Dual Roles for Spt5 in Pre-mRNA Processing and Transcription Elongation Revealed by Identification of Spt5-Associated Proteins (original) (raw)
Abstract
During transcription elongation, eukaryotic RNA polymerase II (Pol II) must contend with the barrier presented by nucleosomes. The conserved Spt4-Spt5 complex has been proposed to regulate elongation through nucleosomes by Pol II. To help define the mechanism of Spt5 function, we have characterized proteins that coimmunopurify with Spt5. Among these are the general elongation factors TFIIF and TFIIS as well as Spt6 and FACT, factors thought to regulate elongation through nucleosomes. Spt5 also coimmunopurified with the mRNA capping enzyme and cap methyltransferase, and spt4 and spt5 mutations displayed genetic interactions with mutations in capping enzyme genes. Additionally, we found that spt4 and spt5 mutations lead to accumulation of unspliced pre-mRNA. Spt5 also copurified with several previously unstudied proteins; we demonstrate that one of these is encoded by a new member of the SPT gene family. Finally, by immunoprecipitating these factors we found evidence that Spt5 participates in at least three Pol II complexes. These observations provide new evidence of roles for Spt4-Spt5 in pre-mRNA processing and transcription elongation.
Synthesis of mRNA is a multistep process of transcription and pre-mRNA processing. The study of mRNA synthesis has proceeded largely through a reductionist approach, with mRNA production viewed as a series of reactions connected by their substrates and products. It is becoming clear that many RNA-processing reactions occur during transcription elongation (reviewed in references 34 and 45).
However, our understanding of the elongation phase of transcription is incomplete. In vitro, two general transcription elongation factors, TFIIF and TFIIS, are sufficient to stimulate in vivo rates of elongation on naked DNA templates (25). In contrast, elongation on nucleosome-bound templates is inefficient, even in the presence of TFIIF and TFIIS, suggesting a requirement for other factors (9, 25, 26). Several factors have been implicated in the regulation of transcription elongation through chromatin. Among these is the yeast Spt4-Spt5 complex, known as DSIF in human cells (21, 57). DSIF/Spt4-Spt5 can inhibit and promote elongation of RNA polymerase II (Pol II) on cellular genes and is required for the stimulation of transcription elongation by human immunodeficiency virus type 1 Tat in vitro (24, 28, 57, 64). A second elongation factor, Spt6, interacts genetically with SPT4, SPT5, and TFIIS and also promotes Tat function in vitro (21, 51, 64). Consistent with their playing a role in elongation, chromatin immunoprecipitation experiments show that the Spt5 and Spt6 proteins associate with transcribed genes in yeast and Drosophila (2, 27, 44). Finally, genetic and biochemical studies of Spt4, Spt5, and Spt6 in yeast have led to the proposal that they function by affecting chromatin structure (6, 51). A third protein complex, FACT, composed of the human Spt16 and SSRP1 proteins, promotes elongation by Pol II through nucleosomes in vitro (40, 41). Its yeast homolog, the CP (or SPN) complex, is composed of two tightly associated subunits, Pob3 and Cdc68/Spt16 (the name Cdc68 will be used here to avoid confusion of Spt6 with Spt16), as well as a weakly associated HMG box protein Nhp6 (7, 8, 18). Mutations in SPT4, SPT5, SPT6, CDC68, POB3, and NHP6 lead to similar mutant phenotypes, and these genes also display numerous genetic interactions with each other (reviewed in references 22 and 60). Thus, although direct evidence is lacking, the overlapping genetic and biochemical behaviors of these Spt proteins suggest that they may collaborate to carry out a common or overlapping set of functions in vivo.
Recent observations suggest a functional interplay between Spt4-Spt5 and the C-terminal heptapeptide repeats (CTD) of Pol II. The CTD serves as a scaffold for factors involved in transcription and processing. For example, the mRNA capping enzyme, polyadenylation factors, and certain splicing proteins all associate with the CTD of transcribing RNA Pol II. Furthermore, perturbation of the CTD or addition of CTD peptides affects splicing in vitro and in vivo, suggesting that the CTD may affect the efficiency of processing reactions (reviewed in references 34 and 45). Biochemical studies show that P-TEFb, a CTD kinase that regulates elongation, works in conjunction with DSIF and possibly FACT (56, 58). In addition, we have recently shown that SPT4 and SPT5 display an extensive set of genetic interactions with the CTD and enzymes that modify the CTD's phosphorylation status, including protein kinases similar to P-TEFb (31, 39). Finally, the human and Schizosaccharomyces pombe Spt5 proteins interact with the capping enzyme (43, 59). These studies show that Spt4-Spt5 is a candidate for an elongation regulator that mediates interactions between the elongating polymerase and processing events linked to the CTD.
A mechanistic understanding of Spt4-Spt5 function requires a knowledge of the proteins that associate with this complex. Here we describe affinity purification of Spt5 from yeast extracts. Using mass spectrometry, we identified a large number of proteins that copurified with Spt5. Many of these interactions were subsequently verified by coimmunoprecipitation and genetic analysis. We show that Spt5 associates with Pol II and the general elongation factors TFIIF and TFIIS, as well as with Spt6, Cdc68, and Pob3. Furthermore, we demonstrate that Spt5 coimmunopurifies with the yeast capping enzyme and cap methyltransferase and that spt4 and spt5 mutations cause splicing defects in yeast. In addition, we show that Spt5 copurifies and genetically interacts with a recently identified Spt6-interacting protein, Iws1. Through extensive coimmunoprecipitation analyses we provide evidence that Spt5 participates in at least three different protein complexes with Pol II. These observations provide new evidence of close connections between pre-mRNA processing and transcription elongation and suggest important roles for Spt4-Spt5 in both processes.
MATERIALS AND METHODS
Media and genetic methods.
Strain construction and other genetic manipulations were carried out by standard methods (46). Yeast media were made as described previously (46). All GHY and FY Saccharomyces cerevisiae strains used in this study (Table 1) are isogenic to S288C (61). C-terminal tagging of Tfg1, Fcp1, and Iws1 with 13 copies of the Myc epitope was performed by a PCR-based method (33). The dilution spotting growth assay (Fig. 4) was carried out as described previously (31), and the results presented are representative of those obtained for multiple transformants in at least two separate transformations. IWS1 was mutagenized by PCR amplifying the _IWS1_-MYC::TRP1 allele from genomic DNA by using primers OGH268 (5′-CAGCGGCCGCCCAAATGCCAGATCATTG) and OGH269 (5′-GTGCGGCCGCTGGGTATCGAATCCAAGC). This PCR product, composed of the Iws1 open reading frame fused to the Myc tag, followed by TRP1 and then by sequences 3′ to IWS1, was transformed into strain FY119, and Trp+ transformants were screened for mutant phenotypes. Integration of the PCR product at the IWS1 locus was confirmed by genetic linkage analysis and plasmid complementation. Strains carrying Spt4 tagged at its C terminus with three copies of the Flag epitope were derived from a strain provided by Steve Hahn.
TABLE 1.
