The Yeast Split-Ubiquitin Membrane Protein Two-Hybrid Screen Identifies BAP31 as a Regulator of the Turnover of Endoplasmic Reticulum-Associated Protein Tyrosine Phosphatase-Like B (original) (raw)
Abstract
In the past decade, traditional yeast two-hybrid techniques have identified a plethora of interactions among soluble proteins operating within diverse cellular pathways. The discovery of associations between membrane proteins by genetic approaches, on the other hand, is less well established due to technical limitations. Recently, a split-ubiquitin system was developed to overcome this barrier, but so far, this system has been limited to the analysis of known membrane protein interactions. Here, we constructed unique split-ubiquitin-linked cDNA libraries and provide details for implementing this system to screen for binding partners of a bait protein, in this case BAP31. BAP31 is a resident integral protein of the endoplasmic reticulum, where it operates as a chaperone or cargo receptor and regulator of apoptosis. Here we describe a novel human member of the protein tyrosine phosphatase-like B (PTPLB) family, an integral protein of the endoplasmic reticulum membrane with four membrane-spanning alpha helices, as a BAP31-interacting protein. PTPLB turns over rapidly through degradation by the proteasome system. Comparisons of mouse cells with a deletion of Bap31 or reconstituted with human BAP31 indicate that BAP31 is required to maintain PTPLB, consistent with a chaperone or quality control function for BAP31 in the endoplasmic reticulum membrane.
The identification of interactions between different proteins is essential for working out protein functions and signaling pathways in vivo. Prominent among the various techniques available for such discoveries are screens utilizing the yeast two-hybrid system (15, 50). This system, however, requires translocation of the binding protein complex to the nucleus for activation of a reporter gene. Thus, this system can only be used to identify interactions between freely diffusing protein domains. The variant three-hybrid system identifies ternary protein complex components (58) or inhibitors of protein interactions (48), including interactions between ligands and receptors (31), proteins and RNA (39), and proteins and DNA (25).
To address the problem of interacting proteins that are tethered at cellular membranes, Fabio Rossi et al. developed a system in eukaryotic cells based on β-galactosidase complementation (37). However, this system is difficult to implement for library screening, as the β-galactosidase gene is the only means of selection. To overcome this problem, Ami Aronheim et al. developed a novel Sos recruitment system, in which the human Ras guanyl nucleotide exchange factor Sos can substitute for the Saccharomyces cerevisiae Ras guanyl nucleotide exchange factor Cdc25 to allow cell survival and proliferation (3, 4). Several variants of this system were also developed recently (5, 20, 21). Briefly, the bait is a plasma membrane-localized protein, whereas library proteins are fused to human Sos, cdc25, or activated Ras. A survival signal occurs if the candidate fusion proteins associate with the bait and recruit Sos, cdc25, or activated Ras to the plasma membrane. However, the survival signals will be activated without bait if the fusion protein is a plasma membrane protein. Clearly, this system cannot be used to identify interactions between two integral membrane proteins.
The split-ubiquitin system developed in Saccharomyces cerevisiae by Johnsson and Varshavsky (24, 29) overcomes this problem. It is based on the reassembly of the N- and C-terminal halves (Nub and Cub, respectively) of ubiquitin if each is fused to proteins that interact. The reassembled quasi-native ubiquitin is recognized by ubiquitin-specific proteases, which then cleave the C-terminally attached polypeptide (i.e., a reporter) from Cub and thereby provide an immediate readout of the Nub-Cub reassociation. NubG (Ile-13 to Gly), a mutant of Nub, has very low intrinsic affinity for Cub and therefore can interact with Cub only if both ubiquitin fragments are linked to proteins that interact. By including Ura3p as a reporter that is released from Cub by ubiquitin-specific protease, the molecular environment and transient interaction of membrane proteins have been studied (12, 56). Stagljar et al. (40) further optimized this technique by attaching PLV (protein A-LexA-VP16) to the C terminus of Cub as the reporter molecule. PLV is liberated upon cleavage from Cub and activates the lacZ and HIS3 reporter genes, thereby providing a potentially useful tool for the screening of interactions between membrane proteins. To date, however, this system has been limited to the analysis of interactions between known proteins (32, 41). Here we provide a detailed description of cDNA libraries expressing proteins fused to NubG and their use to screen for proteins that interact with a resident integral protein of the endoplasmic reticulum (ER), BAP31.
BAP31, an evolutionarily conserved and ubiquitously expressed 28-kDa polytopic integral protein, was originally described as contributing to B-cell receptor activation (1, 26). Its location in the ER (2, 34), however, argued that its contribution to this activation might arise indirectly through an involvement in the egress of membrane-integrated protein complexes from this organelle. Consistent with this, BAP31 has been implicated in export of major histocompatibility complex class I molecules from the ER (40) and in the control of maturation or trafficking of cystic fibrosis transmembrane conductance regulator (28). BAP31 is part of a large hetero-oligomeric complex at the ER membrane that comprises specific components of the actomyosin complex (10) and includes the closely related BAP29 (1, 26) as well as a predicted ion channel protein of the ER called A4 (54). BAP31 and BAP29 are comprised of three membrane-spanning segments and a 13-kDa cytosolic tail containing an extended coiled-coil region that mediates both homo- and heterotypic protein-protein interactions. In addition to its predicted role in regulating egress of membrane complexes from the ER, BAP31 is a BCL-2/BCL-XL-associated protein and plays an important role in apoptosis, both as a regulator of procaspase-8L processing (7) and as a cleavage target of caspase-8 (34, 35).
