimulated p63γ-dependent luciferase activity in a dose-dependent manner. This stimulation was due neither to the change of p63 level when SSRP1 was overexpressed (left panel of Figure 1D), nor to a general effect of SSRP1 on transcription, because SSRP1 failed to produce this stimulation in the absence of the exogenous p63γ (Figure 1B) or in the presence of ΔN-p63γ, which retained a residual transcriptional activity (Figure 1C) (Yang et al., 1998) and differentially regulated some p53 target genes (Dohn et al., 2001). This also indicates that the N-terminal domain of p63 is essential for the stimulation of p63γ activity by SSRP1. In addition, a deletion mutant of SSRP1 retaining only the N-terminal region failed to enhance p63γ-dependent transcription activity (Figure 1B). Similar results were also reproduced in p53 null MEFs (data not shown). Hence, these results indicate that SSRP1 can positively regulate the transcription activity of p63γ and this regulation requires an intact N-terminal domain of p63.
Fig. 1. SSRP1 enhances the transcriptional activity of p63γ and p73α, but not of p53 and ΔN-p63γ. (A) SSRP1 stimulates the transcriptional activity of p63 and p73, but not p53. H1299 cells were transfected with plasmids encoding p53 (50 ng), p63γ (50 ng) or p73α (50 ng) alone or with SSRP1 (750 ng) as indicated, along with reporters as described in Materials and methods. At 48 h post-transfection, cells were harvested for luciferase and β-gal assays. Luciferase activity was normalized by the internal β-gal activity and expressed in fold increase (each column represents the mean activity from three independent assays, and bars show standard deviation). (B) SSRP1, but not its N-terminal fragment, stimulates the p63γ-dependent luciferase activity. H1299 cells were transfected with pCDNA-Myc-p63γ (50 ng), pCDNA3-Flag-SSRP1 (1× 250 ng and 2× 500 ng) and pCDNA3-Flag-SSRP1/1–242 (500 ng) as indicated. Luciferase and β-gal assays were conducted as described above. Luciferase activity was expressed in arbitrary units here. (C) SSRP1 does not stimulate the ΔN-p63γ-dependent luciferase activity. The same transfection was carried out as that described above, except that pCDNA3-Myc-ΔN-p63γ (50 ng) was also used here in comparison with p63γ. A 50 ng aliquot of pCDNA3-Myc-p63γ and 500 ng of pCDNA3-Flag-SSRP1 were used in this experiment. The results shown in (A–C) were reproduced using p53 null H1299 cells and MEFs. (D) Western blot analysis of expression of exogenous p63γ and SSRP1. H1299 cells were transfected as described in the above panels, except that triplicate plates were used in each lane for detection of proteins. A 150 µg aliquot of proteins was loaded directly on an SDS gel. Flag-SSRP1 (F-SSRP1), Flag-SSRP1/1–242 (F-SSRP1/1–242) or Myc-p63γ was detected by western blot analyses using monoclonal anti-Flag or anti-Myc antibodies.
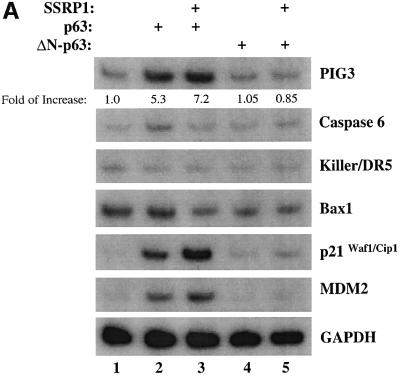
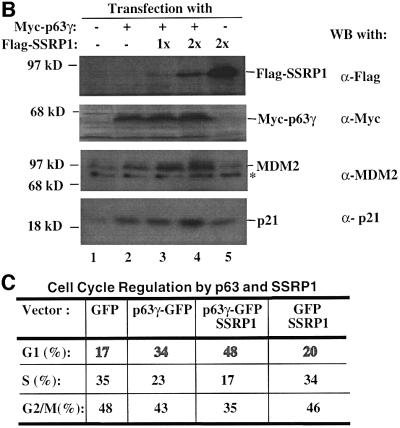
To confirm the above observation further, we wanted to determine the effect of SSRP1 on the expression of some endogenous p63 target genes. It has been shown that p63 can induce expression of several p53 target genes (Yang and McKeon, 2000; Irwin and Kaelin, 2001). First, we analyzed the transcript levels of six p53 target genes, PIG3 (Polyak et al., 1997), caspase 6, Killer/DR5 (Burns et al., 2001), Bax1 (Miyashita and Reed, 1995), p21_waf1/cip1_ (el-Deiry et al., 1993; Harper et al., 1993) and MDM2 (Wu et al., 1993), using semi-quantitative RT–PCR assays after introducing Myc-p63γ or Myc-ΔN-p63γ alone or together with SSRP1 into H1299 cells. As shown in Figure 2A, p63γ induced the mRNA expression of PIG3, p21_waf1/cip1_ and MDM2, but not of caspase 6, Killer/DR5 and Bax1 (lane 1), as expected (Dohn et al., 2001). In contrast, ΔN-p63γ had no apparent effect on the expression of these genes (lanes 4 and 5). In accordance with the results of Figure 1, SSRP1, when co-expressed with p63γ, but not with ΔN-p63γ, further enhanced the expression of the above p63-responsive genes (lane 3). Consistently, SSRP1, when co-expressed with p63γ, also increased the expression of the p21_waf1/cip1_ and MDM2 proteins (Figure 2B, lanes 3 and 4). However, in the absence of p63γ, this increase was significantly reduced (lane 5). These results not only demonstrate that SSRP1 can cooperate with p63 in stimulating expression of endogenous p63 target genes, but also indicate that this effect is not non-specific as SSRP1 did not affect the expression of these genes in the presence of ΔN-p63. Taken together, the results from Figures 1 and 2 demonstrate that SSRP1 can activate the transcriptional activity of p63.
Fig. 2. SSRP1 stimulates the p63γ-dependent expression of endogenous p53 target genes. (A) RNA transcript analysis. H1299 cells (three 60 mm plates of cells for each lane) were transfected with 500 ng of pCDNA3-Myc-p63γ or pCDNA3-Myc-ΔN-p63γ alone or together with 2 µg of pCDNA3-Flag-SSRP1, as indicated on top. At 48 h post-transfection, cells were harvested for RNA preparation. RT–PCRs for six p53-responsive genes were conducted as described in Materials and methods and as indicated on the right. (B) Western blot analysis. The same transfection as described above was carried out except that only p63γ was used with 1 µg (1×) and 2 µg (2×) of SSRP1. Cells were harvested for preparation of cell lysates. Cell lysates containing 150 µg of proteins were loaded directly onto an SDS gel, followed by western blot analyses using antibodies as indicated on the right. (C) Cell cycle analysis of transfected p53 null MEFs as described above, except that GFP and GFP–p63γ were used.
