Strand pairing by Rad54 and Rad51 is enhanced by chromatin (original) (raw)
Abstract
We investigated the role of chromatin in the catalysis of homologous strand pairing by Rad54 and Rad51. Rad54 is related to the ATPase subunits of chromatin-remodeling factors, whereas Rad51 is related to bacterial RecA. In the absence of superhelical tension, we found that the efficiency of strand pairing with chromatin is >100-fold higher than that with naked DNA. In addition, we observed that Rad54 and Rad51 function cooperatively in the ATP-dependent remodeling of chromatin. These findings indicate that Rad54 and Rad51 have evolved to function with chromatin, the natural substrate, rather than with naked DNA.
Keywords: Rad54, Rad51, chromatin, homologous recombination
In the eukaryotic nucleus, chromatin is an integral component of processes that use DNA. The packaging of DNA into chromatin is essential for the compaction and organization of nuclear DNA, but it also influences the functions of factors that interact with DNA. For instance, chromatin represses the basal transcription process, and sequence-specific DNA-binding activators along with coactivators (which include chromatin-remodeling factors and histone-modifying proteins) function to counteract chromatin-mediated transcriptional repression. Because chromatin is the natural substrate for DNA-using processes in the nucleus, it will ultimately be necessary to understand how chromatin affects each of these phenomena. To this end, we have undertaken a biochemical analysis of homologous recombination in chromatin.
Homologous recombination occurs in the repair of DNA double-strand breaks as well as during meiosis. Genetic studies in Saccharomyces cerevisiae led to the identification of the RAD52 epistasis group of genes (which includes RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, MRE11, and XRS2) as components of the recombinational repair pathway (Game 1983; Petrini et al. 1997; Kanaar et al. 1998; Paques and Haber 1999; Cromie et al. 2001; Masson and West 2001). These genes are conserved from yeast to humans. A central protein in this pathway is Rad51, which is related to the bacterial RecA protein. Both Rad51 and RecA are able to mediate strand invasion and annealing to yield a D loop, which is a key step in the recombination process. In this reaction, Rad51 (or RecA) forms a nucleoprotein filament on single-stranded DNA in the presence of ATP, and this filament is used for homologous pairing with a double-stranded DNA molecule. The efficiency of strand pairing by Rad51 (between single-stranded DNA and homologous duplex DNA) has been shown to be stimulated by the presence of additional factors such as RP-A (Sugiyama et al. 1997; Baumann and West 1999), the Rad55–Rad57 heterodimer (Sung 1997a), Rad52 (Sung 1997b; Benson et al. 1998; New et al. 1998; Shinohara and Ogawa 1998), and Rad54 (Petukhova et al. 1998, 1999; Mazin et al. 2000a,b; Van Komen et al. 2000).
To study homologous recombination in the context of chromatin, we focused on the ability of purified recombinant Rad51 and Rad54 to catalyze D-loop formation between single-stranded DNA and homologous double-stranded DNA that is packaged into chromatin. The function of Rad54 in chromatin is of particular interest because it is a member of the Snf2-like family of ATPases (Gorbalenya and Koonin 1993; Eisen et al. 1995). The Snf2-like family includes proteins such as Swi2/Snf2, Sth1, ISWI, Ino80, and Mi-2/CHD3/CHD4, which are the ATPase subunits of chromatin-remodeling factors that catalyze the mobilization of nucleosomes (e.g., see Flaus and Owen-Hughes 2001; Fyodorov and Kadonaga 2001; Becker and Hörz 2002). It thus seemed possible that Rad54 would be important for homologous recombination in chromatin. We therefore sought to investigate whether purified Rad51 and Rad54 can mediate D-loop formation with chromatin.