Strains
| Strain | Genotype | Source |
|---|---|---|
| GHY13 | MATa_his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 spt5-194 | Hartzog lab collection |
| GHY92 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 spt5-242 | Hartzog lab collection |
| GHY96 | _MAT_α _his3_Δ_200 lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 spt4-3 | Hartzog lab collection |
| GHY379 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 spt5-194 | Hartzog lab collection |
| GHY524 | MATa_his4-912_δ _lys2-128_δ leu2_Δ_1 spt4_Δ_2::HIS3 | Hartzog lab collection |
| GHY594 | MAT α _his3_Δ_200 lys2-128_δ ura3-52 spt5-194 | Hartzog lab collection |
| GHY605 | MATa_his4-912_δ _lys2-128_δ leu2_Δ_1 HA-SPT6 | Hartzog lab collection |
| GHY611 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 trp1_Δ_63 SPT5-MYC | Hartzog lab collection |
| GHY617 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 SPT5-FLAG | Hartzog lab collection |
| GHY1073 | _MAT_α _his4-912_δ _lys2-128_δ trp1_Δ_63 spt5-4 | Hartzog lab collection |
| GHY1199 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 iws1-7-MYC::TRP1 | This study |
| GHY1200 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 iws1-13-MYC::TRP1 | This study |
| GHY1202 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 SPT5-FLAG trp1_Δ_63 iws1-13-MYC::TRP1 | This study |
| GHY1207 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 SPT5-FLAG FCP1-MYC::TRP1 | This study |
| GHY1208 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 SPT5-FLAG TFG1-MYC::TRP1 | This study |
| GHY1280 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 SPT5-FLAG HA-SPT6 | This study |
| GHY1300 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 trp1_Δ_63 SPT5-FLAG HA-SPT6 IWS1-MYC::TRP1 | This study |
| GHY1320 | MATa_his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 HA-SPT6 IWS1-MYC::TRP1 | This study |
| GHY1324 | MATalys2(-128_δ or Δ_0) leu2(Δ_0_ or Δ_1_) ura3(Δ_0_ or -52) trp1_Δ_63 HA-SPT6 IWS1-MYC::TRP1 SPT4-FLAG::KanMX | This study |
| GHY1325 | MAT_α his4-912_δ lys2(-128_δ or Δ_0) leu2(Δ_0 or Δ_1) ura3(Δ_0 or -52_) trp1_Δ_63 IWS1-MYC::TRP1 SPT4-FLAG::KanMX | This study |
| DK186 | MATahis3-11 can1-100 trp1-1 leu2-3,112 ura3-52 ade2-1 bar1 Gal+ | Doug Kellogg |
| FY119 | _MAT_α _his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 | Fred Winston |
| FY120 | MATa_his4-912_δ _lys2-128_δ leu2_Δ_1 ura3-52 | Fred Winston |
| FY602 | MATa_his3_Δ_200 lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 | Fred Winston |
| SRY4-1a | MATaura3-52 ade2-101 his3 his7 prp4-1 | Manny Ares |
| OY43 | MATa_his4-912_δ _lys2-128_δ spt5-4 | Fred Winston |
| OY163 | MATa_his3 lys2-128_δ ura3 ceg1-250 | This study |
| OY215 | MATa_his3 lys2-128_δ ura3 ceg1-250 spt5-194 | This study |
| OY167 | MATa_his3 lys2-128_δ ura3 trp1_Δ_63 ceg1-250 spt4-3 | This study |
| YSB427 | _MAT_α _ura3-52 leu2_Δ_1 trp1_Δ_63 abd1_Δ::TRP1 [pRS316-_ABD1_] | Steve Buratowski |
| YSB244 | MATahis3_Δ_200 leu2-3,112 ura3-52 ceg1_Δ_1::HIS3 [pRS316-_CEG1_] | Steve Buratowski |
| OY204 | MATa_his3_Δ_200 lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 spt4-3 cet1_Δ_1::TRP1 [pRS316-_CET1_] | This study |
| OY205 | MATa_his3_Δ_200 lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 spt5-242 cet1_Δ_1::TRP1 [pRS316-_CET1_] | This study |
| OY207 | MATa_his3_Δ_200 lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 spt5-194 cet1_Δ_1::TRP1 [pRS316-_CET1_] | This study |
| OY227 | MATa_his3_Δ_200 lys2-128_δ leu2_Δ_1 ura3-52 trp1_Δ_63 cet1_Δ_1::TRP1 [pRS316-_CET1_] | This study |
| OY240 | _MAT_α _his4-912_δ his3_Δ_200 leu2 ura3 trp1 can1 HA-SPT6 CDC68-MYC::KanMX | This study |
| DY6460 | _MAT_α ade2 can1 his3 leu2 trp1 ura3 POB3-MYC::KanMX NHP6A-HA::URA3 | David Stillman |
| DY6529 | _MAT_α ade2 can1 his3 leu2 trp1 ura3 CDC68-MYC::KanMX NHP6A-HA::URA3 | David Stillman |
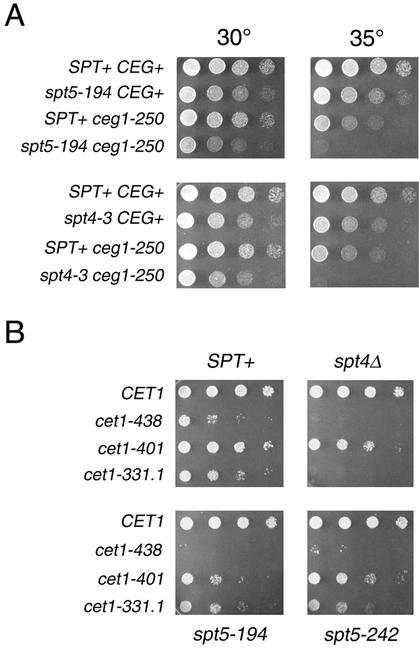
FIG. 4.
SPT4 and SPT5 display genetic interactions with capping enzyme. (A) Strains with the indicated genotypes were assayed by serial dilution onto YPD plates, incubated for 2 days at the indicated temperatures, and photographed. Note that the restrictive temperature for the _ceg1_-250 single mutant is 37°C (19). Strains: wild type, FY120; _spt5_-194, GHY594; _spt4_-3, GHY96; _ceg1_-250, OY163; _spt5_-_194 ceg1_-250, OY215; _spt4_-_3 ceg1_-250, OY167. (B) spt cet1Δ mutants carrying a URA3 CET1 plasmid were transformed with a series of HIS3 cet1 plasmids as indicated. Transformants were grown to saturation in SC-histidine medium and assayed by serial dilution on 5-fluoroorotic acid plates, which allows growth only of cells that have lost the URA3 CET1 plasmid. Plates were incubated for 2 days at 30°C and photographed. Strains: cet1Δ1::TRP, OY227; spt4Δ cet1Δ1::TRP1, OY204; _spt5_-194 cet1Δ1::TRP1, OY207; _spt5_-242 cet1Δ1::TRP1, OY205.
Plasmids.
The CET1 plasmids pRS316-CET1 and pRS313-CET1 and derivatives of pRS313-CET1 carrying cet1 mutations (52) and the pRS315-based plasmids pSB995 (HA-Abd1) and pSB996 (HA-Ceg1) (29) have been described previously. Plasmid pYST138 was used to create the TUB2 probe for Northern blot analysis (50). Plasmid pGH193, used to create the HIS4 probe for Northern blot analysis, is a Bluescript derivative containing the HIS4 open reading frame.
Immunoprecipitation and immunoblotting.
Cells were grown to mid-log phase in yeast extract-peptone-dextrose (YPD) unless otherwise noted, harvested, and frozen in liquid nitrogen. Frozen cell pellets were ground into a fine powder under liquid nitrogen with a mortar and pestle. Protein lysates were prepared as previously described in lysis buffer (30 mM HEPES [pH 7.4], 200 mM potassium acetate, 1 mM magnesium acetate, 1 mM EGTA, 0.05% Tween 20, 10% glycerol) containing protease inhibitors (31). For immunoprecipitations with the antihemagglutinin (anti-HA) polyclonal or anti-Myc 9E10 antibodies, 6 μg of purified immunoglobulin G was prebound to 12 μl of protein A-agarose beads (Bio-Rad) overnight at 4°C. The beads were washed three times with 0.5 ml of lysis buffer, and immunoprecipitations were performed as previously described (31). Proteins were eluted from the beads with lysis buffer containing 1.0 M potassium acetate. Immunoblotting was performed as previously described (31). Approximately 1% of the total crude extracts was immunoblotted for comparison to eluates. The anti-Spt4 and anti-Spt5 polyclonal antibodies, the anti-Myc antibody (9E10; Santa Cruz Biotechnology), and the anti-Pol II antibodies B3 and 8WG16 have been previously described (15, 21, 53). The affinity-purified anti-HA rabbit polyclonal antibody was produced as previously described (37). The anti-Spt6 antibody was a gift from Clyde Denis (14). The anti-TFIIS antibody was a gift from Caroline Kane. The anti-Abd1 antibody was a gift from Steve Buratowski (52). The anti-Cdc68 and anti-Pob3 antibodies were gifts from Tim Formosa (63). The anti-Nhp6 antibody was a gift from David Stillman.
In contrast to the data presented here, we and others were previously unable to detect robust Spt5-Spt6 interactions in yeast by coimmunoprecipitation (21, 51). We have found that Spt5 and Spt6 only weakly coimmunoprecipitate from extracts of yeast cells prepared by bead beating, as was reported previously (21, 51). In contrast, Spt5 and Spt6 show a robust interaction when coimmunoprecipitated from extracts prepared by grinding in a mortar and pestle under liquid nitrogen (see Fig. 3 and 4) (D. L. Lindstrom and G. A. Hartzog, unpublished data).
FIG. 3.