SSRP1 enhances p63γ-induced G1 arrest and apoptosis
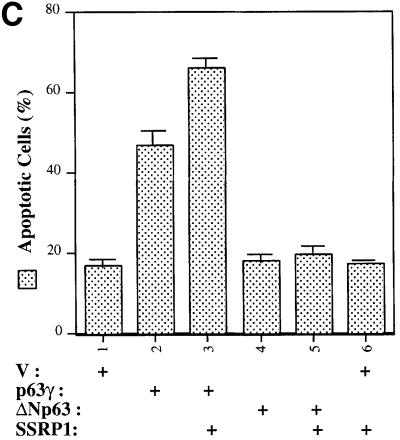
It is known that p63 can induce G1 arrest and apoptosis of some mammalian cells (Osada et al., 1998; Yang et al., 1998). Thus, we wanted to determine whether SSRP1 influences p63-induced G1 arrest and apoptosis. p53 null MEFs were transfected with mammalian expression plasmids encoding green fluorescent protein (GFP), GFP– p63γ or Flag-SSRP1 alone or together. Fluorescence-activated cell sorting (FACS) analysis of the transfected cells showed that p63γ retained 34% of the cells in the G1 phase during the cell cycle in comparison with 17% of the GFP-expressing cells (Figure 2C). Ectopic expression of SSRP1 resulted in 48% of the transfected cells in the G1 phase, indicating that SSRP1 further increases p63-dependent G1 arrest.
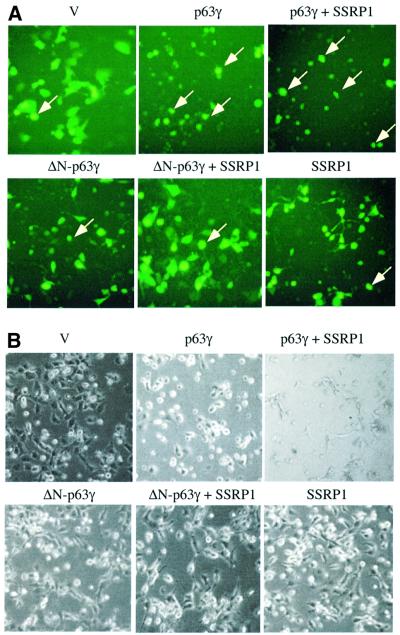
For the apoptotic analysis, H1299 cells were co-transfected with mammalian expression plasmids encoding Myc-p63γ, ΔN-p63γ or Flag-SSRP1 in the presence of the GFP expression vector. Following our previously established method (Zeng et al., 1999, 2000), floating round and shrunken cells expressing GFP were counted as apoptotic cells under a fluorescent microscope. Repre sentative images of the transfected cells are shown in Figure 3A and total cells are shown in Figure 3B. As expected, cells expressing p63γ, but not ΔN-p63γ, underwent drastic apoptosis. About 46% of p63γ-transfected cells died, whereas <17% of ΔN-p63γ-transfected cells died, which is equivalent to that of GFP-transfected cells (Figure 3C). Overexpression of SSRP1 with p63γ caused an ∼43% increase of apoptotic cells (∼66% of transfected cells died). In contrast, overexpression of SSRP1 with ΔN-p63γ resulted in only a marginal increase (∼10%) of apoptotic cells (∼19% of transfected cells died). SSRP1 alone did not cause significant apoptosis in comparison with GFP-transfected cells. These results demonstrate that SSRP1 can augment p63-induced apoptosis as well as G1 arrest, which is consistent with the stimulation of the p63 transcriptional activity by this HMG-1-like protein.
Fig. 3. SSRP1 augments p63γ-induced apoptosis. (A and B) As indicated on the top of each panel, pCDNA3 plasmids encoding no protein as a control (V, 2 µg), p63γ (0.5 µg), ΔN-p63γ (0.5 µg) and/or SSRP1 (1 µg), together with the pGFP plasmid (200 ng), were introduced into 105 H1299 cells/35 mm plate using LipofectAmine as described in Materials and methods. (A) At 32 h post-transfection, cells (200 in total) expressing GFP were counted under a fluorescence microscope using a blind approach, with morphologically round and shrunken cells (arrows) identified as apoptotic. (B) Images of total cells in the same views. (C) The percentage of apoptotic cells, with the standard error indicated by a bar above each column.
SSRP1 associates with p63γ in cells
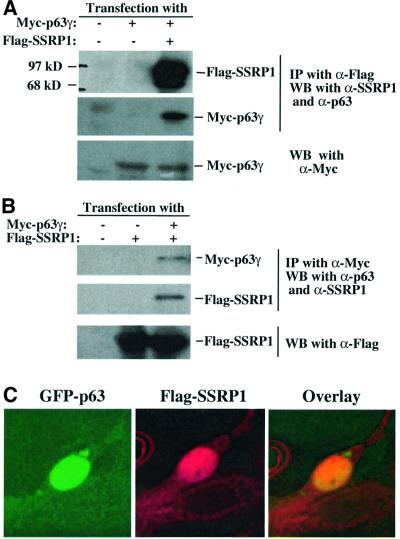
The finding that SSRP1 cooperatively enhances the p63 activity suggests that this HMG-1-like protein may interact with p63. To test this idea, we introduced Myc-p63γ and Flag-SSRP1 alone or together into H1299 cells as described above. Proteins from transfected cell lysates were detected by co-immunoprecipitation with monoclonal antibodies against either Myc or Flag epitopes, followed by western blot with polyclonal antibodies against p63 or SSRP1, or monoclonal antibodies against Flag or Myc. The results (Figure 4) showed that both Flag-SSRP1 and Myc-p63γ were co-immunoprecipitated with either anti-Flag (Figure 4A) or anti-Myc antibodies (Figure 4B). This was not a non-specific cross-reaction between the antibodies and the proteins, because anti-Flag antibodies did not pull down the Myc-p63γ protein in the absence of Flag-SSRP1, and anti-Myc antibodies did not bring down the Flag-SSRP1 protein in the absence of Myc-p63γ (the middle lane of Figure 4A and B). These two proteins must interact with each other in the nucleus, as Flag-SSRP1 and GFP–p63γ co-localized there (Figure 4C).
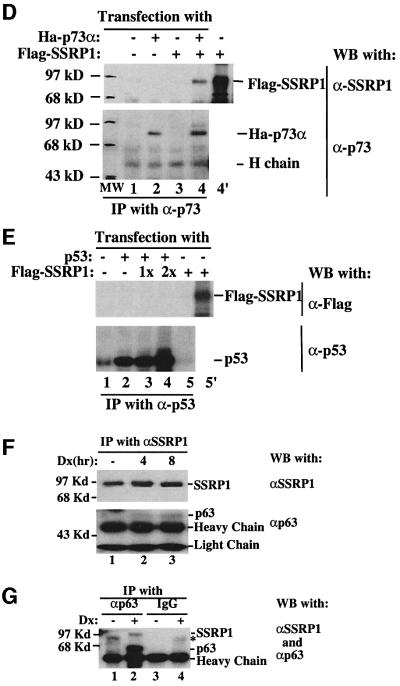
Fig. 4. SSRP1 interacts with p63γ and p73α, but not p53, in cells. H1299 cells (106/60 mm plate, three plates each lane) were transfected with plasmids encoding no protein (3 µg), Myc-p63γ (1 µg), Ha-p73α (1 µg), p53 (1 µg) or Flag-SSRP1 (2 µg) either alone or together as indicated. At 30 h post-transfection, cells were harvested for preparation of cell lysates. A 300 µg aliquot of the lysates was used for immunoprecipitation with antibodies against Myc, Flag, p53 or p73, followed by western blotting with antibodies as indicated on the right. The results with the anti-Flag antibodies used for immunoprecipitation are shown in (A), while those with the anti-Myc antibodies are shown in (B). (C) The co-localization of GFP–p63γ with Flag-SSRP1 in the nucleus. Immunofluorescent staining of transfected H1299 cells was conducted as described in Materials and methods. Stained cells were examined under a deconvolution microscope. A representative image is shown here. The results of immunoprecipiation–western blot analysis for p73–SSRP1 interaction in cells is shown in (D) and (E), while that for p53–SSRP1 is shown in (F) and (G).