Results and Discussion
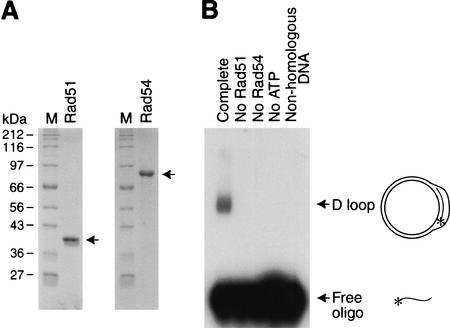
To study the biochemical properties of Rad51 and Rad54, we synthesized Drosophila Rad51 and Rad54 (with C-terminal Flag tags) in Sf9 cells by using a baculovirus expression system, and then purified the proteins to near homogeneity by FLAG immunoaffinity chromatography (Fig. 1A). We tested the ability of these factors to mediate D-loop formation between a radiolabeled, single-stranded oligonucleotide (termed DL2; 135 nt) and a homologous, double-stranded plasmid (pU6LNS; 3291 bp; Pazin et al. 1997). In this reaction, Rad51 assembles onto the single-stranded oligonucleotide in the presence of ATP to give a nucleoprotein filament, and then Rad54 interacts with the Rad51–oligonucleotide complex and facilitates the strand-pairing reaction (Petukhova et al. 1998, 1999; Tan et al. 1999; Mazin et al. 2000a,b). These experiments revealed that purified recombinant Drosophila Rad51 and Rad54 can catalyze the formation of D loops in a manner that is dependent on Rad51, Rad54, ATP, and homologous plasmid DNA (Fig. 1B).
Figure 1.
Drosophila Rad51 and Rad54 mediate D-loop formation. (A) Synthesis and purification of Drosophila Rad51 and Rad54. Flag-tagged Drosophila Rad51 and Rad54 were synthesized in Sf9 cells by using a baculovirus expression vector and affinity-purified with monoclonal antibodies that recognize the Flag epitope. The proteins were subjected to 10% polyacrylamide–SDS gel electrophoresis. The proteins were visualized by staining with Coomassie Brilliant Blue R-250. (B) Formation of D loops with purified Drosophila Rad51 and Rad54 proteins. In the Complete reaction, Rad51 was preincubated with radiolabeled DL2 oligonucleotide in the presence of ATP at 27°C for 20 min; Rad54 and a homologous supercoiled plasmid DNA (pU6LNS) were added; and then the reaction was allowed to proceed at 27°C for 4 min. The resulting DNA was deproteinized, and the samples were subjected to agarose gel electrophoresis and autoradiography. Other reactions either were missing a single component, as indicated, or contained an equivalent mass of nonhomologous DNA. The final concentrations of the reaction components were as follows: Rad51, 200 nM; Rad54, 46 nM; ATP, 2 mM; DL2 oligonucleotide, 1 nM; and pU6LNS, 4 nM.
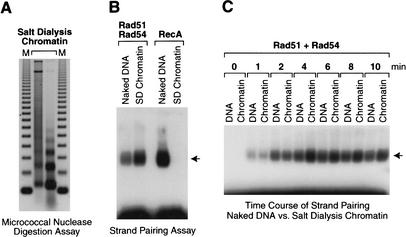
Next, we tested the ability of Rad51 and Rad54 to catalyze D-loop formation in chromatin. In these experiments, we reconstituted chromatin by salt dialysis techniques. The salt dialysis chromatin (SD chromatin) was prepared by gradually decreasing the salt concentration in a mixture of plasmid DNA and purified core histones from Drosophila embryos, and fully reconstituted chromatin was separated from partially reconstituted chromatin by sucrose gradient sedimentation. Micrococcal nuclease digestion analysis of the chromatin samples revealed that the salt dialysis chromatin consisted of closely packed arrays of nucleosomes (Fig. 2A). We then performed D-loop reactions with the SD chromatin. These experiments revealed that Rad51 and Rad54 are able to form D loops with SD chromatin at an efficiency that is slightly higher than that obtained with naked DNA (Fig. 2B). Moreover, the rate of D-loop formation by Rad51 and Rad54 with chromatin is similar to that seen with naked DNA (Fig. 2C). In contrast, the Escherichia coli RecA protein is able to mediate D-loop formation with naked DNA, but not with chromatin (Fig. 2B). Thus, these experiments, which were performed with completely purified components, show that Rad51 in cooperation with Rad54 can mediate D-loop formation with chromatin with comparable efficiency and kinetics as with DNA, whereas the bacterial recombinase RecA is unable to mediate strand pairing with chromatin. The inability of RecA to function with chromatin is consistent with previous studies carried out with mononucleosomes (Kotani and Kmiec 1994), and further suggests that RecA is lacking a chromatin-specific function that is present in Rad51 and/or Rad54. In this regard, we tested whether Rad54 could stimulate D-loop formation in chromatin by RecA, but did not observe any activity (data not shown).