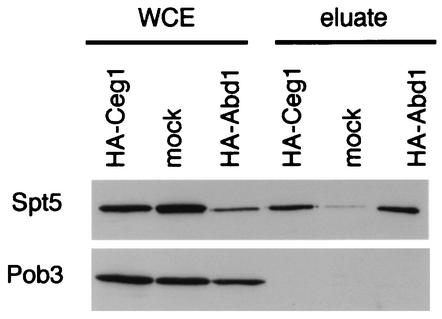
Spt5 but not Pob3 associates with capping enzyme and cap methyltransferase. Anti-HA immunoprecipitations from extracts of HA-Ceg1, HA-Abd1, and untagged (mock) strains are shown. Bound proteins were eluted with 1.0 M potassium acetate, separated by SDS-PAGE, and blotted for Spt5 and Pob3. WCE, 30 μg of whole-cell extract. Eluate, 1.0 M potassium acetate elutions. Strains: HA-Ceg1, YSB244/pSB996; HA-Abd1, YSB427/pSB995; untagged, YSB244/pRS316-CEG1.
Gel filtration chromatography.
Gel filtration chromatography was performed on a Superose 6 column (Pharmacia) equilibrated in lysis buffer (described above). For fractionation of Spt5-Flag complexes, Spt5-Flag immunoprecipitates derived from ∼10 mg of cleared lysate were injected onto the column, and 0.5-ml fractions were collected as previously reported (49). Samples (250 μl) of each fraction were trichloroacetic acid precipitated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose for immunoblotting.
Affinity purification of Spt5-Flag.
Spt5-Flag was affinity purified by a modification of a published method (54). Cleared whole-cell lysates (∼350 mg) were prepared from GHY617 (Spt5-Flag) or GHY611 (mock) as previously described (31) and batch bound to 600 μl of anti-Flag M2 agarose beads for 2 h at 4°C. The supernatant was collected, and the beads were washed four times with 25 volumes of lysis buffer. Bound proteins were eluted twice by addition of 1.5 ml of lysis buffer supplemented with 500 μg of Flag peptide (Research Genetics) per ml. The beads were washed three times with 10 ml of lysis buffer, and the original supernatant was reapplied to the beads for a second round of immunoprecipitation. Eluates from both rounds of immunoprecipitation were pooled and batch bound to 200 μl of Bio-Rex 70 cation exchange resin (Bio-Rad) for 10 min at 4°C. The resin was packed into a 1-ml syringe barrel and washed with 10 volumes of lysis buffer. The column was eluted stepwise with 1.2 ml each of lysis buffer containing 0.4 and 1.0 M potassium acetate.
Mass spectrometry.
To prepare samples for mass spectrometry, Bio-Rex 70 eluates were mixed with 4 volumes of methanol, 1 volume of chloroform, and 3 volumes of water. The phases were separated by centrifugation, and the upper phase was transferred and precipitated with 3 volumes of methanol. The precipitates were pelleted and dried under vacuum. Protein samples were subjected to tryptic digestion and mass spectrometry as previously described (32).
Analysis of RNA.
RNA isolation and primer extension were performed as previously described (4), using 10 μg of total RNA/sample. The sequence of the primer used to detect U3A and U3B was 5′-CCAAGTTGGATTCAGTGGCTC. Reverse transcription-PCR (RT-PCR) was performed as previously described (13). One microgram of total RNA per 10-μl sample was used for reverse transcription (see primer sequences below). The cDNA product was precipitated and suspended in 10 μl of H2O, and 1 μl was used to seed a 20-μl PCR mixture spiked with ∼100 fmol 32P end-labeled reverse primer. PCR conditions were as follows: 94°C for 5 min followed by 20 cycles of 94°C for 1 min, annealing temperature (see below) for 15 s, and 72°C for 1 min; the last cycle was followed by a final incubation at 72°C for 10 min. RT-PCR primers and annealing temperatures were as follows: for RPL26A, forward primer 5′-GGTAAGATTTGTTGAAACTCG, reverse primer 5′-GCTTTTCTGTCCTTGTCCAAA, and a 54°C annealing temperature; for RPS27B, forward primer 5′-TGAAACGACTTTCGTTTTCG, reverse primer 5′-CCTTACCACCGGTTGGAGTA, and a 50°C annealing temperature; for RPS25A, forward primer 5′-CCCAAATTCTACTAGAGTTCGG, reverse primer 5′-TAGCTTGCTTGGAGTGCTTG, and a 52°C annealing temperature; and for SED1, forward primer 5′-AGAGGCTCCAACCACTGCTA, reverse primer 5′-ATAGCAACACCAGCCAAACC, and a 58°C annealing temperature. PCR products were fractionated on 6% native polyacrylamide gels and visualized by autoradiography. Northern blot analysis was performed as described previously (50). The amount of RNA in each lane was normalized to TUB2.
RESULTS
Identification of proteins that copurify with Spt5.
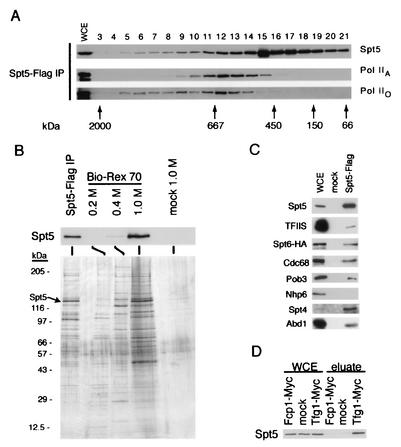
Previous studies have shown that Spt4-Spt5 coimmunopurifies and genetically interacts with Pol II and the Paf1 complex (38, 49). To further characterize the interaction of Spt5 with Pol II, we separated Spt5-Flag immunoprecipitates by gel filtration and analyzed the eluates by immunoblotting (Fig. 1A). Consistent with the idea that Spt5 assembles into high-molecular-weight complexes with Pol IIA and Pol IIO, we observed a broad distribution of high-molecular-weight forms of these proteins. Similarly, Mueller and Jaehning have previously observed Pol II and Spt5 cofractionating in high-molecular-weight complexes with affinity-purified Paf1 (38).
FIG. 1.
Identification of Spt5-associated proteins. (A) Anti-Flag immunoprecipitates from extracts of an Spt5-Flag strain (GHY617) were size fractionated by gel filtration, and fractions were analyzed by blotting with antibodies specific for Spt5, the hypophosphorylated form of Rpb1 (Pol IIA), and the hyperphosphorylated form of Rpb1 (Pol IIO). Control experiments demonstrated that coprecipitation of both Pol II isoforms with Spt5-Flag was specific (reference 31 and data not shown). (B) Spt5-Flag purification. Extracts of an Spt5-Flag strain and an untagged strain (mock) were incubated with anti-Flag M2 agarose beads. Spt5-Flag complexes were competitively eluted from the beads with Flag peptide and further fractionated on a Bio-Rex 70 column. Fractions were analyzed by silver staining and blotting for Spt5. Upper panel, anti-Spt5 Western blot. Lower panel, silver-stained gel. (C) Anti-Flag immunoprecipitations from extracts of Spt5-Flag and untagged (mock) strains. Proteins were eluted with Flag peptide, separated by SDS-PAGE, and blotted with the indicated antibodies. WCE, 30 μg of whole-cell extract of Spt5-Flag strain. Note that in this panel the relative proportion of whole-cell extract to eluate is five times higher than in other immunoprecipitations. (D) Anti-Myc immunoprecipitations from extracts of strains carrying Fcp1-Myc or Tfg1-Myc and a strain lacking the Myc tag (mock). Proteins were eluted in 1.0 M potassium acetate, fractionated by SDS-PAGE, and blotted for Spt5. Strains: Spt5-Flag, GHY617; Fcp1-Myc, GHY1207; Tfg1-Myc, GHY1208; mock in panels A to C, GHY611; mock in panel D, GHY617.
To determine if proteins other than Pol II and the Paf1 complex associate with Spt5, we immunoprecipitated Spt5-Flag from yeast extracts, competitively eluted it from anti-Flag beads with the Flag peptide, and further fractionated it on a Bio-Rex 70 column. Immunoblotting demonstrated that Spt5 bound to Bio-Rex 70 in buffer containing 0.2 M potassium acetate, that a small amount of Spt5 eluted in 0.4 M potassium acetate, and that the bulk of Spt5 was eluted by 1.0 M potassium acetate (Fig. 1B, upper panel, and data not shown). Silver staining showed that many proteins copurified with Spt5-Flag (Fig. 1B, lower panel). To control for nonspecific binding to the anti-Flag beads, this experiment was repeated with an equal amount of an extract of a strain that lacked the Flag epitope. We did not observe Spt5 by immunoblotting or significant silver staining of other protein bands in these mock purified samples (Fig. 1B).