To determine whether SSRP1 also binds to p73 and p53, transfection assays using H1299 cells, followed by immunoprecipitation–western blot analysis, were carried out with mammalian expression vectors encoding p53 or p73α in the presence or absence of the SSRP1 expression plasmid. Like p63γ, p73α also interacted with SSRP1, whereas p53 did not appear to associate with SSRP1 in cells (Figure 4D and E). These results are consistent with the result of Figure 1A, suggesting that SSRP1 appears to be specific for activation of p63 and p73, but not p53.
Next, we determined whether endogenous SSRP1 and p63 can bind to each other. Because of the low level of p63 in cells, we treated human embryonic kidney epithelial 293 cells with 0.25 µM doxorubicin, which recently was shown to induce p63 (Flores et al., 2002). Cells were harvested at different times post-treatment for immunoprecipitation–western blot analysis using antibodies against p63 and SSRP1. As shown in Figure 4, p63 was indeed co-immunoprecipitated with SSRP1 by the anti-SSRP1 antibody after doxorubicin treatment (Figure 4F), and the reverse was also true with the anti-p63 antibody (Figure 4G). Although the anti-p63 antibody can recognize all the isoforms of p63 (Oncogene Science), SSRP1 must only associate with the N-terminal intact isoform in cells because the N-terminus of p63 is required for its interaction with SSRP1 in vitro (Figure 5A) and for functional activation of p63 by SSRP1 in cells (Figure 1C). Based upon the size of the immunoprecipitated p63, we speculated that this isoform might be p63γ, but this requires further confirmation using isoform-specific antibodies, which is not available yet. Taken together, these results demonstrate that SSRP1 and p63γ are able to associate with each other in cells.
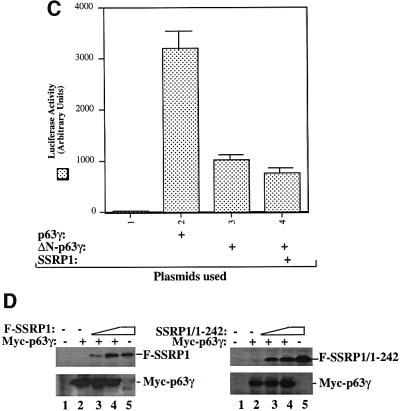
Fig. 5. SSRP1 binds directly to p63γ but not ΔN-p63γ in vitro. (A) GST fusion protein association assays were conducted as described in Materials and methods. A 300 ng aliquot of His-p63γ or His-ΔN-p63γ proteins was incubated with GST alone or GST–SSRP1 or deletion mutant fusion proteins that bound on the glutathione–Sepharose 12B beads (containing ∼1 µg of proteins). The GST fusion proteins used in this assay were stained with Coomassie Blue (the top panel). Bound p63 proteins were detected by western blotting using anti-p63 antibodies as shown in the two bottom panels. Thirty percent of ΔN-p63γ input was loaded directly onto the SDS gel as a reference. (B) The p63-binding fragment of SSRP1 impeded SSRP1-mediated stimulation of p63 activity. H1299 cells were transfected with plasmids encoding Myc-p63γ (50 ng) alone or with Flag-SSRP1 (500 ng) or Flag-250–490 (the p63-binding domain of SSRP1) (500 ng) along with reporters, and luciferase assays were carried out as described in Figure 1. (C) Western blot analysis of the levels of Myc-p63γ, Flag-SSRP1 and Flag-250–490 expressed in cells using antibodies against Myc and Flag, respectively. The asterisk denotes the degraded form of SSRP1 in lane 5.
SSRP1 interacts with the N-terminal domain of p63γ
To demonstrate further that SSRP1 binds directly to p63γ and also to define their binding domains, we carried out a set of GST fusion protein association assays. GST was fused with either the full-length SSRP1 or its three deletion mutants as shown in Figure 5A (top panel). These fusion proteins were immobilized on glutathione– Sepharose 12B beads and incubated with bacterially expressed and purified p63γ or ΔN-p63γ proteins. After washing, bound proteins were detected by western blotting with anti-p63 antibodies, which recognize both the intact and N-terminally deleted isoforms of p63 (middle and bottom panels). The result showed that p63γ, but not ΔN-p63γ, was retained on the GST–SSRP1- and GST– SSRP1 central domain-containing beads, indicating that p63γ binds to the central domain of SSRP1 directly. This binding requires the N-terminal domain of p63, as none of the GST–SSRP1 fusion proteins was able to retain any detectable ΔN-p63γ (bottom panel). This is consistent with the functional studies as described above (Figures 1 and 2). Also, the central domain of SSRP1 appeared to bind to p63γ with a higher affinity than did wild-type SSRP1, which was reproducible in vitro, although the rationale for this remains to be clarified. In summary, the result from this in vitro mapping study demonstrates that SSRP1 binds directly to p63γ and this binding requires the N-terminal domain of p63γ and the central domain of SSRP1.
Because the middle region (amino acids 235–475) of SSRP1 binds directly to p63γ, it is possible that overexpression of this domain may alleviate the SSRP1-mediated stimulation of p63 transcriptional activity. To test this idea, we performed a transient transfection– luciferase assay by introducing p63γ alone or with SSRP1 or SSRP1 amino acids 250–490 into H1299 cells as described above. Again, SSRP1 stimulated p63γ-dependent luciferase activity (Figure 5B). In striking contrast, the p63-binding fragment of SSRP1 not only failed to stimulate p63 activity, but also, conversely, reduced p63 activity (Figure 5B), suggesting that this fragment may play a dominant-negative role probably by competing with endogenous SSRP1 for p63. The reduction of p63 activity was not due to the difference in the protein level, as both wild-type and deletion mutant SSRP1s are expressed equivalently and the p63 level did not change significantly, regardless of which form of SSRP1 was co-expressed with p63 (Figure 5C). These results confirm that SSRP1 is important for the activation of p63-dependent transcription in cells.