Figure 2.
Rad51 and Rad54, but not RecA, are able to mediate D-loop formation with chromatin. (A) Micrococcal nuclease digestion analysis of chromatin reconstituted from purified components by salt dialysis. Purified Drosophila core histones were reconstituted into chromatin by using salt dialysis techniques (Jeong et al. 1991). The samples were subjected to partial digestion with two different concentrations of micrococcal nuclease and subsequently deproteinized. The resulting DNA fragments were resolved by agarose gel electrophoresis and visualized by staining with ethidium bromide. The mass markers (M) are the 123-bp DNA ladder (GIBCO-BRL). (B) Comparison of the ability of Rad51 + Rad54 versus RecA to mediate D-loop formation with either naked DNA or salt dialysis chromatin. Reactions with Rad51 and Rad54 were performed as in Figure 1B with naked DNA or salt dialysis chromatin (SD Chromatin), except that the final concentration of Rad54 was 28 nM and that of DNA or chromatin was 1 nM. Reactions with RecA were performed in an analogous manner by incubation of purified Escherichia coli RecA with radiolabeled DL2 oligonucleotide at 27°C for 20 min, followed by the addition of plasmid DNA and incubation at 27°C for an additional 20 min. The final concentration of RecA protein was 870 nM. (C) Kinetics of D-loop formation with naked DNA and chromatin. Reactions were performed as in B, except that they were allowed to proceed for the indicated times after the addition of Rad54 and homologous DNA.
The bulk of the eukaryotic genome appears to possess little superhelical tension (Sinden et al. 1980; Giaever and Wang 1988), and we therefore sought to investigate the effect of torsional stress upon D-loop formation by Rad51 and Rad54. To this end, we relaxed the salt dialysis chromatin with purified topoisomerase I (Fig. 3A). The salt dialysis chromatin was reconstituted by using supercoiled plasmid DNA in the absence of topoisomerases. Under these conditions, the DNA remains chemically unchanged, as no phosphodiester bonds are broken. Hence, in the absence of topoisomerase I, the numbers of supercoils in the naked DNA and chromatin (which was deproteinized prior to electrophoresis) are essentially identical (Fig. 3A, cf. lanes 1 and 3). When topoisomerase I is added to the chromatin, the unconstrained supercoils are relaxed, but upon deproteinization, the resulting DNA exhibits supercoils that are caused by the wrapping of the DNA in nucleosomes, because the wrapping of the DNA around each histone octamer constrains approximately one negative supercoil (Fig. 3A, cf. lanes 3,4, and 5; Germond et al. 1975; Simpson et al. 1985).
Figure 3.
Chromatin enhances D-loop formation by Rad51 and Rad54 in the absence of superhelical tension. (A) Relaxation of DNA and chromatin by topoisomerase I. Plasmid DNA and chromatin (reconstituted by salt dialysis) were relaxed with purified recombinant Drosophila topoisomerase I (catalytic fragment). An aliquot of each of the samples was deproteinized and subjected to 1% agarose gel electrophoresis in the presence of 5 μM chloroquine followed by staining with ethidium bromide. ++ indicates twice the topoisomerase I concentration as that used in the + lanes. (B) D-loop formation with relaxed chromatin. D-loop reactions were performed as in Figure 1B, with equimolar amounts of the DNA and chromatin samples shown in A. The topoisomerase I remained in the samples throughout the strand-pairing reactions.
We then performed strand-pairing reactions with DNA and salt dialysis chromatin in the absence or presence of topoisomerase I (Fig. 3B). With naked DNA, we observed a >100-fold reduction in the efficiency of D-loop formation upon relaxation of the template with topoisomerase I (Fig. 3B, cf. lanes 1 and 2). Notably, this >100-fold decrease in strand-pairing efficiency is much more pronounced than the twofold reduction seen with yeast Rad51, Rad54, and RPA (Van Komen et al. 2000). This difference could potentially be due to the use of yeast (Van Komen et al. 2000) versus Drosophila (this study) factors, the presence (Van Komen et al. 2000) or absence (this study) of RPA, the length of the single-stranded DNA (5386 nt, Van Komen et al. 2000; 135 nt, this study), and/or the concentration of Rad51 in the reaction medium (1500 nM, Van Komen et al. 2000; 200 nM, this study). Note, however, that we did not observe stimulation of D-loop formation by purified RPA in our reactions (data not shown). In contrast to the effects seen with naked DNA, relaxation of the chromatin by topoisomerase I has little effect on the efficiency of D-loop formation by Rad51 and Rad54. Thus, in the absence of superhelical tension, strand pairing by Rad51 and Rad54 occurs with higher efficiency in chromatin than in naked DNA (Fig. 3B, cf. lanes 2 and 4).