To identify proteins that copurified with Spt5, the 1.0 M eluates of the mock- and affinity-purified samples from the Bio-Rex 70 column were subjected to direct analysis of large protein complexes (DALPC) by mass spectrometry (32). In DALPC, a purified sample of a protein complex is proteolysed without prior separation of the constituent proteins, and the resulting peptides are fractionated by high-pressure liquid chromatography and analyzed on an electrospray-equipped triple-quadrupole mass spectrometer. By using the SEQUEST program, acquired masses were correlated with peptide sequences predicted from the genomic sequence. This approach has previously been used to identify subunits of the ribosome, proteosome, and other protein complexes (32, 55).
Peptides from many proteins were found in the Spt5-Flag fractions. To focus on those most likely to associate with Spt5 in vivo, we excluded known cytoplasmic proteins from our analysis, since Spt5 is nuclear (50), and we also discarded heat shock proteins, ribosomal proteins, and translation factors, as these are often found as contaminants in proteomic studies (23). Of the proteins that met these criteria, nine were also found in the mock-purified fraction and were also excluded from further analysis. The 92 proteins in 1.0 M Spt5-Flag fraction that pass all of these criteria are presented in Table 2. Recently, Ho et al. described the purification of a large set of protein complexes by using Flag affinity chromatography (23). In addition to ribosomal proteins, eight proteins that were frequent contaminants in their protein complexes appeared in our data set and are indicated in Table 2. Although some of the proteins in Table 2 have not been studied and lack strong homologs, we were able to group many by previously reported functions or homology to proteins of known function, and many of these are implicated in transcription.
TABLE 2.
Proteins that copurified with Spt5-Flag
| Protein type and gene | No. of peptides | Note(s)a |
|---|---|---|
| Polymerase subunits | ||
| RPO21 | 33 | B, RPB1 |
| RPB2 | 24 | A, B |
| RPB3 | 3 | B |
| RPB4 | 6 | B |
| RPB5 | 4 | B, shared subunit with Pol I and Pol III |
| RPO26 | 2 | B, RPB6, shared subunit with Pol I and Pol III |
| RPB7 | 1 | B |
| RPB8 | 2 | A, B, shared subunit with Pol I and Pol III |
| RPB11 | 2 | B |
| RPB12 | 1 | Shared subunit with Pol I and Pol III |
| General transcription factors | ||
| SUA7 | 1 | TFIIB |
| TFG1 | 9 | B, TFIIF, Rap74 homolog |
| TFG2 | 3 | B, TFIIF |
| ANC1 | 3 | B, TFIIF |
| DST1 | 3 | TFIIS |
| TAF60 | 1 | |
| Transcription elongation | ||
| IKI3 | 1 | ELP1, elongator subunit |
| NHP6A | 1 | CP/SPN subunit |
| NHP6B | 1 | CP/SPN subunit |
| CDC68 | 1 | CP/SPN subunit |
| SPT5 | 41 | B |
| SPT6 | 17 | |
| THO1 | 1 | Elongation-associated recombination |
| mRNA capping | ||
| ABD1 | 1 | B, cap methyltransferase |
| CEG1 | 6 | Cap guanylyltransferase |
| CET1 | 12 | Cap triphosphatase |
| Termination/3′ end formation | ||
| CFT2 | 1 | CFII component |
| PAB1 | 2 | A, poly(A)-binding protein |
| PAP1 | 1 | Poly(A) polymerase |
| REF2 | 1 | 3′ end processing |
| RNA14 | 2 | CF1 component |
| RNA15 | 1 | CF1 component |
| SUB1 | 2 | 3′ end processing |
| RNA processing | ||
| CBF5 | 2 | rRNA processing |
| DBP3 | 2 | RNA helicase |
| DBP9 | 1 | rRNA processing |
| DBP10 | 2 | RNA helicase |
| DIM1 | 2 | rRNA processing |
| HCA4 | 2 | rRNA processing |
| MRT4 | 1 | mRNA turnover |
| MTR4 | 3 | mRNA transport |
| NOP2 | 8 | rRNA processing |
| ROK1 | 1 | RNA helicase |
| RRP5 | 6 | rRNA processing |
| SEN1 | 1 | tRNA splicing |
| YRA1 | 6 | mRNA transport |
| YRA2 | 1 | mRNA transport |
| Other polymerases | ||
| POL1 | 1 | Alpha DNA polymerase |
| POL2 | 2 | Epsilon DNA polymerase |
| RPA49 | 5 | RNA polymerase I |
| RPA135 | 5 | RNA polymerase I |
| RPA190 | 11 | RNA polymerase I |
| RPC40 | 1 | Shared subunit of RNA Pol I and Pol III |
| Casein kinase II | ||
| CKA1 | 1 | Catalytic subunit |
| CKA2 | 1 | Catalytic subunit |
| CKB1 | 1 | Regulatory subunit |
| CKB2 | 2 | Regulatory subunit |
| Other proteins | ||
| AMD1 | 1 | AMP deaminase |
| BBC1 | 3 | SH3 domain |
| BRE5 | 2 | RRM motif |
| CRP1 | 5 | Binds cruciform DNA |
| DOT6 | 1 | |
| IMD1 | 3 | A, IMP dehydrogenase |
| IMD4 | 3 | Similarity to IMP dehydrogenase |
| IWS1 | 9 | YPR133C, this work |
| KRE30 | 5 | ABC superfamily |
| KRE33 | 3 | |
| LOC1 | 2 | A |
| MAP2 | 1 | A, methionyl aminopeptidase |
| NOG1 | 1 | GTPase |
| NOP1 | 1 | |
| NOP7 | 2 | Pescadillo homology |
| NOP14 | 1 | |
| NSA1 | 1 | |
| PUF6 | 1 | PUF family member |
| RAI1 | 1 | |
| RFC1 | 1 | DNA replication |
| RLI1 | 2 | RNase L inhibitor |
| RRP1 | 1 | RNA binding protein |
| SMC2 | 1 | A, chromosome condensation |
| SMC3 | 1 | Chromosome condensation |
| STM1 | 1 | A, nucleic acid-binding protein |
| SVL3 | 2 | |
| TOM1 | 1 | Ubiquitin-protein ligase |
| VIP1 | 4 | |
| YDR266C | 1 | RING finger domain |
| YDR365C | 2 | |
| YFR024C | 1 | SH3 domain |
| YGR054W | 2 | 20% identity to Prt1 |
| YHR197W | 1 | NuA3 HAT complex |
| YJR014W | 1 | |
| YOR262W | 1 | |
| YPR169W | 1 |
Four potential protein complexes containing Spt4-Spt5 were described in two recent large-scale proteomic studies of yeast proteins (20, 23). Only one of these complexes is composed predominantly of proteins implicated in transcription (complex 145 in reference 20). The composition of this complex was derived from the combined results of tandem affinity purifications of Abd1 and several Pol II subunits, and 15 of the 19 proteins in the complex are represented in our data (Table 2). No independent methods were used to confirm the composition of this complex. Spurious copurification of proteins is a common concern in proteomic studies (23), and it is likely that some of the proteins reported as Spt5 associated in Table 2 or by Gavin et al. (20) do not specifically associate with Spt5-Flag. The remainder of this report is devoted to the analysis of the association of Spt5 with proteins reported in Table 2 that are implicated in Pol II transcription and pre-mRNA processing.
Pol II and general transcription factors.
The general transcription elongation factor TFIIS and at least 10 of the 12 Pol II subunits copurified with Spt5 (Table 2). This is consistent with previous observations of Spt5-Pol II coimmunoprecipitation and of genetic interactions of SPT4 and SPT5 with DST1, RPO21, and RPB2, which encode TFIIS and the two largest subunits of Pol II (21, 31, 58) (Fig. 1A and C). Because DALPC is not quantitative, protein stoichiometry cannot be determined (32). To examine the relationship between Spt5 and TFIIS, we probed blots of Spt5-Flag immunoprecipitates and found that TFIIS had coprecipitated (Fig. 1C). However, in contrast to Pol II, TFIIS did not cofractionate with Spt5 on a gel filtration column (data not shown). Thus, TFIIS likely associates with Spt5 in a transient or indirect manner.