Protein tyrosine phosphatase-like (PTPL) is a newly identified, universally expressed, and very conserved family, including PTPLA (30, 45) and Pepino/Pasticcino2 (6, 17). These proteins contain a sequence highly similar to a conserved protein tyrosine phosphatase (PTP) catalytic domain except for the change of a conserved arginine (R) to a proline (P), rendering the protein phosphatase inactive and arguing for a role as a potential regulator rather than an executor of phosphatase activity. Mutations of the Pepino/Pasticcino2 gene in Arabidopsis thaliana cause abnormal shoot phenotypes corresponding to slow tumor-like cell proliferations, suggesting that Pepino and Pasticcino2 are general negative regulators of plant cell division and proliferation. The other two pasticcino members, PAS1 and PAS3, have functions similar to those of Pepino/Pasticcino2 (13, 49). To date, the function and regulation of PTPL proteins are unclear, as there have been no reports describing the protein expression of any PTPL family members. Here, we identified a new member of this family, PTPLB, as a candidate BAP31-interacting protein in the yeast split-ubiquitin screen. Newly synthesized PTPLB, an integral membrane protein with four membrane-spanning alpha helices, localizes at the ER, where it rapidly turns over via the proteasome system. This turnover is further enhanced in the absence of BAP31, indicating a quality control function for BAP31 consistent with its proposed role in the proper assembly of integral proteins at the ER.
MATERIALS AND METHODS
Antibodies and reagents.
The following antibodies were used: calnexin monoclonal antibody (BD Biosciences, Mississauga, Canada); chicken or rabbit anti-human BAP31 (35, 36); anti-Flag M2 monoclonal antibody (Sigma); antihemagglutinin (HA) 12CA5 monoclonal antibody (Babco, Richmond, Calif.); anti-c-Myc monoclonal antibody 9E10 (Babco); rabbit anti-human TOM20 (8); rabbit anti-γ-actin (gift from P. Braun); rabbit immunoglobulin G-horseradish peroxidase conjugate (Cappel, ICN Pharmaceuticals). 3-Aminotriazole was purchased from Sigma; MG132 and lactacystin were obtained from Calbiochem.
Construction of bait plasmids.
For pRSB (Cup1-BAP31-Cub-protein A-LexA-VP16), full-length human BAP31 with an XhoI-NsiI-BsiwI linker at the 5′ end and an XhoI linker at the 3′ end was generated by PCR with primers 5′-TCCGCTCGAGATGCATCTCTCTCTCTCGTACGATGAGTCTGCAGTGGACT and 5′-TCCGCTCGAGCTCTTCCTTCTTGTCCAT. The resulting PCR product was used to replace Δ_wbp1_ in pRS305 (Δwbp1-Cub-PLV) (32, 41) at the XhoI sites. The yeast Cup1 promoter was created by PCR with primers 5′-CCAATGCATTGGATCCCATTACCGACATTTG and 5′-CCACGTACGGCTTGATATCGAATTCGT and inserted into the NsiI and BsiwI sites upstream of BAP31. The resulting plasmid, termed pRSB, was linearized at the unique NsiI site and integrated into the Cup1 gene of S. cerevisiae L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ), and clones (termed pRSB-L40) were selected on leucine plates. The expression of the BAP31-Cub-PLV fusion protein was verified by Western blot analysis with antibodies against both BAP31 and protein A.
For construction of pRSLN (Cup1-cDNA-NubG) and pRSLC (Cup1-NubG-cDNA), Cup1-NubG with linkers EcoRI and SalI at the 5′ end and BglII and XhoI at the 3′ end of NubG was generated by PCR from pRS314 (Cup1-NubG-ALG5) (32, 41) with primers 5′-TCGGCGGTGGCGGCCGCTCTAGAAC and 5′-AACTGCAGCTCGAGGAGAGAGAGATCTCCACCAGGGATCCCTTC. The resulting PCR products were ligated into pRS314 (NubG-ALG5 deleted). Library cDNA fragments were ligated to the BglII and XhoI or EcoRI and SalI sites, leading to the formation of pRSLC and pRSLN, respectively.
Construction of PTPLB and its mutants.
The full-length human PTPLB cDNA obtained from the screen in this study was ligated into the pcDNA3.1 vector with sequences specifying the hemagglutinin (HA) (YPYDVPDYA) or Myc (EQKLISEEDL) epitope tags introduced at the C terminus immediately upstream of the ER retention signal KKFE by PCR. The long Myc tag (MASMEQKLISEEDLNNG) was added at the N terminus of PTPLB-HA. The 11 internal lysines of PTPLB were mutated to arginines step by step by standard overlay mutant primer PCR. A PTPLB-green fluorescent protein (GFP) fluorescence fusion protein expression plasmid was constructed by inserting DNA encoding PTPLB into the pEGFP-N2 vector with the EcoRI and BamHI sites. The authenticity of all constructions was verified by sequence analysis.
Split-ubiquitin two-hybrid cDNA library synthesis.