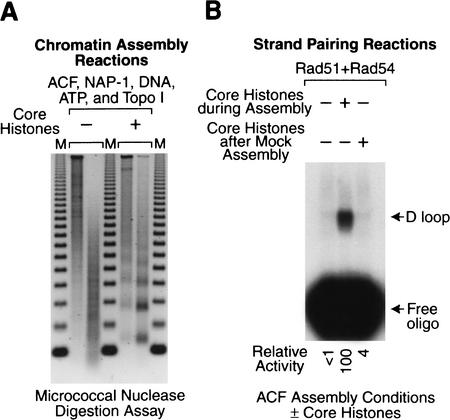
To test further whether chromatin is important for D-loop formation by Rad54 and Rad51, we used a different experimental approach. Instead of relaxing preassembled chromatin, as shown in Figure 3, we examined the effect of chromatin assembly on the efficiency of D-loop formation. To this end, we assembled relaxed DNA into chromatin by using purified recombinant ACF, purified recombinant NAP-1, purified core histones, relaxed plasmid DNA, and ATP in the presence of purified topoisomerase I (Ito et al. 1999). As a control, the core histones were omitted from the reaction. The assembly reaction products were analyzed by the micrococcal nuclease digestion assay (Fig. 4A). Then, in parallel, these samples were used as substrates for strand pairing by Rad54 and Rad51. These experiments revealed that the addition of core histones during chromatin assembly results in a >100-fold enhancement of strand pairing (Fig. 4B, cf. left and center lanes). In contrast, the addition of core histones after a mock assembly reaction (carried out in the absence of histones) did not stimulate D-loop formation (Fig. 3B, right lane). These results indicate that strand pairing is enhanced by chromatin but not nonnucleosomal core histones. [It was also potentially relevant that ACF contains the ISWI ATPase, which is related to the Rad54 protein. We therefore tested whether ACF and/or the NAP-1 core histone chaperone affects the efficiency of the D-loop reaction with salt dialysis chromatin, which is prepared in the absence of ACF or NAP-1, but did not see any effect (data not shown).] Thus, these findings indicate that the packaging of relaxed DNA into chromatin results in a >100-fold stimulation of D-loop formation by Rad54 and Rad51.
Figure 4.
The packaging of relaxed DNA into chromatin facilitates strand pairing by Rad51 and Rad54. (A) ACF-mediated chromatin assembly. Chromatin assembly reactions were performed with purified ACF, NAP-1, topoisomerase I, plasmid DNA, and ATP in the presence or absence of purified core histones, as indicated. The reaction products were subjected to micrococcal nuclease digestion analysis. (B) Strand-pairing reactions. The samples in A were used in strand-pairing reactions with purified Drosophila Rad51 and Rad54 along with the DL2 oligonucleotide. D-loop reactions were performed as in Figure 3B, except that the final concentration of Rad54 was 27 nM. The effect of nonnucleosomal histones on strand pairing was also tested by the addition of core histones (the same amount as that used in the center lane) to the DNA after mock chromatin assembly (with ACF, NAP-1, DNA, ATP, and topoisomerase I in the absence of core histones) and immediately prior to the strand-pairing reactions (right lane).