All three subunits of transcription initiation and elongation factor TFIIF copurified with Spt5 (Table 2). To confirm these data, we immunoprecipitated a Myc epitope-tagged allele of Tfg1 and observed that Spt5 specifically coimmunoprecipitated with it (Fig. 1D). TFIIF interacts biochemically with the CTD phosphatase Fcp1, and an fcp1 mutation interacts genetically with spt4 and spt5 mutations (3, 31). Furthermore, human Fcp1 associates weakly with a complex that includes Spt5 (42). Therefore, even though it was not found in our mass spectrometry data set, we immunoprecipitated a Myc-tagged form of Fcp1 to determine whether it coimmunopurifies with Spt5 (Fig. 1D). We failed to detect Spt5-Fcp1 coimmunoprecipitation and conclude that Fcp1 does not associate with Spt5 under these conditions, whereas the largest subunit of TFIIF specifically coimmunopurifies with Spt5.
Spt proteins.
The identification of Spt6, Cdc68, and Nhp6A/B in the Flag-Spt5 purification was of particular interest, as these proteins, like Spt4-Spt5, have been proposed to facilitate elongation through nucleosomes (7, 18, 21, 41). Immunoblot analysis of Spt5-Flag immunoprecipitates confirmed the coimmunopurification of Spt6 and Cdc68 but not Nhp6, which only weakly associates with Cdc68 under the moderate salt conditions used here (7, 18) (Fig. 1C). Cdc68 also interacts strongly with Pob3 (8, 63). Consistent with this, we found that Pob3 also coimmunoprecipitated with Spt5-Flag (Fig. 1C). In addition, like Spt5, Pob3 eluted across a broad size range when Spt5-Flag immunoprecipitates were separated by gel filtration (data not shown).
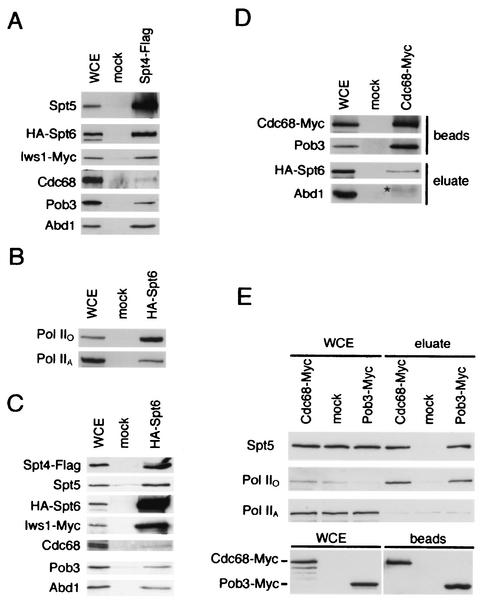
We have previously shown that Spt5 binds to Spt4 (21, 57). Although Spt4 was not found by mass spectrometry, it clearly coprecipitated with Spt5-Flag (Fig. 1C). Spt4's absence probably reflects its small size and highlights the qualitative nature of DALPC. When we immunoprecipitated a Flag-tagged derivative of Spt4 from yeast extracts, we found that it coprecipitated Spt5, Pob3-Cdc68, Spt6, Pol IIO, and Pol IIA but not Nhp6 (Fig. 2A and data not shown). We also size fractionated these Spt4-Flag immunoprecipitates by gel filtration and found that Spt4 is associated with high-molecular-weight forms of Spt5, Spt6, and Pob3 (data not shown).
FIG. 2.
Immunoprecipitation of Spt4, Spt6, Cdc68, and Pob3. (A) Anti-Flag immunoprecipitation from lysates of an Spt4-Flag strain and an untagged strain (mock). Bound proteins were eluted from the beads in 1.0 M potassium acetate, fractionated by SDS-PAGE, and Western blotted for the indicated proteins. Strains: mock, GHY1320; Spt4-Flag, GHY1324. (B) Anti-HA immunoprecipitations from lysates of an HA-Spt6 strain and an untagged strain were performed as for panel A. Strains: mock, GHY611; HA-Spt6, GHY605. WCE, 30 μg of whole-cell extract. (C) Anti-HA immunoprecipitations from lysates of a HA-Spt6 and an untagged strain were performed as for panel A. Strains: mock, GHY1325; HA-Spt6, GHY1324. WCE, 60 μg of whole-cell extract. (D) Anti-Myc immunoprecipitations from extracts of Cdc68-Myc and untagged (mock) strains were performed as for panel A. Top panel, 60 μg of whole-cell extract and 10% of the anti-Myc beads from the immunoprecipitations were separated by SDS-PAGE and blotted for the presence of Cdc68-Myc and Pob3. In contrast to the other proteins analyzed in panels D and E, Cdc68-Pob3 complexes are stable in 1.0 M potassium acetate and therefore remain bound to the beads (data not shown). Bottom panel, 60 μg of whole-cell extract and 1 M potassium acetate eluates from the anti-Myc beads were separated by SDS-PAGE and blotted for the presence of HA-Spt6 and Abd1. The asterisk indicates a weak cross-reactivity to immunoglobulin G migrating near the expected position of Abd1 in the gel. Strains: Cdc68-Myc, OY240; mock, GHY605. (E) Anti-Myc immunoprecipitations from extracts of Pob3-Myc, Cdc68-Myc, and untagged (mock) strains were performed as for panel A. Top panel, 30 μg of whole-cell lysates and 1 M potassium acetate eluates were separated by SDS-PAGE and probed for the indicated proteins. Bottom panel, 30 μg of whole-cell extract and 10% of the anti-Myc beads from the immunoprecipitations were separated by SDS-PAGE and blotted with an anti-Myc antibody for the presence of Cdc68-Myc and Pob3-Myc. Strains used: Pob3-Myc, DY6460; Cdc68-Myc, DY6529; untagged, DK186.
Consistent with the data above, when we immunoprecipitated an HA-tagged derivative of Spt6 from yeast extracts, we found Spt4, Spt5, Cdc68, Pob3, Pol IIA, and Pol IIO in the immunoprecipitates (Fig. 2B and C). The difference in the strength of the Spt5-Spt6 coprecipitation reported here and that observed previously (21, 51) can be explained by differences in methods of extract preparation (see Materials and Methods). Consistent with these observations, when we immunoprecipitated a Myc-tagged derivative of Cdc68 from yeast extracts, we found both Spt5 and Spt6 in the precipitates (Fig. 2D and E). Interestingly, and in contrast to Spt4, Spt5, and Spt6, both Pob3 and Cdc68 specifically coimmunoprecipitated with Pol IIO but not with Pol IIA (Fig. 2E). Thus, we conclude that Spt4, Spt5, Spt6, and Cdc68-Pob3 coimmunopurify with each other and Pol II. We further conclude that while Cdc68-Pob3 appears to associate only with the hyperphosphorylated form of Pol II, Spt4-Spt5 and Spt6 associate with both Pol IIA and Pol IIO. Finally, in the immunoprecipitations of Spt4, Spt5, and Spt6, we observed some variability in the fraction of Pob3 that coprecipitated compared to the fraction of Cdc68 that coprecipitated (Fig. 1C and 2A and B and data not shown). These differences likely represent experiment-to-experiment variability, as they were not consistently observed.
Spt4-Spt5 and RNA processing.
Both components of the yeast mRNA capping enzyme, Ceg1 and Cet1 (48), copurified with Spt5 (Table 2). We also identified Abd1, the cap methyltransferase, which binds Pol II independently of the capping enzyme (35). To confirm these interactions, we performed anti-HA immunoprecipitations of strains carrying epitope-tagged alleles of Ceg1 or Abd1 (Fig. 3). Spt5 specifically coimmunoprecipitated with both HA-Ceg1 and HA-Abd1. In contrast, Pob3 did not coprecipitate with either HA-Ceg1 or HA-Abd1 (Fig. 3). In reciprocal experiments, we observed that Abd1 coimmunoprecipitated with Spt4-Flag, Spt5-Flag, and HA-Spt6 (Fig. 1C and 2A and C). We also probed Cdc68-Myc immunoprecipitates for Abd1 and did not observe significant coimmunoprecipitation (Fig. 2D [compare to Fig. 1C and 2A and C; note that in comparison to the other immunoprecipitations, fivefold more extract relative to the eluate was loaded on the gel in Fig. 1C]). However, because we observe a weak cross-reactivity to immunoglobulin G migrating near the expected position of Abd1 in the gel, we cannot rule out a very weak interaction between Abd1 and Cdc68. We conclude that Spt4, Spt5, and Spt6 associate with the yeast capping enzyme and cap methyltransferase, whereas Cdc68-Pob3 does not appreciably associate with these proteins.