Total mRNAs from HeLa cell lines were used as templates to synthesize the first-strand cDNAs with Superscript II reverse transcriptase and random primers with XhoI or SalI linkers. The first-strand cDNAs were methylated because CH3-dCTP was present during its synthesis and radiolabeled with tritium as a tracer. The quantity of the first strand was measured by the total amount of radioactivity and the amount of trichloroacetic acid-precipitable radioactivity. For the second-strand synthesis, RNase H was used to nick the template mRNAs that hybridized with the first-strand cDNAs, and the digested small fragments of the template mRNAs were used as random primers to synthesize the second-strand cDNAs with Escherichia coli DNA polymerase I and DNA ligase. [α-33P]dATP served as a radioactivity tracer to monitor the quantity of the second-strand synthesis via trichloroacetic acid precipitation.
Alkaline agarose gel electrophoresis was used to monitor the quality of the double-stranded 33P-labeled cDNAs. The double-stranded cDNAs were modified by blunt-end extension with T4 DNA polymerase, BglII, or EcoRI adapter ligation and 3′-end SalI or XhoI linker digestion. Finally, the cDNAs were fractionated by 10 to 30% sucrose density gradient centrifugation, 30 drops of each fractions were collected from the bottom of the centrifuge tubes, and 30 μl of each fraction was precipitated by trichloroacetic acid and assessed by counting 3H and 33P in the scintillation fluid. Following assessment by alkaline agarose gel electrophoresis, four fractions providing pools of double-stranded cDNAs in the range of average sizes of 2 to 13 kb were selected to ligate into plasmid vectors. Each fraction ligation mixture was used to electroporate into competent E. coli HB101 cells. For each fraction, five million transformants were grown on 15-cm plates containing Luria-Bertani broth with ampicillin, colonies were scraped into Luria-Bertani medium and frozen in 20% glycerol.
Split-ubiquitin yeast two-hybrid screening.
Each fraction of the cDNA library in pRSLNs and pRSLCs was transformed into the bait yeast strain (pRSB-L40) by the lithium acetate transformation method (53). Clones were selected on leucine-tryptophan-histidine triple selection plates in the presence of 0.2 mM CuSO4 and 5 mM 3-aminotriazole. Fifty independent clones from each of the libraries were randomly picked. Plasmids were rescued from these yeast clones according to standard protocols and electroporated into E. coli strain DH5α. Isolated plasmids were characterized by DNA sequence analysis. Nucleotide sequences were compared to existing sequence databases by use of the BLAST program. Selected candidates were transformed into empty L40 or pRSB-L40 for further analysis.
RACE.
Poly(A)-containing mRNA from HeLa cells was isolated with the Oligotex direct mRNA kit (Qiagen). Both 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE were performed with the SMART RACE cDNA amplification kit (Clontech) (51).
Northern blot analysis.
Northern blot membranes with mRNA from various human tissue specimens (multiple tissue Northern blots; Clontech) were hybridized according to standard protocols (51, 52). The radiolabeled probes corresponded to the entire open reading frames of the human PTPLB or β-actin gene.
Immunofluorescence.
Cells were grown on glass coverslips for 24 h, washed in phosphate-buffered saline and fixed in 4% paraformaldehyde. The cells were briefly permeabilized in phosphate-buffered saline-10% fetal calf serum-0.2% Triton X-100 and then treated with blocking solution (phosphate-buffered saline containing 10% fetal calf serum and 0.1% Triton X-100). Antibody incubations were conducted in blocking solution for 1 h at room temperature with the indicated primary antibodies and either goat anti-mouse immunoglobulin G or goat anti-rabbit immunoglobulin G secondary antibodies coupled to Alexa 488 (green) or Alexa 594 (red) (Molecular Probes). Cells were visualized by confocal or conventional fluorescence microscopy.
Immunoprecipitation and immunoblots.
Yeast cells were lysed in 50 μl of 1.85 M NaOH per 3 _A_546 units and incubated on ice for 10 min. An equal volume of 50% trichloroacetic acid was added, and proteins were recovered by centrifugation for 5 min. The pellet was suspended in 50 μl of sodium dodecyl sulfate (SDS) sample buffer containing 8 M urea and combined with 20 μl of 1 M Tris base and incubated for 1.5 h at 37°C. Eukaryotic cells were lysed in buffer containing 1% 3-[(cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 150 mM NaCl, 50 mM Tris-HCl (pH 7.3), 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml. Lysates were assayed for protein content with the Bio-Rad reagent. After preclearing for 30 min with protein G-Sepharose (Pharmacia), lysates were incubated with antibodies for 90 min at 4°C. Immune complexes bound to protein G-Sepharose were washed four times with lysis buffer and eluted in SDS sample buffer. All samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Cell culture.
Cos-1 cells were maintained in minimal essential medium (Gibco) supplemented with 10% fetal calf serum and 1% penicillin-streptomycin at 37°C in humidified 5% CO2. Embryoid epithelium-like (differentiated) _Bap31_-null embryonic stem (ES) cell lines expressing either neo or neo and human BAP31-Flag (7) were grown at 37°C and 5% CO2 on knockout Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 100 U of streptomycin sulfate and penicillin per ml, 15% fetal bovine serum, 2 mM l-glutamine, 1 mM nonessential amino acids, and 1 mM β-mercaptoethanol.
Turnover of PTPLB in vivo.