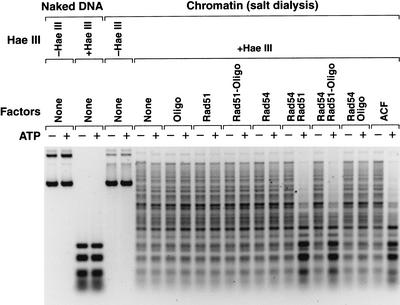
Lastly, because Rad54 is related to the ATPase subunit of chromatin-remodeling complexes, we investigated whether Rad54 possesses chromatin-remodeling activity. We therefore tested the ability of Rad54 and/or Rad51 to facilitate the access of a restriction enzyme (_Hae_III) to DNA packaged into nucleosome arrays (Fig. 5). ACF was used as a positive control. This type of restriction-enzyme accessibility assay has been used for the analysis of chromatin remodeling in vivo (Almer et al. 1986), the biochemical purification of the CHRAC chromatin-remodeling factor (Varga-Weisz et al. 1997), the characterization of the INO80.com remodeling complex (Shen et al. 2000), and the comparative analysis of six chromatin-remodeling complexes (ySWI/SNF, yRSC, hSWI/SNF, xMi-2, dCHRAC, dNURF; Boyer et al. 2000). As shown in Figure 5, neither Rad54 alone nor Rad51 alone exhibited any detectable chromatin-remodeling activity in the absence or presence of the DL2 oligonucleotide. In sharp contrast, we observed that Rad54 and Rad51 function cooperatively in the ATP-dependent remodeling of chromatin. The ability of Rad54 and Rad51 to rearrange chromatin structure is consistent with their ability to catalyze strand pairing with chromatin. It is also notable that Rad54 requires the presence of Rad51 to function as a chromatin-remodeling factor.
Figure 5.
Rad54 and Rad51 function cooperatively in the remodeling of chromatin. Restriction enzyme accessibility assays were carried out with naked DNA or chromatin (salt dialysis reconstitution), the indicated factors, and the restriction enzyme _Hae_III (15 units, GIBCO-BRL) in the same reaction medium used for D-loop reactions. The reactions were incubated at 27°C for 1 h. The samples were deproteinized and subjected to electrophoresis on a 1% agarose gel. The DNA was visualized by staining with ethidium bromide. The final concentrations of the components, which were included as indicated, were as follows: plasmid DNA or chromatin, 2 nM; DL2 oligonucleotide, 1 nM; ATP, 2 mM; Rad51, 200 nM; Rad54, 46 nM; and ACF, 3 nM. The amount of remodeling observed increases with the concentration of the factors (Rad51 and Rad54) as well as with the reaction time (data not shown). Reactions containing DL2 oligonucleotide and Rad51 were preincubated at 27°C for 20 min. There are 14 _Hae_III sites in the pU6LNS plasmid, one of which is in the homologous pairing site.
In conclusion, these studies have revealed that D-loop formation by Rad54 and Rad51 occurs with >100-fold higher efficiency with chromatin relative to naked DNA in the absence of superhelical torsion. In addition, Rad54 and Rad51 act cooperatively in the ATP-dependent remodeling of chromatin. This ability of Rad54 and Rad51 to alter chromatin structure is likely to be related to their chromatin-specific function in the strand-pairing reaction. These findings provide an example of optimized function of eukaryotic DNA-using proteins in chromatin. Moreover, it is possible that the use of chromatin templates, instead of naked DNA templates, might similarly increase the efficiency of targeted homologous recombination in vivo.
Materials and methods
Synthesis and purification of proteins
Full-length cDNA clones that encode Drosophila Rad51 and Rad54 were obtained from Research Genetics and were subcloned into pFastBac1 (GIBCO-BRL). Sequences that encode the Flag epitope tag (DYKDDDDK) were introduced into both constructs at the 3′ end of the coding sequences. The Drosophila homolog of RAD54 has been termed okra (Ghabrial et al. 1998) and DmRAD54 (Kooistra et al. 1997, 1999). In this study, we refer to Drosophila Rad54 protein as Rad54.