These observations are consistent with the finding of Spt5-capping enzyme interactions in humans and S. pombe (43, 59). Although Spt5 has mild effects on the in vitro activity of the cap guanyltransferase in humans, it does not affect the in vitro activity of the S. pombe enzyme (43, 59). Neither set of observations indicates whether Spt5 and the capping enzyme functionally interact in vivo. We used genetic analysis to begin to address this issue. _spt5_-194 and _spt4_-3 strains were crossed to a strain carrying the Ts− _ceg1_-250 allele (10). _spt5_-_194 ceg1_-250 and _spt4_-_3 ceg1_-250 double mutants both showed a decrease in their restrictive temperature, indicating an interaction between these genes (Fig. 4A). We used a plasmid shuffle assay to test interactions between SPT4, SPT5, and CET1 and found that spt cet1 double mutants displayed allele-specific synthetic lethality and poor growth phenotypes (Fig. 4B). Thus, Spt5 interacts genetically and coimmunopurifies with the capping enzyme in yeast, suggesting a functional interaction between these proteins in vivo.
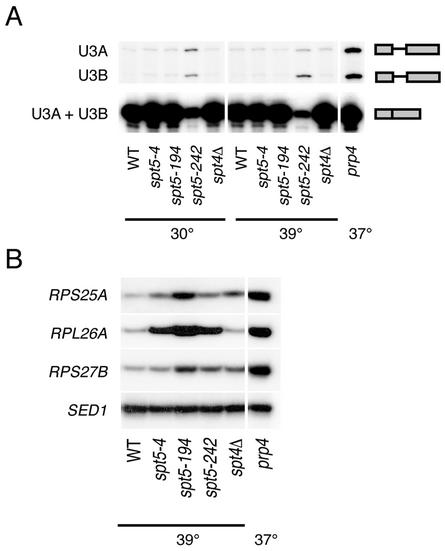
Given the interactions between Spt5 and the capping enzyme in yeast and humans and previous studies showing that the 5′ cap influences the efficiency of subsequent steps of pre-mRNA processing (16, 19, 47), we asked whether spt mutations affect pre-mRNA splicing. RNA was extracted from a series of spt4 and spt5 mutants and from a strain carrying a mutation in the essential splicing factor PRP4 (5). Because the spt4Δ, _spt5_-4, and _spt5_-194 mutants grow poorly or are inviable at elevated temperatures, RNA was prepared from cells that were grown at 30°C and then shifted to 39°C for 45 min prior to harvest. Although the cold-sensitive _spt5_-242 mutant does not have an obvious growth defect on rich media at elevated temperatures, it is Spt− at both 30 and 37°C (G. A. Hartzog, unpublished data). Thus, the _spt5_-242 mutation causes mutant phenotypes at all temperatures tested. We first performed primer extension analysis of the closely related U3A and U3B snRNAs and observed unspliced U3 RNA in the _spt5_-242 and prp4 strains (Fig. 5A). Next, we used an RT-PCR assay to monitor levels of unspliced RPS25A, RPL26A, and RPS27B pre-mRNAs in the spt4 and spt5 mutants (Fig. 5B). We observed strong accumulation of unspliced RPL26A and moderate accumulation of unspliced RPS25A for all three spt5 mutants, as well as moderate accumulation of unspliced RPS27B in the _spt5_-194 and _spt5_-242 mutants (Fig. 5B). We also observed small but reproducible accumulation of unspliced RPS25A and RPS27B RNAs in the spt4Δ mutant (Fig. 5B). Thus, spt4 and spt5 mutations lead to accumulation of unspliced forms of several yeast genes.
FIG. 5.
Splicing defects in spt4 and spt5 mutants. RNA was prepared from strains with the indicated genotypes either grown at 30°C or grown at 30°C and then shifted to 39°C for 45 min prior to harvest. The prp4 strain was grown at 30°C and then shifted to 37°C for 1 h prior to harvest. (A) Primer extension analysis of the U3A and U3B snRNAs. Note that mature U3A and U3B snRNAs are indistinguishable in this assay. In contrast, the unspliced U3A and U3B pre-snRNAs give products that differ by 27 nucleotides (4). (B) RT-PCR analysis for unspliced forms of the RPS25A, RPL26A, and RPS27B genes. Unspliced pre-mRNAs were specifically amplified by using one primer chosen from intron sequences and another chosen from the second exon. As a loading control, SED1, which does not contain an intron, was also analyzed by RT-PCR. Control reactions confirmed that the PCRs were performed in the linear range and that the PCR products were dependent upon reverse transcriptase (data not shown). Strains: wild type (WT), FY120; _spt5_-4, OY43; _spt5_-194, GHY379; _spt5_-242, GHY92; spt4Δ, GHY524; prp4, SRY4-1a.
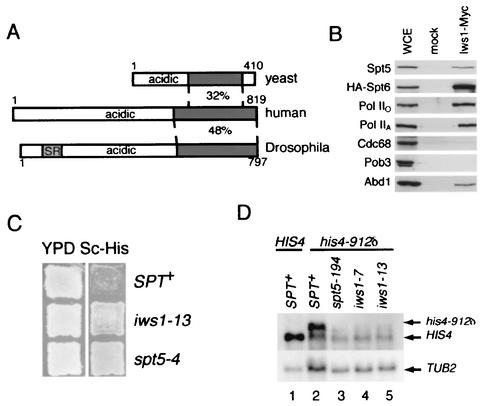
Characterization of Iws1.
We initially chose proteins from the mass spectrometry data for further analysis based on their connection to Pol II transcription and processing. To examine whether previously uncharacterized proteins in this data set might also be involved in transcription or processing, we chose to study YPR133C, an essential gene of unknown function (62). While this work was in progress, YPR133C was identified as an Spt6-interacting protein and named Iws1 (30). BLAST searches revealed metazoan homologs of Iws1, none of which has previously been characterized (Fig. 6A). These proteins all have acidic N termini and a conserved C-terminal domain. The predicted Drosophila Iws1 protein has a region enriched in serine-arginine dipeptides. This motif is found in a number of proteins that associate with Pol II and that have roles in pre-mRNA splicing (34). We tagged Iws1 with the Myc epitope, performed anti-Myc immunoprecipitations, and observed that Spt5, Spt6, Abd1, Pol IIO, and Pol IIA specifically coimmunoprecipitated with Iws1-Myc (Fig. 6B). We also found Iws1 in Spt4 and Spt6 immunoprecipitates (Fig. 2A and C). In contrast, neither Cdc68 nor Pob3 coprecipitated with Iws1-Myc (Fig. 6B).
FIG. 6.
IWS1/YPR133C encodes a conserved Spt5-associated protein required for normal transcription. (A) Alignment of Iws1 and homologs. The regions of highest identity between the proteins are marked by shading, and the percent amino acid identity is noted. The N-terminal SR repeats in the Drosophila homolog are also noted by shading. The accession numbers for the human and Drosophila homologs are BAA91402 and AAF48587, respectively. Alignments and percent amino acid identities were determined by BLAST searches (1). (B) Anti-Myc immunoprecipitations were performed from extracts of an Iws1-Myc strain and an untagged (mock) strain. Bound proteins were eluted with 1.0 M potassium acetate, separated by SDS-PAGE, and blotted for Spt5. Strains: Iws1-Myc, GHY1300; mock, GHY1280. WCE, 30 μg of whole-cell extract of GHY1300. (C) iws1 mutants display an Spt− phenotype. Cells were replica plated to the indicated media and grown for 2 days at 30°C. Strains: Spt+, GHY611; _iws1_-13, GHY1202; _spt5_-4, GHY1073. (D) The Spt− phenotypes of _iws1_-7 and _iws1_-13 cells are due to altered transcription. Top panel, Northern blot analysis of HIS4 and _his4_-912δ RNA derived from strains with the indicated genotypes. RNA from a _spt5_-_194 his4_-912δ strain was included for comparison to the _iws1 his4_-912δ strains. Bottom panel, as a loading control, the blot was stripped and then rehybridized with a TUB2 probe. Strains: lane 1, FY602; lane 2, FY119; lane 3, GHY13; lane 4, GHY1199; lane 5, GHY1200.