PTPLB-HA and its mutants were analyzed by pulse-chase labeling followed by immunoprecipitation. At 20 h posttransfection with the indicated vectors, Cos-1 cell cultures were washed with a medium that lacks methionine and cysteine (Gibco) and labeled for 1 h in the same medium containing 200 μCi of [35S]methionine and [35S]cysteine per ml. Where present, the proteasome inhibitor MG132 (100 μM) was added after 30 min. Following the 1-h pulse-labeling, either the cells were harvested (chase time zero) or the medium was replaced with complete medium with or without MG132 and incubated for the indicated time periods (chase). Cell lysates were precleared with protein G-Sepharose, and the labeled proteins in the supernatant were incubated with anti-HA antibody. Immunocomplexes were collected with protein G-Sepharose and subjected to SDS-PAGE, and the 35S-labeled products were detected by autoradiography and recorded with a PhosphorImager (Fuji).
Phosphatase assay.
The standard reaction mixture which contained equivalent amounts of PTP1B-DA, PTP1B, PTPLB or PTPLB-PR protein, 50 mM Tris-HCl, 2% glycerol, 0.01% Triton X-100 (pH 8.0), and 10 mM _p_-nitrophenyl phosphate (_p_NPP) in a total of 1 ml performed at 37°C for 20 min and then stopping the reaction by adding NaOH to a final concentration of 1 M. An increase in the amount of _p_-nitrophenol was monitored by measuring the absorbance at 410 nm on an Ultraspec 4000 (Pharmacia) spectrophotometer.
Flow cytometry analysis.
_Bap31_-null and _Bap31_-null/BAP31-Flag cells were transfected with plasmid pcDNA3.1 (control vector), pEGFP-N2 (Invitrogen), or pPTPLB-GFP. After 36 h, cell suspensions were fixed with 1% paraformaldehyde in phosphate-buffered saline for 30 min. Flow cytometric measurements were performed with a Becton Dickinson FACSCalibur system.
RESULTS
Identification of the BAP31-associated protein PTPLB in a split-ubiquitin two-hybrid screen.
As illustrated in Fig. 1A, full-length human BAP31 was fused to the N terminus of Cub-PLV, and the bait fusion protein was termed BAP31-Cub-PLV. For screening, cDNA libraries encoding HeLa cell mRNA open reading frames fused at either the 3′ or 5′ end of NubG were also created. In situations where BAP31-protein interactions liberate PLV by ubiquitin-specific protease, PLV enters the nucleus and interacts with LexA-binding sites, leading to activation of transcription of the lacZ and HIS3 reporter genes. To minimize background arising from nonspecific release of PLV, we ligated the Cu2+-responsive Cup1 yeast promoter to the 5′ end of BAP31-Cub-PLV and integrated this structure into the Cup1 genomic locus of L40 yeast cells instead of using a high-copy-number 2μm plasmid for bait expression. The bait protein expression of BAP31-Cub-PLV was verified by Western blotting with antibodies against protein A or BAP31 (data not shown). Even so, with rabbit immunoglobulin G-horseradish peroxidase, which recognizes the protein A epitope of PLV, used to study the cleavage of BAP31-Cub-PLV, it was evident that a background cleavage of BAP31-Cub-PLV could release trace amounts of PLV, which caused histidine selection leakage and activation of the His3 reporter gene. We therefore used 3-aminotriazole, a competitive inhibitor of the imidazole glycerolphosphate dehydratase involved in histidine biosynthesis (27), to inhibit the basic leakage but not the strong activation of the histidine reporter gene. We used 0.2 mM Cu2+ to induce the bait BAP31-Cub-PLV and NubG library protein expression because both bait and library protein expression depend on the Cup1 promoter (see Materials and Methods). Background expression of BAP31-Cub-PLV is caused by Cu2+ contamination in reagents and solutions.
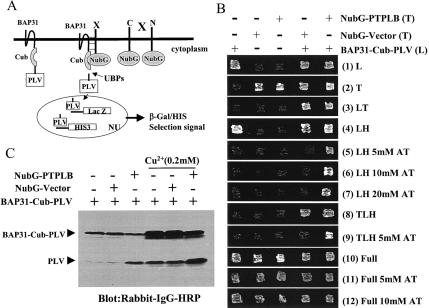
FIG. 1.
Identification of the BAP31-associated protein PTPLB in the novel split-ubiquitin yeast two-hybrid screen. (A) Diagrammatic representation of the split-ubiquitin two-hybrid system. Polypeptides that interact with the bait protein bring together the C-terminal fragment (Cub) and a modified N-terminal fragment (NubG, isoleucine 13 replaced by glycine) of ubiquitin, allowing ubiquitin-specific proteases (UBPs) to liberate PLV (protein A, LexA, and VP16) from Cub, which then is free to enter the nucleus (NU) and activate transcription of the lacZ and HIS3 reporter genes. (B) Growth of yeast L40 cells expressing BAP31-Cub-PLV, NubG, or NubG-PTPLB fusion proteins alone or together on various selective agar plates. BAP31-Cub-PLV in a leucine selection vector was integrated at the Cup1 genetic site in L40 yeast cells (pRSB-L40). NubG and NubG-PTPLB were constructed into a 2μm plasmid with tryptophan selection and transfected into L40 or pRSB-L40 yeast cells. The yeast strains containing various constructs were inoculated on agar plates with different selections. All the agar plates contained 0.2 mM Cu2+ with or without 3-aminotriazole (AT). (C) Western blot analysis of yeast L40 cell lines expressing BAP31-Cub-PLV fusion protein with or without coexpression of the NubG or NubG-PTPLB vector in the presence or absence of added Cu2+. Cells were grown to the logarithmic phase in relative selection medium. Proteins were extracted and analyzed by Western blotting with rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP) conjugate. L, lysine; T, tryptophan; H, histidine; full, no selection.