SSRP1 co-resides with p63γ at the p53RE site of the endogenous promoters
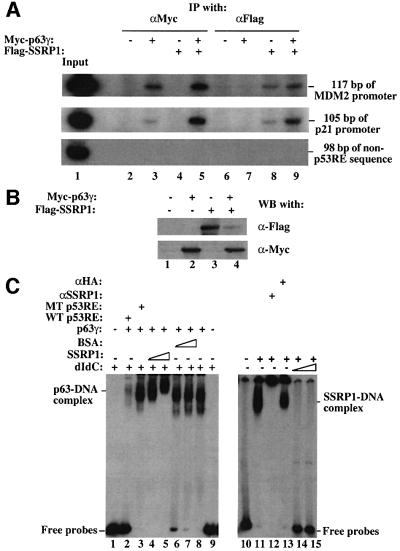
After defining the interaction between SSRP1 and p63γ, we then tested whether SSRP1 associates with p63γ at the p53RE-containing promoter region of endogenous target genes. The MDM2 and p21waf1/cip1 promoters were selected for this experiment, as p63γ-dependent MDM2 or p21waf1/cip1 expression was stimulated markedly by SSRP1 in cells (Figure 2). We conducted a ChIP assay using antibodies against Flag or Myc, following transfection of H1299 cells with Myc-p63γ and Flag-SSRP1 expression vectors alone or together as described in Figure 2. After immunoprecipitation, PCRs were carried out with pairs of primers designed to encompass the p53RE motif of the MDM2 promoter or of the p21waf1/cip1 promoter (see Materials and methods), generating a PCR product of 117 bp (MDM2 promoter) or 105 bp (p21waf1/cip1 promoter). As shown in Figure 6, antibodies against Myc-p63γ immunoprecipitated these p53RE motif-containing PCR products (Figure 6, lanes 3 and 5) only when Myc-p63γ was expressed (Figure 6B). This indicates that Myc-p63γ binds to the p53RE sequence of these promoters in cells. This binding apparently increased in the presence of Flag-SSRP1 (compare lane 3 with lane 5). This difference was not due to the change of p63γ level, because the p63γ level did not change whether Flag-SSRP1 was expressed or not (Figure 6B). Also, the antibody against Flag-SSRP1 immunoprecipitated the same PCR product (Figure 6A, lanes 7 and 8) when Flag-SSRP1 was expressed (Figure 6B). Although this antibody also pulled down some PCR products of these promoters in the absence of exogenous p63γ (lane 7), more PCR products were detected in the presence of exogenous p63γ (lane 8), suggesting that more SSRP1 proteins are recruited by p63γ to the p53RE sites in cells. The association of SSRP1 with the p53RE-containing region without exogenous p63γ might be due to the interaction of SSRP1 with endogenous p63 proteins, because SSRP1 did not bind specifically to the p53RE motif in vitro (Figure 6C), or partially due to the non-specific binding of SSRP1 to chromatin (Rottgers et al., 2000). The latter appeared to be less likely because when using a pair of primers encompassing a DNA sequence 2.5 kb upstream from the p53RE motif of the same MDM2 promoter for PCR, neither anti-Myc nor anti-Flag antibodies were able to immunoprecipitate the PCR product of this non-p53RE DNA sequence (Figure 6A). Therefore, these results indicate that SSRP1 can complex with p63γ at the endogenous p53RE-containing MDM2 and p21waf1/cip1 promoters, suggesting that SSRP1 may mediate p63γ-dependent transcription through direct interaction with this activator.
Fig. 6. SSRP1 co-resides with p63γ at the p53RE sites of the endogenous MDM2 and p21_waf1_ promoters. (A) ChIP analyses. H1299 cells were transfected with plasmids encoding no protein (3 µg), or with Myc-p63γ (1 µg) and/or Flag-SSRP1 (2 µg), as indicated on top. At 36 h post-transfection, cells were treated with a 1% formaldehyde solution for ChIP assays as described in Materials and methods. PCR products of 117 bp (top) encompassing the p53RE motif of the MDM2 promoter, 105 bp (middle) of the p21_waf1/cip1_ promoter or 98 bp (bottom) of the non-p53RE sequence 2.5 kb upstream from the p53RE site of the MDM2 promoter were tested from the immunoprecipitates using antibodies against either Myc or Flag, as indicated on top. (B) Western blot analysis of the same transfected cells. Cell lysates containing 150 µg of proteins were loaded onto an SDS gel. Myc-p63γ and Flag-SSRP1 were detected with antibodies against Myc and Flag. (C) SSRP1 directly binds to the p53RE-bound p63γ. EMSAs were carried out as described in Materials and methods. In the reactions in lanes 1–9, 300 ng of poly(dIdC), 100 ng of wild-type (lane 2) or mutant (lane 3) p53RE oligomers, 500 ng of p63γ, 200 and 400 ng of SSRP1 (lanes 4 and 5), and 200 and 400 ng of BSA (lanes 6 and 7) were used as indicated on top. In the reactions in lanes 10–15, 400 ng of SSRP1 and 500 ng of anti-SSRP1 or anti-HA peptide antibodies were used in the presence or absence of 100 or 200 ng of poly(dIdC) as indicated on top.
To test whether SSRP1 is able to associate directly with the DNA-bound p63γ, we performed electrophoretic gel mobility shift assays (EMSAs) using 32P-labeled p53RE-containing DNA probes derived from the MDM2 promoter and purified recombinant p63γ and SSRP1 proteins. As expected (Zeng et al., 2001b), the result in Figure 6C showed that p63γ bound to these DNA fragments. Interestingly, addition of increasing amount of SSRP1 but not bovine serum albumin (BSA) supershifted the p63γ–DNA complexes. This supershifting must be caused by the association of SSRP1 with p63γ, because SSRP1 alone was not able to bind to the radiolabeled p53RE oligomers in the presence of 100 ng of poly(dIdC) (lanes 14 and 15). Note, in the reactions of lanes 4 and 5 where the supershifting was observed, 300 ng of poly(dIdC) were used. These protein–DNA complexes were specific for the p53RE sequence, as non-labeled wild-type, but not mutant, p53RE-containing oligomers (20-fold in molar ratio) significantly reduced the p63γ–DNA complex (compare lane 2 with lane 3). SSRP1 was only able to bind to the p53RE oligomers non-specifically in the absence of poly(dIdC), as poly(dIdC) (∼25-fold more than the radiolabeled oligomers in molar ratio) readily eliminated this binding (lanes 14 and 15). Thus, these results together with results from the ChIP analyses (Figure 6A) demonstrate that SSRP1 directly interacts with p63γ at the p53RE-containing promoters to facilitate p63γ transcriptional activity.
Discussion
Although it is clear that p63 plays an essential role in development and displays p53-like transcription activity (Levrero et al., 2000; Yang and McKeon, 2000; Irwin and Kaelin, 2001), how this transcriptional activator is regulated has not been explored intensively since its discovery in 1998 (Osada et al., 1998; Yang et al., 1998). In this study, we have identified the HMG-1-like protein SSRP1 as a positive regulator of p63. First, overexpression of SSRP1 remarkably stimulated the transcriptional activity of p63γ as well as p63γ-dependent expression of the endo genous p53 target genes PIG3, MDM2 and p21waf1/cip1. Consistently, SSRP1 enhanced p63γ-induced apoptosis and G1 arrest. Also, SSRP1 interacted with p63γ both in vitro and in cells. Domain mapping revealed that the central region of SSRP1 and N-terminal domain of p63γ are essential for the interaction between these proteins. In accordance with the observation that SSRP1 was not able to bind to the N-terminally deleted ΔN-p63γ protein, SSRP1 did not appear to affect the residual transcription activity of ΔN-p63γ either. Using ChIP assays, SSRP1 was found to co-reside with p63γ at the p53RE motif-containing region of the endogenous MDM2 and p21waf1/cip1 promoters. Although only the results with p63γ are presented in this study, we also observed that SSRP1 positively affected the activity of p63α and p63β (data not shown). This is reasonable because they all share the same N-terminal SSRP1-binding domain. Similarly, SSRP1 also stimulated the activity of p73α (Figure 1A), whose N-terminus shares 34% homology with that of p63. Taken together, these results demonstrate that SSRP1 can act as a co-activator of p63 to mediate p63-dependent transcription.