To genetically characterize IWS1, we isolated temperature-sensitive alleles of Myc-tagged IWS1. Interestingly, we found that these mutants displayed a Spt− phenotype (Fig. 6C). spt mutations were originally identified by virtue of their ability to suppress transcription defects caused by insertion of the long terminal repeat of the Ty retrotransposon into certain promoters (60). For example, in Spt+ cells carrying the _his4_-912δ mutation, the HIS4 transcript is longer than normal and translationally nonfunctional, rendering the cells His− (60). In a spt mutant, this defect is suppressed and the strain reverts to a His+ phenotype (60) (Fig. 6C). Northern blot analysis showed that the Spt− phenotype of the iws1 mutants was due to altered transcription of the _his4_-912δ gene (Fig. 6D). Finally, when we crossed an _iws1_-13 strain with spt4Δ or _spt5_-194 mutants, we found that neither the _iws1_-13 spt4Δ nor the _iws1_-_13 spt5_-194 double mutant was viable (data not shown). Thus, Iws1 is a conserved protein that coimmunopurifies with Spt5 and Pol II, displays an Spt− phenotype when altered by mutation, and causes synthetic lethality when combined with spt4 or spt5 mutations, results indicative of roles in Spt4-Spt5 function and Pol II transcription.
DISCUSSION
To determine the roles that the Spt4-Spt5 complex plays in gene expression, we sought to identify proteins that associate, directly or indirectly, as members of a protein complex with Spt5. Combining affinity purification and mass spectrometry, we identified a large number of proteins that specifically coimmunopurified with Spt5-Flag. We further confirmed a number of these observations by coimmunoprecipitation and genetic analysis. Consistent with Spt5's role in transcription elongation, we found evidence of copurification of Pol II and many transcription elongation factors. In contrast, we recovered only two peptides for initiation factors that function exclusively at the promoter, one for TFIIB and one for Taf60 (Table 2). No peptides for SRB/mediator subunits or other general initiation factors were recovered. Thus, these data strongly support the model that Spt5 functions as an elongation factor in vivo.
The Spt4-Spt5 complex influences RNA processing in vivo.
The copurification of capping factors with Spt5 is consistent with previous observations with human and S. pombe cells. However, the effect of Spt5 on capping enzyme activity in vitro is weak (43, 59), and the relevance of the Spt5-capping enzyme interaction is therefore uncertain. We confirmed the association of Spt5 with the capping enzyme and also found that Spt5 coimmunopurifies with Abd1, the cap methyltransferase, which does not directly associate with the capping enzyme (35) (Fig. 3). We also found that SPT4 and SPT5 interact genetically with both CET1 and CEG1 (Fig. 5). These observations strongly suggest an in vivo role for Spt4-Spt5 in pre-mRNA capping.
We also observed accumulation of several intron-containing RNAs in spt mutants (Fig. 4). In preliminary studies using splicing-sensitive DNA microarrays (12), we have observed similar effects of spt mutations for at least half of all intron-containing genes in yeast (T. A. Burckin and G. A. Hartzog, unpublished results). This may indicate a role for Spt4-Spt5 in splicing or possibly in the nuclear degradation of unspliced pre-mRNAs. We did not detect any splicing factors in our mass spectrometry data, and thus we have no evidence for a direct interaction of Spt4-Spt5 with the splicing machinery. Even if Spt4-Spt5 does not play a direct role in splicing, it is possible that spt4 and spt5 mutations indirectly lead to splicing defects as a consequence of defects in elongation or pre-mRNA capping (17, 19, 47). Regardless of the mechanism, our observations suggest an important role for Spt4-Spt5 in pre-mRNA processing.
Is Spt5 a component of more than one complex?
When Spt5-Flag immunoprecipitates were subjected to gel filtration chromatography, Spt5 was broadly distributed across the eluates, suggesting that it assembles into several large complexes (Fig. 1). Preliminary results indicate that treatment of Spt5-Flag complexes with RNase does not affect association of Pol II, Spt6, Cdc68-Pob3, or Iws1, indicating that their interactions with Spt5, although not necessarily direct, are likely mediated by protein interactions (D. L. Lindstrom and G. A. Hartzog, unpublished results). An interesting question is whether these Spt proteins form a complex before association with Pol II or are recruited individually to elongating polymerase. For example, the association of Spt4, Spt5, Spt6, and Iws1 with both Pol IIA and Pol IIO suggests that they may associate with Pol II prior to and during processive elongation (31) (Fig. 1A and 2B). This model is consistent with chromatin immunoprecipitation studies with yeast and Drosophila (2, 27, 44). In contrast, the capping enzyme and cap methyltransferase are recruited specifically to the phosphorylated CTD of Pol IIO (11, 36, 68), suggesting that they may be recruited to Pol II separately from Spt4-Spt5, Spt6, and Iws1. Similarly, we have found that Cdc68 and Pob3 associate with Pol IIO but not Pol IIA (Fig. 2E). Furthermore, the failure of Cdc68 or Pob3 to coimmunoprecipitate with HA-Ceg1 or HA-Abd1 (Fig. 3) and the at best weak coprecipitation of Abd1 with Cdc68-Myc (Fig. 2D and data not shown) suggest that recruitment of the capping machinery and Cdc68-Pob3 are also distinct events.
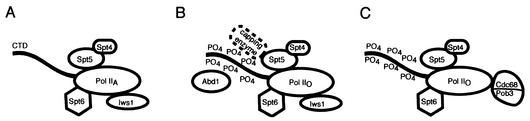
In summary, we have provided evidence that Spt4-Spt5 participates in three or more Pol II complexes. The first is a Pol IIA complex with Spt4, Spt5, Spt6, and Iws1 (Fig. 7A). The second is a Pol IIO complex with Spt4, Spt5, Spt6, Iws1, and Abd1 (Fig. 7B). This complex likely includes the yeast capping enzyme (i.e., Ceg1-Cet1), as we have found that Spt5 coprecipitates with HA-Ceg1 (Fig. 3). However, the potential association of the capping enzyme with Spt4, Spt6, and Iws1 remains to be tested. The third complex (Fig. 7C) includes Pol IIO, Spt4, Spt5, Spt6, and Cdc68-Pob3 but lacks Iws1 and the capping apparatus. We cannot rule out the possibility of other, lower-abundance or unstable complexes. Although it is intriguing to speculate that these complexes may share temporal relationships during elongation, this idea remains to be tested.
FIG. 7.
Model of Spt5 complexes. This model is based on coimmunoprecipitation and purification data presented in Fig. 1, 2, 3, and 6 and on previous studies demonstrating that Abd1 and the capping enzyme interact specifically with Pol IIO (11, 36, 68). The presence of Spt4, Spt6, and Iws1 in the same complex as the capping enzyme remains to be tested directly. For this reason, the capping enzyme complex in panel B is indicated with a dashed line.
None of our data indicate whether these Spt proteins directly interact or are indirectly associated with each other, perhaps with Pol II as an intermediate. However, based upon their shared genetic behaviors, Spt4, Spt5, Spt6, Cdc68, Pob3, and Nhp6 have been proposed to perform similar or overlapping functions in transcription elongation in the context of chromatin (reviewed in references 22, 60, and 66). Combinations of spt4, spt5, and spt6 mutations display unlinked noncomplementation and synthetic lethality, behaviors often observed for genes encoding interacting proteins (51). CDC68, POB3, and NHP6 also display genetic interactions with SPT4 and SPT5, and recent biochemical studies suggest that these proteins may have overlapping functions (7, 18, 41, 56). To date, evidence for Spt5-Spt6 binding has been weak and there has been no evidence that Spt5 physically interacts, either directly or indirectly, with Cdc68-Pob3. Our observations that Spt5, Spt6, Cdc68, and Pob3 coimmunopurify with each other and with Pol IIO are consistent with their genetic interactions and suggest that these proteins cooperate with each other to regulate transcription elongation in the context of chromatin.
Other proteins that associate with Spt5.