L40 yeast cells integrated with the construct expressing BAP31-Cub-PLV, termed pRSB-L40, can grow on leucine-histidine double selection plates but not when the plates contain 5 to 20 mM 3-aminotriazole (Fig. 1B, lanes 1 of panels 4 to 7). Thus, we transfected library cDNAs into pRSB-L40 yeast cell lines and made selection on leucine-tryptophan-histidine triple selection plates with 5 mM 3-aminotriazole. As a result, we identified multiple independent cDNAs encoding a novel full-length PTPLB protein. In order to verify this hit, NubG-PTPLB or NubG-mock in the 2μm tryptophan selection plasmid was transfected back into L40 or pRSB-L40. The coexpression of both BAP31-Cub-PLV and NubG-mock or BAP31-Cub-PLV and NubG-PTPLB resulted in yeast growth on leucine-tryptophan-histidine triple selection plates (panel 8), but only BAP31-Cub-PLV plus NubG-PTPLB grew on triple selection plates containing 5 mM 3-AT (panel 9). All the transfected yeast cell lines grew normally on the full (no selection) plates even with 10 mM 3-aminotriazole (panels 10 to 12). Thus, the positive clones selected on triple selection plates containing 5 mM 3-aminotriazole were due to the association between BAP31 and PTPLB. Furthermore, Western blot analysis showed that cleavage of the BAP31-Cub-PLV reporter protein was dependent on coexpression of NubG-PTPLB but not the NubG vector (Fig. 1C).
Cloning and characterization of PTPLB.
As illustrated in Fig. 2A, two fragments of the _PTPLB_-encoding open reading frame, each containing a translation stop codon, were obtained. By 3′-RACE and 5′-RACE PCR, 1.2-kb and 43-bp overlapping extension fragments, respectively, were recovered and a 4,304-bp full-length cDNA of PTPLB was generated by overlapping the PCR-generated sequences with overlaps from the GenBank expressed sequence tag (EST) database. According to a homology search of the putative amino acid sequences of PTPLB in GenBank, PTPLB had significant identities over the entire protein with mouse PTPLB (96%; unpublished data), human PTPLA (66%) (45), and Arabidopsis PAS2/PEPINO (41%) (6, 17).
FIG. 2.
Molecular cloning of human PTPLB cDNAs. (A) Representative clones covering the full-length cDNA of the PTPLB gene are shown. An asterisk indicates the translation stop codon. The open box represents the open reading frame identified in the two-hybrid screen; the 5′ and 3′ untranslated sequences were determined by 5′- and 3′-RACE and expressed sequence tag human GenBank overlaps. (B) PTPLB is composed of six exons. The locations of the six exons are indicated. (C) Proposed topology of PTPLB in the ER membrane, as predicted by hydropathy, the “positive inside” rule, and the known cytosolic disposition of C-terminal ER retention signals (see reference 34). (D) Nucleotide and deduced amino acid sequences of human PTPLB. The amino acid sequences are shown in the single-letter code. The translation stop codon is indicated by an asterisk. The PTPase homology motifs, HCXXGXXRS and an aspartic acid (D), are indicated by bold letters, and the proline that replaces the catalytic arginine is bold and underlined. The C-terminal ER retention motif, KKFE, is in bold. Four transmembrane segments are predicted by hydropathy plot analysis, performed with the TMHMM software, and the sequences are indicated by the heavy underlines.
The chromosomal location of PTPLB was obtained by Blast search of the cDNA against the human genome in GenBank. PTPLB contains six exons located in chromosome 3; 3,416-bp exon 6 corresponds to 79% of the whole PTPLB cDNA and contains both the translation stop codon and the polyadenylation signal sequence (Fig. 2B). The 4.3-kb cDNA of PTPLB encodes a 254-amino-acid protein (Fig. 2D). A hydropathy analysis of the deduced PTPLB amino acid sequence revealed four putative membrane-spanning α-helices, with a disposition in the membrane as depicted in Fig. 2C based on rules for orientation of the first transmembrane segment (see reference 34). Like BAP31, PTPLB lacks a transient signal sequence and contains a canonical, cytosol-disposed KKXX ER retention signal at its extreme C terminus. All PTPs contain the conserved motif (I/V)HCXXGXXR(S/T), which corresponds to the catalytic site. Mutation of the cysteine or arginine residues abolishes the catalytic activities of PTPs. Similar to other PTPL members, PTPLB also contains the PTP motif except for substitution of the conserved arginine (R) to a proline (P), rendering the protein phosphatase inactive. The PTP motif of PTPLB, however, retains the conserved aspartic acid residue (D), which is also necessary for PTP catalytic activity (Fig. 2D).