How does SSRP1 cooperate with p63 to stimulate its activity? Because SSRP1 directly binds to p63 and co-resides with this activator at the p63RE-containing promoter region in cells, one testable hypothesis would be that SSRP1, though not binding specifically to the p63RE promoter, might consolidate the association of p63 with its responsive DNA promoter. Two lines of evidence are correlated with this model. First, SSRP1 is capable of binding to bent DNA through its C-terminal HMG box domain (amino acids 539–614) (Bruhn et al., 1992; Yarnell et al., 2001), while its central domain (amino acids 235–475) binds to the N-terminal domain of p63 (Figure 5A). Secondly, SSRP1 can bind to DNA non-specifically with a low affinity (Figure 6C) and has been shown to associate with nucleosomes in yeast cells (Formosa et al., 2001) and with chromatin in vitro (Orphanides et al., 1999; Lichota and Grasser, 2001). Hence, by simultaneously associating with p63 and DNA, SSRP1 might stabilize the p63–DNA interaction on chromatin, as indicated in our ChIP assays (Figure 6A). One additional piece of evidence supporting this assumption is that SSRP1 has been shown to enhance the interaction of the transcriptional activator SRF with its responsive DNA element (Spencer et al., 1999). Therefore, SSRP1 may utilize its DNA-binding property to regulate the transcription activity of specific transcriptional activators.
The other possibility would be that SSRP1 might serve as an anchor to recruit and initiate the assembly of the chromatin-remodeling complex. It was shown recently that the yeast SSRP1 homolog Pob3 associates with ySpt16 and the small HMG box proteins Nhp6a and Nhp6b to form a nucleosome-recognizing complex (Brewster et al., 2001; Formosa et al., 2001); it is believed that the function of the yeast Pob3–Nhp complex is equivalent to that of the human SSRP1 (Brewster et al., 2001; Formosa et al., 2001). Genetic studies in yeast showed that Nhp deficiency in combination with mutations impairing the Swi/Snf chromatin-remodeling complex caused severe impairment of transcription (Formosa et al., 2001). Thus, it is possible that SSRP1, once bound to p63, might bring a chromatin-remodeling complex to the promoter, although evidence showing direct SSRP1– Swi/Snf interaction is needed. The third possibility is that SSRP1, once bound to p63, might recruit other transcriptional regulators such as p300/CBP or TAFs to communicate indirectly or directly with the RNA polymerase II initiation complex on chromatin. All of these hypotheses may not necessarily exclude one another, although more studies are necessary for a better understanding of the detailed mechanism for SSRP1-mediated regulation of p63’s transcriptional activity.
The other question would be whether the enhancement of p63 activity by SSRP1 is due partly to the positive effect of SSRP1 on transcriptional elongation (Orphanides et al., 1999). Although it has been shown that SSRP1 can form a heterodimer with Spt16 to facilitate the transcription elongation on chromatin templates in vitro (Orphanides et al., 1999), we also isolated an SSRP1-associated protein complex free of Spt16 from HeLa nuclear extracts (Y.Jin and H.Lu, unpublished observations), suggesting that SSRP1 might also function independently of Spt16. In addition, as mentioned previously, SSRP1 binds directly to p63 in vitro (Figure 5A) and co-localizes with p63 on the p63RE region (Figure 6). Furthermore, without exogenous p63γ, ectopic expression of SSRP1 was unable to markedly induce cell growth arrest (Figure 2C), apoptosis (Figure 3) and the expression of the p63 target genes (Figure 2A and B). Finally, SSRP1 was unable to elevate the residual transcription activity of ΔN-p63γ (Figure 1), indicating that the effect of SSRP1 on p63γ activity might not be due to its general role in transcriptional elongation (Orphanides et al., 1999). Thus, SSRP1 can activate p63 activity specifically, besides its general role in transcription elongation (Orphanides et al., 1999). Nevertheless, it would be interesting to test whether Spt16 joins SSRP1 as a co-activator of transcription.
Is SSRP1 unique for p63 regulation among the p53 family members? Although our previous study showed that SSRP1 was one of the five subunits in the Ser392 kinase complex that enhanced p53 activity by phosphorylating its Ser392 residue in vitro (Keller et al., 2001), the result presented here (Figure 1A) shows that SSRP1, when overexpressed, stimulates the transcriptional activity of p63 and p73, but not of p53 in cells. Consistently, SSRP1 associated with p63γ and p73α, but not p53, in cells (Figure 4). These studies suggest that SSRP1 may utilize distinct mechanisms to modulate the function of the p53 family members. On the one hand, SSRP1 modulates p53 activity by associating with CK2 and converting this kinase into a form that prefers p53 as a substrate in response to UV irradiation. On the other hand, SSRP1 can also regulate the activity of p63 and possibly p73, but not p53, by interacting directly with the proteins and enhancing their ability to bind to the promoters. Biochemical and mutagenic analyses of the interaction between SSRP1 and the p53 family members is in progress and will provide insights into the molecular mechanism underlying how SSRP1 distinguishes the N-termini of the p53 family members. Also, it is important to study whether SSRP1 regulates the activity of p63 and p73 in response to intracellular or external signals.
Materials and methods
Plasmids and antibodies
The pCDNA3-Myc-ΔN-p63γ and pCDNA3-Myc-p63γ plasmids were constructed using ΔN-p63α cDNA obtained from David Sidransky (Johns Hopkins Medical School, Baltimore) (Trink et al., 1998) and the RT–PCR product of p63γ with primers 5′-TCCCCCGGGGATGTCCCAGAGCACACAG-3′ and 5′-CGGGATCCTGGGTACACTGATCGGTT-3′ (inverse), respectively. The p63γ sequence was confirmed by automatic sequencing. pCDNA3-Flag-SSRP1 and pCDNA3-Flag-SSRP1/1–242 plasmids were constructed using the pET-His-SSRP1 plasmid that was described previously (Keller et al., 2001). pEGFP-C1 plasmid was purchased from Gibco-BRL. The monoclonal and polyclonal anti-p63 antibodies, which recognize both p63γ and ΔN-p63γ, were purchased from Santa Cruz and Oncogene, respectively. Monoclonal anti-Flag antibodies were purchased from Sigma. Monoclonal anti-Myc antibodies were produced in this laboratory. Polyclonal anti-PIG8 and anti-p21_waf1_ antibodies were purchased from Santa Cruz. Polyclonal anti-MDM2 antibodies were as described previously (Zeng et al., 1999).
Buffers
Lysis buffer consisted of 50 mM Tris–HCl pH 8.0, 0.5% NP-40, 1 mM EDTA, 150 mM NaCl and 1 mM phenylmethylsulfonyl fluoride (PMSF). SNNTE buffer contained 50 mM Tris–HCl pH 7.4, 5 mM EDTA, 1% NP-40, 500 mM NaCl and 5% sucrose. RIPA was comprised of 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS and 1% (w/v) sodium deoxycholate. Buffer C 100 (BC100) included 20 mM Tris–HCl pH 7.9, 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 4 mM MgCl2, 0.2 mM PMSF, 1 mM dithiothreitol (DTT) and 0.25 µg/ml of pepstatin A.