In this work we aimed to comprehensively identify proteins that copurify with Spt5. In a recent large-scale proteomic study, a complex containing Spt4, Spt5, TFIIF, Abd1, and Pol II was identified, although it was not independently verified by other methods (complex 145 in reference 20). Many of the proteins that we have shown to associate with Spt5 here, including Spt6, Cdc68, Pob3, Iws1, and the capping enzyme, were not found in that study. Conversely, our mass spectrometry data did not include three proteins, Aos1, YDL115C, and YHL021C, that were identified in complex 145. Similarly, Krogan et al. (30) used a TAP-Tag approach to identify a network of protein interactions that overlap with those we have reported here and elsewhere (49). While Krogan et al. did not identify the capping apparatus in their work, they did note interactions between Cdc68-Pob3 and several proteins not observed here, using lower-salt conditions than we have used. Previous purifications of Spt4-Spt5 from human cells have been based upon functional assays of Pol II transcription. DSIF, the human Spt4-Spt5 complex, was purified based on its ability to inhibit transcription in the presence of the protein kinase inhibitor DRB (57). Subsequent work identified another multiprotein complex, NELF, which is required for DSIF's repressive activity. The identities of two NELF subunits, RD and WHSC2, have been reported, but neither has an obvious yeast homolog (65, 67). Protein complexes that stimulate Tat activity in vitro have been partially purified from HeLa cells and reported to contain Spt4-Spt5, P-TEFb, Tat-SF1, nucleolin, XP-E, Pol II, the small subunit of TFIIF (equivalent to Tfg2) and other novel polypeptides (28, 42, 64). Other than Pol II and TFIIF, we have not identified homologs of these proteins here. A number of proteins known to associate with Spt5 are not included in our data, reflecting technical pitfalls of complex purifications. For example, we recently demonstrated genetic and biochemical interactions between Spt4-Spt5 and the Paf1 complex (49). While we identified Paf1 complex members in the Spt5-Flag fractions, each was also present in the mock purification, possibly due to a Flag-mimetic epitope in Rtf1 (D. L. Lindstrom and G. A. Hartzog, unpublished results).
Many of the Spt5-associated proteins that we identified have not been previously characterized or are known to function in processes other than Pol II transcription and pre-mRNA processing. Although these proteins may have nonspecifically copurified with Spt5-Flag, our characterization of Iws1 (Fig. 6) suggests that some of these factors are involved in Pol II transcription and/or are likely to associate and functionally interact with Spt5. Thus, analysis of these other putative Spt5-associated proteins may reveal further clues to Spt4-Spt5 function.
Acknowledgments
This work was supported by a grant from the NIH to G.A.H. (GM60479) and by NIH Yeast Resource Center grant RR11823-05 to J.R.Y.
We thank Steve Buratowski, Tim Formosa, David Stillman, Caroline Kane, Steve Hahn, Clyde Denis, Fred Winston, and Doug Kellogg for gifts of strains, plasmids, and antibodies. We thank Manny Ares and Caroline Kane for sharing information prior to publication. We thank Manny Ares, John Tamkun, Fred Winston, Craig Kaplan, and members of the Hartzog lab for helpful discussions and their comments on the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215**:**403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14**:**2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault, J., R. S. Chambers, M. S. Kobor, Y. Ho, M. Cartier, D. Bolotin, B. Andrews, C. M. Kane, and J. Greenblatt. 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94**:**14300-14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ares, M., Jr., and A. H. Igel. 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 4**:**2132-2145. [DOI] [PubMed] [Google Scholar]
- 5.Banroques, J., and J. N. Abelson. 1989. PRP4: a protein of the yeast U4/U6 small nuclear ribonucleoprotein particle. Mol. Cell. Biol. 9**:**3710-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bortvin, A. L., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272**:**1473-1476. [DOI] [PubMed] [Google Scholar]
- 7.Brewster, N. K., G. C. Johnston, and R. A. Singer. 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21**:**3491-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewster, N. K., G. C. Johnston, and R. A. Singer. 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273**:**21972-21979. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. H., and D. S. Luse. 1997. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem. 272**:**23427-23434. [DOI] [PubMed] [Google Scholar]
- 10.Cho, E. J., C. R. Rodriguez, T. Takagi, and S. Buratowski. 1998. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 12**:**3482-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11**:**3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, T. A., C. W. Sugnet, and M. Ares, Jr. 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296**:**907-910. [DOI] [PubMed] [Google Scholar]
- 13.Davis, C. A., L. Grate, M. Spingola, and M. Ares, Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28**:**1700-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denis, C. L., M. P. Draper, H. Y. Liu, T. Malvar, R. C. Vallari, and W. J. Cook. 1994. The yeast CCR4 protein is neither regulated by nor associated with the SPT6 and SPT10 proteins and forms a functionally distinct complex from that of the SNF/SWI transcription factors. Genetics 138**:**1005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5**:**3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaherty, S. M., P. Fortes, E. Izaurralde, I. W. Mattaj, and G. M. Gilmartin. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 94**:**11893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong, Y. W., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414**:**929-933. [DOI] [PubMed] [Google Scholar]
- 18.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20**:**3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fresco, L. D., and S. Buratowski. 1996. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA 2**:**584-596. [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415**:**141-147. [DOI] [PubMed] [Google Scholar]
- 21.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12**:**357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartzog, G. H., J. L. Speer, and D. L. Lindstrom. 2002. Transcript elongation on a nucleoprotein template. Biochim. Biophys. Acta 1577**:**276-286. [DOI] [PubMed] [Google Scholar]
- 23.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415**:**180-183. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20**:**2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izban, M. G., and D. S. Luse. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267**:**13647-13655. [PubMed] [Google Scholar]
- 26.Izban, M. G., and D. S. Luse. 1991. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 5**:**683-696. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14**:**2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. B., Y. Yamaguchi, T. Wada, H. Handa, and P. A. Sharp. 1999. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19**:**5960-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14**:**2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22**:**6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159**:**487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17**:**676-682. [DOI] [PubMed] [Google Scholar]
- 33.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14**:**953-961. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416**:**499-506. [DOI] [PubMed] [Google Scholar]
- 35.Mao, X., B. Schwer, and S. Shuman. 1995. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 15**:**4167-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11**:**3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortensen, E. M., H. McDonald, J. Yates III, and D. R. Kellogg. 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 13**:**2091-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22**:**1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray, S., R. Udupa, S. Yao, G. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21**:**4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92**:**105-116. [DOI] [PubMed] [Google Scholar]
- 41.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400**:**284-288. [DOI] [PubMed] [Google Scholar]
- 42.Parada, C. A., and R. G. Roeder. 1999. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18**:**3688-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei, Y., and S. Shuman. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277**:**19639-19648. [DOI] [PubMed] [Google Scholar]
- 44.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9**:**799-809. [DOI] [PubMed] [Google Scholar]
- 45.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108**:**501-512. [DOI] [PubMed] [Google Scholar]
- 46.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schwer, B., and S. Shuman. 1996. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA 2**:**574-583. [PMC free article] [PubMed] [Google Scholar]
- 48.Shuman, S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66**:**1-40. [DOI] [PubMed] [Google Scholar]
- 49.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21**:**1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson, M. S., E. A. Malone, and F. Winston. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11**:**3009-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson, M. S., and F. Winston. 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132**:**325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takase, Y., T. Takagi, P. B. Komarnitsky, and S. Buratowski. 2000. The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol. Cell. Biol. 20**:**9307-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, N., T. Steinberg, D. Aronson, and R. Burgess. 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 264**:**11511-11520. [PubMed] [Google Scholar]
- 54.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the ISWI subfamily of ATP-dependent chromatin remodeling factors in S. cerevisiae. Genes Dev. 13**:**686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11**:**3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell 5**:**1067-1072. [DOI] [PubMed] [Google Scholar]
- 57.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12**:**343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17**:**7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen, Y., and A. J. Shatkin. 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 13**:**1774-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winston, F. 1992. Analysis of SPT genes: a genetic approach towards analysis of TFIID, histones and other transcription factors of yeast, p. 1271-1293. In S. L. McKnight and K. R. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient S. cerevisiae strains that are isogenic to S288C. Yeast 11**:**53-55. [DOI] [PubMed] [Google Scholar]
- 62.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285**:**901-906. [DOI] [PubMed] [Google Scholar]
- 63.Wittmeyer, J., L. Joss, and T. Formosa. 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38**:**8961-8971. [DOI] [PubMed] [Google Scholar]
- 64.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 277**:**179-197. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293**:**124-127. [DOI] [PubMed] [Google Scholar]
- 66.Yamaguchi, Y., T. Narita, N. Inukai, T. Wada, and H. Handa. 2001. SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem. (Tokyo) 129**:**185-191. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97**:**41-51. [DOI] [PubMed] [Google Scholar]
- 68.Yue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94**:**12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]