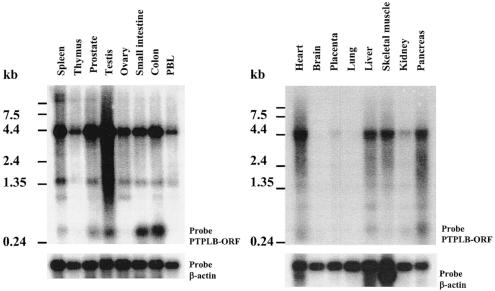
The distribution of PTPLB gene expression in human tissues was examined by Northern blot analysis. As shown in Fig. 3, the PTPLB mRNA (4.3 kb), detected by a probe containing the entire open reading frame, was strongly expressed in testis, spleen, prostate, colon, and heart but was only weakly detected (after a long exposure of blots) in kidney, placenta, brain, and lung (data not shown) and moderately expressed in thymus, ovary, small intestine, peripheral blood leukocytes, liver, skeletal muscle, and pancreas. Thus, PTPLB, a new member of PTPL family, is encoded by a widely expressed 4.3-kb mRNA with a 762-bp open reading frame encoding a 254-amino-acid polypeptide.
FIG. 3.
Northern blot analysis of PTPLB gene expression. The membranes contained 2 μg each of high-quality polyadenylated mRNA isolated from the indicated human tissues and organs. Hybridization was done with the 32P-labeled DNA fragment of the PTPLB open reading frame (upper panels) and a β-actin probe as a control (lower panels). PBL, peripheral blood leukocytes.
Rapid proteasome-mediated degradation of PTPLB.
To date, no members of the PTPL family have been studied at the protein level. Likewise, our initial attempts to detect ectopic expression of PTPLB containing epitope tags by Western blot were unsuccessful. To examine the potential contributions of rapid turnover of PTPLB to this problem, pcDNA3.1 expression vectors were constructed with either a Myc or a HA tag inserted immediately upstream of the KKFE ER retention signal. Cos-1 cells were transiently transfected with these constructs or the control vector, and after 18 h the cells were treated or not with the proteasome inhibitor MG132 (100 μM) for 4 h, stained with anti-Myc and anti-HA antibodies, and examined by fluorescence microscopy. MG132 significantly increased the detectable expression of PTPLB (representative fields shown in Fig. 4A). Similar results were obtained with another proteasome-specific inhibitor, lactacystin (not shown), and time course microscopy analysis of PTPLB expression in the presence of lactacystin in Cos-7 cells at high magnification revealed a progressive transition of PTPLB staining from an ER-like network to a punctate disposition in the cell caused by the inhibitor (representative images shown in Fig. 4B). Similar results were obtained with MG132. Treatment of cells with lactacystin and MG132 has previously been shown to cause the accumulation of proteins that follow the ER-associated degradation pathway at the proteasome (22, 55).
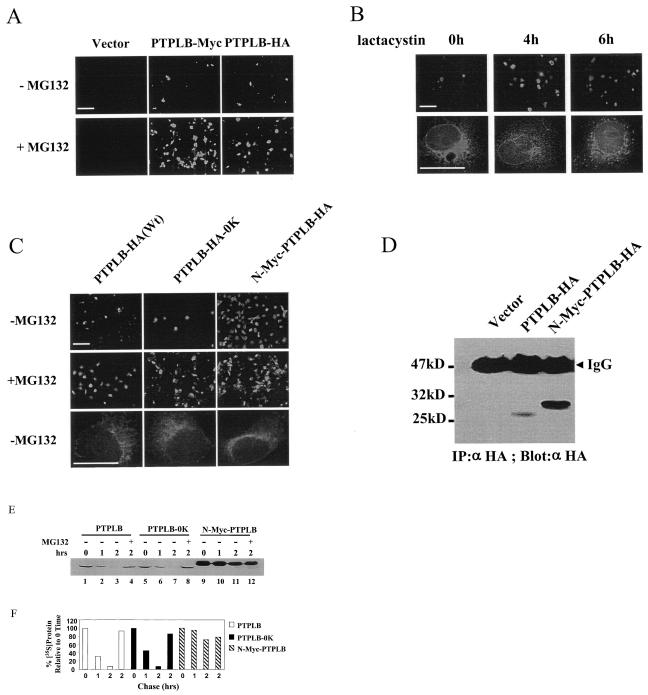
FIG. 4.