Cell culture
Human embryonic kidney 293 cells, lung small cell carcinoma H1299 cells and mouse p53/mdm2 double null embryonic fibroblasts (MEFs; obtained from Guillermina Lozano of M.D. Anderson Cancer Center, Houston) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 50 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a 5% CO2 atmosphere.
Construction and preparation of GST–SSRP1 and GST–SSRP1 deletion fusion proteins
The pGST–SSRP1 fusion protein-expressing plasmids were constructed by inserting the PCR-generated products of SSRP1 into pGEX-KG (Pharmacia) at the _Xba_I sites. The orientation and sequence of the SSRP1 fragments were confirmed by sequencing and western blotting. These GST fusion proteins were expressed and purified from bacteria. The GST–SSRP1/1–242 fusion protein was also utilized as an antigen for generation of polyclonal anti-SSRP1 antibodies.
Purification of His-SSRP1, His-p63γ and His-ΔN-p63γ
His-SSRP1 was expressed and purified from bacteria using Ni-NTA–agarose (Qiagen), as described previously (Keller et al., 2001). His-p63γ and His-ΔN-p63γ were subcloned into the pET-30a vector and expressed and purified from bacteria using Ni-NTA beads. The purified proteins were used for protein association assays and EMSAs as described below.
Transfection and co-immunoprecipitation
H1299 cells (three 60 mm plates) were transfected with 3 µg of the parental pCDNA3, Myc-p63γ, Myc-ΔN-p63γ, pCDNA-Ha-p73α, pCMV-p53 or Flag-SSRP1-expressing plasmids by means of LipofectinAmine (Gibco-BRL). At 6 h post-transfection, fresh DMEM was added to the plates to replace the media containing plasmids. Cells were harvested 36–48 h post-transfection. Cell lysates were prepared as described (Zeng et al., 1999, 2000). Lysates (∼300 µg of proteins) were pre-cleared with 35 µl of protein A–agarose (50% slurry), and then incubated for 3 h at 4°C with fresh protein A–beads (35 µl) and 1 µg of monoclonal anti-Myc or anti-Flag antibodies. The beads were loaded directly onto an SDS–acrylamide gel after vigorously washing twice with lysis buffer, twice with SNNTE and once with RIPA. The co-immunoprecipitated proteins were detected by western blotting using polyclonal anti-SSRP1, anti-p63, anti-Myc or anti-Flag antibodies, and detected by enhanced chemiluminescence (Amersham).
Transient transfection and luciferase assays
p53 null H1299 cells and MEFs (60% confluence in a 12-well plate) were transfected with a pCMV-β-galactoside reporter plasmid (0.1 µg) and a luciferase reporter plasmid (0.1 µg) driven by two copies of the p53RE motif derived from the MDM2 promoter (Wu et al., 1993), together with a combination of different plasmids (total plasmid DNA = 1 µg/well) as indicated in Figure 1, using LipofectAmine (Gibco-BRL). At 48 h post-transfection, cells were harvested for luciferase assays as described previously (Zeng et al., 2001a). Luciferase activity was normalized by a factor of β-gal activity tested in the same assay.
RT–PCR analysis
Total RNA was isolated from H1299 cells transfected with p63, SSRP1 or both (see the section above) using Qiagen RNeasy Mini Kits (Qiagen, Valencia, CA). Reverse transcriptions were performed by incubating 1 µg of total RNA in a 20 µl reverse transcription reaction mixture containing 10 mmol/l dNTPs, 0.5 µg of oligo(dT)15 primer, 26 U of RNase inhibitor and 200 U of murine Molony leukemia virus reverse transcriptase (MLV-RT; Promega Corp, Madison, WI) in 1× reverse transcription buffer (Promega) at 42°C for 1 h. After cDNA synthesis, 1 µl of the cDNA solution was used for a PCR in 20 µl of mixture containing 1× PCR buffer, 60 µmol/l of dNTPs, 1 U of Taq polymerase (Boehringer Mannheim, Germany), 0.5 µmol/l of each primer and 0.2 µCi of [32P]dCTP. PCRs were conducted at 94°C for 30 s, at 55 or 60°C for 30 s, and at 72°C for 2 min for 18–20 cycles. PCR products were resolved on a 6% polyacrylamide gel. The gel was dried, followed by autoradiography. The following primers were used: p21waf1/cip1, 5′-ATGTCAGAACCGGCTGGGGATG-3′, 5′-TTAGGGCTTCCTCTTGGAGAAG-3′; MDM2, 5′-AACCACCTCACAGATTCCAG-3′, 5′-TCAAGGTGACACCTGTTCTC-3′; PIG3, 5′-TGTGCTAATCCATGCAGGAC-3′, 5′-ACTGGTGATCAGACTTCCTC-3′; caspase 6, 5′-TTGGCACTTAACACTGCCAG-3′, 5′-TGGTGTCCAACTTCTGTGTC-3′; GAPDH, 5′-CCACCCATGGCAAATTCCATGGCA-3′, 5′-TCTAGACGGCAGGTCAGGTCCACC-3′; Bax1, 5′-TTCATCCAGGATCGAGCAGG-3′, 5′-AGGAAGTCCAATGTCCAGCC-3′; and Killer/DR5, 5′-ATTGTGTCCACCTGGACACC-3′, 5′-TACGGCTGCAACTGTGACTC-3′.
Cell cycle analysis
p53 null MEFs were transfected with plasmids encoding GFP or GFP–p63γ alone or together with Flag-SSRP1. At 40 h post-transfection, cells were harvested and resuspended in 100 µl of phosphate-buffered saline (PBS), and transferred to a polystyrene tube. Then, 200 µl of pH 7.2 propidium iodide (PI) stain (50 µg/ml PI, 30 µg/ml polyethylene glycol 8000, 2 µg/ml RNase A, 0.1% Triton X-100, 0.38 M NaCl) was added, and the samples were incubated for 10 min. The samples were analyzed for DNA content using a Becton Dickinson FACScan flow cytometer. Data were analyzed by the multicycle software program using the polynomial S-phase algorithm. GFP-positive cells were scored for cell cycle analysis.
Immunofluorescent staining analysis
H1299 cells were transfected with plasmids encoding GFP–p63γ and Flag-SSRP1 and fixed for immunofluorescent staining as described previously (Zeng et al., 1999). Monoclonal anti-Flag (Sigma) and Cy-3-conjugated sheep anti-mouse IgG (Jackson Immuno-Research) antibodies were used for detection of Flag-SSRP1 (red). Cells were examined under a Deltavision deconvolution microscope.
GST fusion protein association assay
The fusion proteins were expressed in Escherichia coli and purified on a glutathione–Sepharose 12B column. Protein–protein association assays were conducted as reported using fusion protein-containing beads (Zeng et al., 1999). Purified p63γ or ΔN-p63γ proteins were incubated with the glutathione–Sepharose 4B beads (50% slurry) containing ∼500 ng of GST–SSRP1, GST–SSRP1/1–242, GST–SSRP1/235–475, GST–SSRP1/471–709 or GST, respectively. At 1 h after incubation at room temperature, the mixtures were washed once in BC100 containing 0.1% NP-40, twice in SNNTE and once in RIPA. Bound proteins were analyzed on a 10% SDS gel and detected by western blotting using the anti-p63 monoclonal antibody to detect p63-GST–SSRP1 interactions.