Recombinant ACF, recombinant NAP-1, and core histones from Drosophila embryos were purified as previously described (Bulger and Kadonaga 1994; Ito et al. 1999). Rad51 and Rad54 proteins containing C-terminal Flag tags were synthesized in Spodoptera frugiperda (Sf9) cells. The proteins were affinity-purified essentially as described for Flag-tagged ACF (Ito et al. 1999), with the following modifications. After incubation of the cell lysate with Flag M2 resin (Sigma), the resin was washed four times with 12 mL each of wash buffer A [20 mM Tris-HCl at pH 7.9, 150 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 15% (vol/vol) glycerol, 0.01% (vol/vol) NP-40, 10 mM β-glycerophosphate, 0.2 mM PMSF, 0.5 mM benzamidine-HCl, 2 μg/mL leupeptin, 1 μg/mL aprotinin] and two times with 12 mL each of wash buffer B [20 mM HEPES (K+) at pH 7.6, 50 mM potassium glutamate, 0.2 mM EDTA, 15% (vol/vol) glycerol, 0.01% (vol/vol) NP-40, 1 mM DTT, 10 mM β-glycerophosphate, 0.1 mM PMSF, 0.5 mM benzamidine-HCl, 2 μg/mL leupeptin, 1 μg/mL aprotinin]. The protein was eluted by four successive cycles of addition and removal of 100 μL elution buffer (wash buffer B containing 0.4 mg/mL Flag peptide, Sigma; and 0.5 mg/mL recombinant human insulin, Roche). Protein concentrations were estimated by polyacrylamide–SDS gel electrophoresis and staining with Coomassie Brilliant Blue R-250 along with BSA standards. RecA–His6 protein, which contains a C-terminal His6 tag, was synthesized in E. coli and purified by Ni(II) affinity chromatography under native conditions as described (QIA Expressionist, QIAGEN), except that protein was eluted in the following buffer: 50 mM sodium phosphate at pH 8.0, 100 mM NaCl, 250 mM imidazole, 1 mM benzamidine, 1 mM PMSF, 10 mM 2-mercaptoethanol, 2.5 μg/mL aprotinin, 2.5 μg/mL pepstatin, and 2.5 μg/mL leupeptin. Commercially available RecA (Promega) was also used, and yielded identical results to those seen with the His6-tagged RecA.
D-loop reactions
The pU6LNS plasmid (3291 bp; Pazin et al. 1997) was purified by two successive CsCl isopycnic centrifugation steps. The 135-mer oligonucleotide DL2 (5′-GCAGTTCCCCTGCATAAGGATGAACCGTTT TACAAAGAGAAGCTTAACTGCAAAATTGGGCCAAAATTGGGT CGGATCCATGGAAATAACATATGTGTATCTTTATCTTCCTGTA TGATATAGATAACTAACATC-3′) is complementary to pU6LNS. The nonhomologous DNA control used in Figure 1 is a pFastBac1 derivative, and was also purified by two successive CsCl isopycnic centrifugation steps. The DL2 oligonucleotide was radiolabeled by incubation with T4 polynucleotide kinase (Promega) and [γ-32P]ATP (ICN).
D-loop reactions were performed essentially as described previously (Mazin et al. 2000b). In a standard reaction, Rad51 was incubated with radiolabeled DL2 oligonucleotide in buffered medium (25 mM Tris-acetate at pH 7.5, 10 mM magnesium acetate, 100 μg/mL bovine serum albumin, 1 mM DTT, 2 mM ATP, 3 mM phosphoenolpyruvate, 20 units/mL pyruvate kinase) at 27°C for 20 min. Then, Rad54 and pU6LNS (as plasmid DNA or chromatin) was added, and the mixture was incubated at 27°C for 4 min (unless stated otherwise). The reaction was terminated by the addition of EDTA to 50 mM and SDS to 1% (wt/vol). The sample was treated with proteinase K (500 μg/mL) at 37°C for 10 min, and then 1/10 volume of 20% Ficoll, 0.1% bromphenol blue was added. Lastly, the resulting DNA species were resolved by 1% agarose gel electrophoresis, and the dried gel was subjected to autoradiography. The final concentrations of the reaction components were as follows: Rad51 (200 nM), Rad54 (46 nM), ATP (2 mM), DL2 oligonucleotide (1 nM), and pU6LNS (4 nM). Under these conditions with chromatin templates (with excess chromatin relative to oligonucleotide), ∼7% of the radiolabeled oligonucleotide is incorporated into the D loop. Reactions with RecA were performed in an analogous manner by incubation of purified E. coli RecA with radiolabeled DL2 oligonucleotide at 27°C for 20 min, followed by the addition of plasmid DNA and incubation at 27°C for an additional 20 min. The final concentration of RecA protein was 870 nM.