PTPLB turns over via the ER-associated degradation pathway. (A) Cos-1 cells were transfected with vector alone or vector encoding PTPLB-Myc-KKFE or PTPLB-HA-KKFE, and after 18 h, MG132 (100 μM) was added to the medium for 4 h and the cells were analyzed by immunofluorescence microscopy. (B) Cos-7 cells were transfected with PTPLB-Myc-KKFE, and 18 h later, lactacystin (5 μM) was added for the indicated times. Cells were analyzed by immunofluorescence microscopy with 20× (top panel) or 100× (lower panel) lenses. (C) Cos-1 cells were transfected with vector encoding wild-type PTPLB-HA (Wt), PTPLB-HA-0K, and N-Myc-PTPLB-HA, and after 18 h, MG132 (100 μM) was added to the medium for 4 h and the cells were analyzed by immunofluorescence microscopy with a ×20 or a ×100 lens. The scale bar in the pictures taken at 20× magnification represents 125 μm (top panels), whereas in the pictures taken at 100× magnification it represents 25 μm (lower panels). (D) Cos-1 cells were transfected with the vector, PTPLB-HA, or long N-Myc-PTPLB-HA. After 18 h, cell extracts containing equivalent amounts of protein were assessed by immunoblotting of anti-HA immunoprecipitates (IP) with anti-HA antibodies. (E) Stability of PTPLB and its mutants in vivo. The half-life of wild-type PTPLB, lysine-free PTPLB-0K (lysine residues were replaced with arginines), and N-Myc-PTPLB in Cos-1 cells was measured in a pulse-chase experiment, followed by immunoprecipitation of the labeled proteins as described in Materials and Methods. The proteasome inhibitor MG132 (100 μM) was added at 30 min after the addition of [35S]methionine-[35S]cysteine and was present throughout the 2-h chase period (lanes 4, 8, and 12). (F) Quantitative (PhosphorImager) analysis of the data depicted in panel E after the indicated chase periods in the absence and presence of the proteasome inhibitor. Quantities are relative to the amount of the indicated 35S-labeled PTPLB proteins at time zero.
Most ER-associated degradation substrates become ubiquitinated on lysine residues; ubiquitination acts as a signal for degradation by the proteasome (46, 47). Mutation of all 11 lysine residues in PTPLB to arginine (PTPLB-HA-0K), however, did not change the rapid degradation character of PTPLB detected by both Western blotting and immunofluorescence (Fig. 4C and data not shown). In the case of MyoD, on the other hand, the free NH2 group of the N-terminal residue of the protein rather than internal lysines serves as the essential and sufficient ubiquitin conjugation signal necessary for subsequent degradation of the protein. Addition of multiple copies of a Myc epitope tag to the N-terminal residue of MyoD successfully protected the MyoD protein from degradation (9, 18). Thus, an extended Myc tag was placed at the N-terminal residue of PTPLB and found to significantly increase the accumulation of PTPLB in transfected cells detected by both immunofluorescence and Western blotting (Fig. 4C and D, respectively). Similar to wild-type PTPLB, PTPLB-0K and N-Myc-PTPLB were distributed in the cell in an ER-like network pattern (Fig. 4C, lower panel).
The majority of protein substrates targeted by the ubiquitin system are short-lived. A pulse-chase experiment was carried out in order to assess the stability of the PTPLB protein and its mutants in vivo. As shown in Fig. 4E, the half-life of wild-type PTPLB and the lysine-free mutant (PTPLB-0K) was less than 1 h, and the pulse-labeled protein was barely detectable after 2 h of chase. In contrast, PTPLB containing the extended Myc tag at the N-terminal residue of PTPLB was significantly more stable. Moreover, the proteasomal inhibitor MG132 blocked the degradation of both wild-type PTPLB and PTPLB-0K but did not affect the stability of N-Myc-PTPLB after a 2-h chase (cf. lanes 11 and 12, Fig. 4E). Though not proven, it may be therefore that PTPLB, like MyoD, requires N-terminal ubiquitination. In any event, it appears that PTPLB is delivered to the proteasome for degradation.
PTPLB associates and colocalizes with BAP31 at the ER.
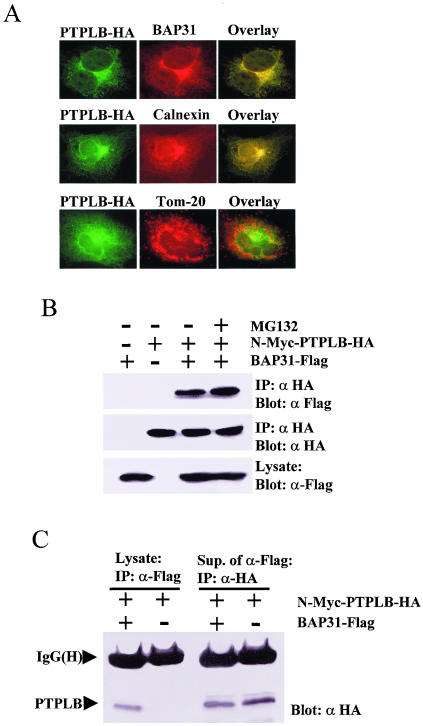
PTPLB-HA was transiently expressed in Cos-1 cells which were treated with MG132 for 4 h before double fluorescence staining with anti-HA antibody and either anti-BAP31, anti-calnexin (an ER marker), or anti-TOM20 (a mitochondrial marker) antibody. PTPLB was demonstrated to colocalize with endogenous BAP31 and calnexin by fluorescence microscopy, whereas TOM20 did not significantly overlap PTPLB (Fig. 5A). Furthermore, interactions between PTPLB and BAP31 could be demonstrated by coimmunoprecipitation. To this end, Cos-1 cells were cotransfected with N-Myc-PTPLB-HA and BAP31-Flag (Materials and Methods), and the association of BAP31 with PTPLB was monitored by immunoblotting of anti-HA immunoprecipitates with antibodies against Flag (Fig. 5B). However, if PTPLB is an ER-associated degradation substrate with which BAP31 only transiently associates to influence its turnover, it would be expected that only a small fraction of the stable N-Myc-PTPLB-HA mutant expressed in cells would be recovered by coimmunoprecipitation with BAP31-Flag. Indeed, following cotransfection of vectors expressing these proteins in Cos-1 cells, Myc-PTPLB-HA was detectable in the BAP31-Flag immunoprecipitate (Fig. 5C, lane 1), but the majority was found in the postimmunoprecipitate supernatant at levels only somewhat less than in the equivalent supernatants derived in the absence of BAP31-Flag (Fig. 5C, cf. lanes 3 and 4). This is in contrast to the constitutive components of the BAP31 complex such as A4, in which the majority of cotransfected A4-HA is recovered in BAP31-Flag immunoprecipitates (54).