Doxorubicin treatment followed by immunoprecipitation–western blotting
HEK 293 cells were treated with 0.25 µM of doxorubicin and harvested at 0, 4 and 8 h after treatment for immunoprecipitation–western blotting using antibodies against SSRP1 and p63, respectively. See the legend of Figure 4F and G for details.
Apoptotic analysis
This analysis was carried out as described previously (Zeng et al., 1999, 2000). H1299 cells (105/35 mm dish) were transfected with a plasmid encoding GFP (0.85 µg of DNA/dish) together with combinations of expression plasmids (total plasmid DNA = 2 µg/dish) as indicated in Figure 3. Transfected cells in cultures were analyzed under a fluorescent microscope and identified by the presence of green fluorescence. Apoptotic cells were identified by their rounded and shrunken morphology in contrast to the spread out morphology of non-apoptotic H1299 cells, counted in a blinded fashion and presented as a percentage out of the total population of fluorescent cells (see Figure 3).
EMSA
EMSA was conducted as described (Zeng et al., 1998). Proteins as indicated in the figure legends were pre-incubated in the presence or absence of antibodies, at 4°C for 15 min, prior to being mixed with a DNA-binding cocktail containing 10 mM HEPES buffer pH 7.5, 4 mM MgCl2, 60 mM NaCl, 0.3 µg of poly(dIdC), 0.1% NP-40, 0.1 mM EDTA and 5′/3′ [32P]end-labeled DNA fragments harboring two copies of the p53RE sequence derived from the MDM2 promoter (5000 c.p.m., 2.0 ng of DNA per assay). The reaction was incubated at room temperature for 30 min, and loaded directly onto a 4% non-denatured gel.
Transient transfection–western blot analysis of endogenous p63 targets
H1299 cells were transfected with pCDNA3-Myc-p63γ or pCDNA3-Flag-SSRP1 alone or together as indicated in Figure 2. Transfected cells were harvested for preparation of cell lysates. Cell lysates containing 150 mg of proteins were loaded directly onto an SDS gel and western blotting was carried out as described previously (Zeng et al., 1999) to detect expression of endogenous MDM2, p21_waf1_ and PIG8, as well as the expression of exogenous p63 and SSRP1 proteins.
Chromatin immunoprecipitation (ChIP)-PCR
H1299 cells were transfected with pcDNA3-Myc-p63γ or flag-SSRP vector alone, or together. ChIP analysis was performed as described (Shang et al., 2000; Szak et al., 2001), with minor modifications. Briefly, cells were cross-linked with a 1% formaldehyde solution in PBS for 36 h after transfection. The cross-linking was stopped by incubating the cells on 0.125 M glycine for 5 min. The cells were then harvested in 1 ml of lysis buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, 5 mM EDTA) containing the protease inhibitors pepstatin A (10 µg/ml), leupeptin (10 µg/ml), DTT (1 mM), and PMSF (10 µg/ml). Cell lysates were sonicated four times for 15 s each at maximal setting to yield chromatin fragments of ∼600 bp as assessed by agarose gel electrophoresis, followed by centrifugation for 10 min to remove the cell debris. Cell lysates were pre-cleared with 50 µl of protein A–Sepharose/2 µg of sheared salmon sperm DNA slurry for 30 min at 4°C. A total of 2 µg of anti-myc or anti-flag antibodies was incubated with the pre-cleared extracts overnight at 4°C followed by incubation with protein A/G–agarose for 2 h at 4°C. The immunocomplexes were washed twice with lysis buffer, four times with immunoprecipitation (IP) wash buffer (100 mM Tris pH 8.5, 50 mM LiCl, 1% NP-40, 1% deoxycholic acid), and twice more with lysis buffer. The precipitates were then extracted twice, each with 150 µl of IP elution buffer (50 mM NaHCO3, 1% SDS). The total eluates were pooled by adding 2 µl of 5 mg/ml RNase A and 12 µl of 5 M NaCl (brought to 0.2 M) and incubated at 65°C for at least 6 h to reverse the formaldehyde cross-linking. DNA fragments were purified by phenol–chloroform extraction and ethanol precipitation, and dissolved in 100 µl of sterile H2O.
DNA samples were then analyzed with 25 or 28 cycles of PCR to amplify mdm2 and p21_waf1_ promoter sequences containing the p53RE sequence, each cycle consisting of denaturation at 94°C for 30 s, annealing at 60°C (MDM2 promoter) or 65°C (p21 promoter) for 30 s, followed by extension at 72°C for 2 min. A total of 0.5 µCi of [32P]dCTP was added in each 50 µl of PCR mixture. The primers for amplifying mdm2 promoter sequence (Wu et al., 1993) are 5′-GGTTGACTCAGCTTTTCCTCTTG-3′ and 5′-GGAAAATGCATGGTTTAAATAGCC-3′ (inverse); the primers for amplifying the p21 promoter sequence (el-Deiry et al., 1993) are 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAGG-3′ (inverse); the primers for amplifying the sequence located 2.5 kb upstream from the p53RE site of the mdm2 promoter (Wu et al., 1993) are 5′-TGAATCTACTCTTGGTGGTCC-3′ and 5′-AAGGAAATTTGGGCTTTCGAC-3′ (inverse). PCR products were resolved on a 6% polyacrylamide gel, dried and exposed on film.
Acknowledgments
Acknowledgements
We thank other members of our laboratory for active discussion, Hunjoo Lee for the purified p63 proteins, and Guillermina Lozano, David Sidransky, Jiandong Chen, Zhimin Yuan, Xinbin Chen, Brian Dynlacht and Maureen Murphy for generously providing us with cells, plasmids and the protocol for ChIP assays. D.M.K. was supported by an NEI pre-doctoral fellowship. This work was partly supported by NIH/NCI and ACS grants to H.L.