Chromatin assembly
The ATP-dependent assembly of chromatin by purified recombinant ACF and NAP-1 was carried out as described (Ito et al. 1999). Chromatin was reconstituted by salt dialysis with purified Drosophila core histones and plasmid DNA, and the resulting minichromosomes were purified by 15% to 50% sucrose gradient sedimentation (Jeong et al. 1991). Micrococcal nuclease digestion was performed as described previously (Pazin et al. 1997).
Acknowledgments
We thank Buyung Santoso, Chin Yan Lim, Alexandra Lusser, Tammy Juven-Gershon, Karl Haushalter, Dmitry Fyodorov, Mark Levenstein, and Tom Boulay for critical reading of the manuscript; Jim Wang for helpful advice; and Dmitry Fyodorov for purified recombinant topoisomerase I. V.A. was supported by a long-term EMBO postdoctoral fellowship (ALTF 22-2000). This work was supported by a grant from the National Institutes of Health (GM58272) to J.T.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jkadonaga@ucsd.edu; FAX (858) 534-0555.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1032102.
References
- Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, West SC. Heteroduplex formation by human Rad51 protein: Effects of DNA end-structure, hRP-A and hRad52. J Mol Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- Becker PB, Hörz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J Biol Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- Bulger M, Kadonaga JT. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods. Mol Genet. 1994;5:241–262. [Google Scholar]
- Cromie GA, Connelly JC, Leach DRF. Recombination at double-strand breaks and DNA ends: Conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodeling. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT. The many faces of chromatin remodeling: SWItching beyond transcription. Cell. 2001;106:523–525. doi: 10.1016/s0092-8674(01)00478-0. [DOI] [PubMed] [Google Scholar]
- Game JC. Radiation-sensitive mutants and repair in yeast. In: Spencer JFT, et al., editors. Yeast genetics: Fundamental and applied aspects. New York: Springer-Verlag; 1983. pp. 105–137. [Google Scholar]
- Germond JE, Hirt B, Oudet P, Gross-Bellard M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, Schüpbach T. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes & Dev. 1998;12:2711–2723. doi: 10.1101/gad.12.17.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AW, Koonin EV. Helicases: Amino acid sequence comparisons and structure–function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function co-operatively in the ATP-dependent catalysis of chromatin assembly. Genes & Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SW, Lauderdale JD, Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J Mol Biol. 1991;222:1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JHJ, van Gent DC. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Kooistra R, Vreeken K, Zonneveld JBM, de Jong A, Eeken JCJ, Osgood CJ, Buerstedde J-M, Lohman PHM, Pastink A. The Drosophila melanogaster RAD54 homolog, DmRAD54, is involved in the repair of radiation damage and recombination. Mol Cell Biol. 1997;17:6097–6104. doi: 10.1128/mcb.17.10.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra R, Pastink A, Zonneveld JBM, Lohman PHM, Eeken JCJ. The Drosophila melanogaster DmRAD54 gene plays a crucial role in double-strand break repair after P-element excision and acts synergistically with Ku70 in the repair of X-ray damage. Mol Cell Biol. 1999;19:6269–6275. doi: 10.1128/mcb.19.9.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani H, Kmiec EB. DNA cruciforms facilitate in vitro strand transfer on nucleosomal templates. Mol Gen Genet. 1994;243:681–690. doi: 10.1007/BF00279578. [DOI] [PubMed] [Google Scholar]
- Masson J-Y, West SC. The Rad51 and Dmc1 recombinases: A non-identical twin relationship. Trends Biochem Sci. 2001;26:131–136. doi: 10.1016/s0968-0004(00)01742-4. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000a;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Bornarth CJ, Solinger JA, Heyer W, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol Cell. 2000b;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Bhargava P, Geiduschek EP, Kadonaga JT. Nucleosome mobility and the maintenance of nucleosome positioning. Science. 1997;276:809–812. doi: 10.1126/science.276.5313.809. [DOI] [PubMed] [Google Scholar]
- Petrini JHJ, Bressan DA, Yao MS. The RAD52 epistasis group in mammalian double strand break repair. Semin Immunol. 1997;9:181–188. doi: 10.1006/smim.1997.0067. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: A model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: Analogous measurements in insects and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997a;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- ————— Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997b;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Tan TL, Essers J, Citterio E, Swagemakers SM, de Wit J, Benson FE, Hoeijmakers JH, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]