References
- Avantaggiati M.L., Ogryzko,V., Gardner,K., Giordano,A., Levine,A.S. and Kelly,K. (1997) Recruitment of p300/CBP in p53-dependent signal pathways. Cell, 89, 1175–1184. [DOI] [PubMed] [Google Scholar]
- Baxevanis A.D. and Landsman,D. (1995) The HMG-1 box protein family: classification and functional relationships. Nucleic Acids Res., 23, 1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster N.K., Johnston,G.C. and Singer,R.A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol., 21, 3491–3502.11313475 [Google Scholar]
- Bruhn S.L., Pil,P.M., Essigmann,J.M., Housman,D.E. and Lippard,S.J. (1992) Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl Acad. Sci. USA, 89, 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns T.F., Bernhard,E.J. and El-Deiry,W.S. (2001) Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene, 20, 4601–4612. [DOI] [PubMed] [Google Scholar]
- Celli J. et al. (1999) Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell, 99, 143–153. [DOI] [PubMed] [Google Scholar]
- Costanzo A. et al. (2002) DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell, 9, 175–186. [DOI] [PubMed] [Google Scholar]
- Dailey L., Yuan,H. and Basilico,C. (1994) Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol. Cell. Biol., 14, 7758–7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn M., Zhang,S. and Chen,X. (2001) p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene, 20, 3193–3205. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey,M., Slagle,B.L., McArthur,M.J., Montgomery,C.A.,Jr, Butel,J.S. and Bradley,A. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356, 215–221. [DOI] [PubMed] [Google Scholar]
- Dyer M.A., Hayes,P.J. and Baron,M.H. (1998) The HMG domain protein SSRP1/PREIIBF is involved in activation of the human embryonic β-like globin gene. Mol. Cell. Biol., 18, 2617–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry W.S. et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Harley,V.R., Pontiggia,A., Goodfellow,P.N., Lovell-Badge,R. and Bianchi,M.E. (1992) SRY, like HMG1, recognizes sharp angles in DNA. EMBO J., 11, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E.R., Tsai,K.Y., Crowley,D., Sengupta,S., Yang,A., McKeon,F. and Jacks,T. (2002) p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature, 416, 560–564. [DOI] [PubMed] [Google Scholar]
- Formosa T., Eriksson,P., Wittmeyer,J., Ginn,J., Yu,Y. and Stillman,D.J. (2001) Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J., 20, 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Amsterdam,A. and Grosschedl,R. (1991) DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev., 5, 2567–2578. [DOI] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Giese,K. and Pagel,J. (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet., 10, 94–100. [DOI] [PubMed] [Google Scholar]
- Gu W., Shi,X.L. and Roeder,R.G. (1997) Synergistic activation of transcription by CBP and p53. Nature, 387, 819–823. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hollstein M. et al. (1994) Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res., 22, 3551–3555. [PMC free article] [PubMed] [Google Scholar]
- Ianakiev P., Kilpatrick,M.W., Toudjarska,I., Basel,D., Beighton,P. and Tsipouras,P. (2000) Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet., 67, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M.S. and Kaelin,W.G. (2001) p53 family update: p73 and p63 develop their own identities. Cell Growth Differ., 12, 337–349. [PubMed] [Google Scholar]
- Jayaraman L., Moorthy,N.C., Murthy,K.G., Manley,J.L., Bustin,M. and Prives,C. (1998) High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev., 12, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D.M. et al. (2001) A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16 and SSRP1. Mol. Cell, 7, 283–292. [DOI] [PubMed] [Google Scholar]
- Ko L.J. and Prives,C. (1996) p53: puzzle and paradigm. Genes Dev., 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- Levrero M., De Laurenzi,V., Costanzo,A., Gong,J., Wang,J.Y. and Melino,G. (2000) The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci., 113, 1661–1670. [DOI] [PubMed] [Google Scholar]
- Lewin B. (1990) Commitment and activation at pol II promoters: a tail of protein–protein interactions. Cell, 61, 1161–1164. [DOI] [PubMed] [Google Scholar]
- Lichota J. and Grasser,K.D. (2001) Differential chromatin association and nucleosome binding of the maize HMGA, HMGB and SSRP1 proteins. Biochemistry, 40, 7860–7867. [DOI] [PubMed] [Google Scholar]
- Lill N.L., Grossman,S.R., Ginsberg,D., DeCaprio,J. and Livingston,D.M. (1997) Binding and modulation of p53 by p300/CBP coactivators. Nature, 387, 823–827. [DOI] [PubMed] [Google Scholar]
- McStay B., Hu,C.H., Pikaard,C.S. and Reeder,R.H. (1991) xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X.laevis. EMBO J., 10, 2297–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A.A., Zheng,B., Wang,X.J., Vogel,H., Roop,D.R. and Bradley,A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature, 398, 708–713. [DOI] [PubMed] [Google Scholar]
- Miyashita T. and Reed,J.C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell, 80, 293–299. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu,W.H., Lane,W.S., Hampsey,M. and Reinberg,D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature, 400, 284–288. [DOI] [PubMed] [Google Scholar]
- Osada M. et al. (1998) Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med., 4, 839–843. [DOI] [PubMed] [Google Scholar]
- Polyak K., Xia,Y., Zweier,J.L., Kinzler,K.W. and Vogelstein,B. (1997) A model for p53-induced apoptosis. Nature, 389, 300–305. [DOI] [PubMed] [Google Scholar]
- Rottgers K., Krohn,N.M., Lichota,J., Stemmer,C., Merkle,T. and Grasser,K.D. (2000) DNA-interactions and nuclear localisation of the chromosomal HMG domain protein SSRP1 from maize. Plant J., 23, 395–405. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Spencer J.A., Baron,M.H. and Olson,E.N. (1999) Cooperative transcriptional activation by serum response factor and the high mobility group protein SSRP1. J. Biol. Chem., 274, 15686–15693. [DOI] [PubMed] [Google Scholar]
- Szak S.T., Mays,D. and Pietenpol,J.A. (2001) Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol., 21, 3375–3386.11313463 [Google Scholar]
- Travis A., Amsterdam,A., Belanger,C. and Grosschedl,R. (1991) LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function [corrected]. Genes Dev., 5, 880–894. [DOI] [PubMed] [Google Scholar]
- Trink B., Okami,K., Wu,L., Sriuranpong,V., Jen,J. and Sidransky,D. (1998) A new human p53 homologue. Nat. Med., 4, 747–748. [DOI] [PubMed] [Google Scholar]
- Watt F. and Molloy,P.L. (1988) High mobility group proteins 1 and 2 stimulate binding of a specific transcription factor to the adenovirus major late promoter. Nucleic Acids Res., 16, 1471–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessagowit V., Mellerio,J.E., Pembroke,A.C. and McGrath,J.A. (2000) Heterozygous germline missense mutation in the p63 gene underlying EEC syndrome. Clin. Exp. Dermatol., 25, 441–443. [DOI] [PubMed] [Google Scholar]
- Wu X., Bayle,J.H., Olson,D. and Levine,A.J. (1993) The p53–mdm-2 autoregulatory feedback loop. Genes Dev., 7, 1126–1132. [DOI] [PubMed] [Google Scholar]
- Yang A. and McKeon,F. (2000) p63 and p73: p53 mimics, menaces and more. Nat. Rev. Mol. Cell. Biol., 1, 199–207. [DOI] [PubMed] [Google Scholar]
- Yang A., Kaghad,M., Wang,Y., Gillett,E., Fleming,M.D., Dotsch,V., Andrews,N.C., Caput,D. and McKeon,F. (1998) p63, a p53 homolog at 3 des multiple products with transactivating, death-inducing and dominant-negative activities. Mol. Cell, 2, 305–316. [DOI] [PubMed] [Google Scholar]
- Yang A. et al. (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature, 398, 714–718. [DOI] [PubMed] [Google Scholar]
- Yarnell A.T., Oh,S., Reinberg,D. and Lippard,S.J. (2001) Interaction of FACT, SSRP1 and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J. Biol. Chem., 276, 25736–25741. [DOI] [PubMed] [Google Scholar]
- Zeng X., Levine,A.J. and Lu,H. (1998) Non-p53 p53RE binding protein, a human transcription factor functionally analogous to p53. Proc. Natl Acad. Sci. USA, 95, 6681–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X. et al. (1999) MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol., 19, 3257–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Li,X., Miller,A., Yuan,Z., Yuan,W., Kwok,R.P., Goodman,R. and Lu,H. (2000) The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol. Cell. Biol., 20, 1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Lee,H., Zhang,Q. and Lu,H. (2001a) p300 does not require its acetylase activity to stimulate p73 function. J. Biol. Chem., 276, 48–52. [DOI] [PubMed] [Google Scholar]
- Zeng X., Zhu,Y. and Lu,H. (2001b) NBP is the p53 homolog p63. Carcinogenesis, 22, 215–219. [DOI] [PubMed] [Google Scholar]