FIG. 5.
PTPLB associates and colocalizes with BAP31 in the ER. (A) Cos-1 cells grown on coverslips were transfected with PTPLB-HA for 18 h, followed by the addition of MG132 to the medium for 4 h, and the cells were double stained with anti-HA antibody (green) and either anti-BAP31 (top panel), anti-calnexin (ER marker, middle panel), or anti-TOM20 (mitochondrion marker, bottom panel) (red), and images were visualized in the red, green, and yellow (overlay) channels. (B) Association of PTPLB with BAP31 in Cos-1 cells. Cos-1 cells were transfected with cDNAs coding for N-Myc-PTPLB-HA and/or BAP31-Flag as indicated. Anti-HA immunoprecipitates (IP) were recovered and resolved by SDS-PAGE, and immunoblots were probed with anti-Flag (top panel) or anti-HA (middle panel). Equivalent samples from cell lysates were immunoblotted with anti-Flag (bottom panel). (C) Small amounts of PTPLB associate with BAP31 in Cos-1 cells. Cos-1 cells were transfected with cDNAs coding for N-Myc-PTPLB-HA with or without BAP31-Flag as indicated. Anti-Flag immunoprecipitates (IP) were recovered and resolved by SDS-PAGE, and immunoblots were probed with anti-HA (lanes 1 and 2). Subsequently, the remaining PTPLB molecules were collected from the BAP31-depleted supernatants with an anti-HA antibody, and the immunoblots were probed with anti-HA (lanes 3 and 4).
PTPLB and PTPLB-PR lack PTPase activity.
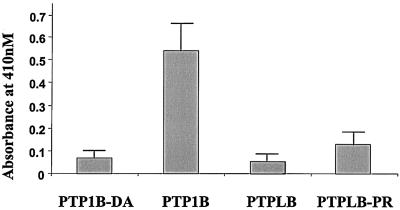
All PTPL family proteins contain a sequence highly similar to a conserved PTP catalytic domain except for the substitution of a conserved arginine (R) with a proline (P). As there have been no reports describing the protein expression of any PTPL family members, it is not known whether PTPL family members can be converted into PTPase-competent enzymes. Thus, we mutated the proline of N-Myc-PTPLB to the required arginine present in the active sites of PTPs and transfected N-Myc-PTPLB, N-Myc-PTPLB-PR, N-Myc-PTP1B (an ER-located PTPase), or N-Myc-PTP1B-DA (a mutant of PTP1B lacking PTPase activity) into Cos-1 cells. The protein expressions were normalized by immunoblotting of the anti-Myc immunoprecipitates with anti-Myc antibody (data not shown), and equal amounts of the proteins were used to detect PTPase activities with the universal small compound substrate _p_NPP (see Materials and Methods). As shown in Fig. 6, neither PTPLB nor PTPLB-PR expressed significant PTPase activity, with the negative control, PTP1B-DA, and the positive control, PTP1B, used as reference standards.
FIG. 6.
Immune complex phosphatase assays. Cos-1 cells were transfected for 18 h with cDNAs coding for N-Myc-PTP1B, N-Myc-PTP1B-D24A, N-Myc-PTPLB, and N-Myc-PTPLB-P101R. The protein expression levels were monitored by immunoblotting of the anti-Myc immunoprecipitates with anti-Myc antibodies, and the abundances of the proteins were adjusted to the same levels. The PTPase activity assay was performed as described in Materials and Methods. The absorbance at 410 nM represents the quantities of the dephosphorylated product of _p_NPP.
BAP31 maintains PTPLB expression.
The rapid turnover of wild-type PTPLB via the proteasome results in a low steady-state level of the protein following expression by transient transfection (Fig. 4). To investigate if BAP31 contributes to the survival of even these low levels of PTPLB, we compared differentiated embryoid epithelium-like mouse ES cell lines with X-linked Bap31 deleted (_Bap31-_null) or with Bap31 deleted and stably expressing human BAP31 (_Bap31_-null/BAP31) (7, 8). The Bap31 and BAP31 expression levels were verified by Western blotting (Fig. 7A). Following transfection of these cells with vectors expressing PTPLB-GFP or GFP, GFP was found to be expressed highly and equally in the two cell lines, whereas PTPLB-GFP was expressed at a level at least fivefold higher in _Bap31_-null/BAP31 cells compared to _Bap31_-null cells (Fig. 7B and C). BAP31 therefore contributes to the levels that can be achieved for this marker of PTPLB expression.
FIG. 7.
BAP31 maintains PTPLB expression levels. (A) Equivalent samples of cell lysates from _Bap31_-null and _Bap31_-null/BAP31 cell lines were analyzed by immunoblotting with anti-BAP31 and anti-γ-actin antibodies, as indicated. (B) The two cell lines were transfected with vectors expressing GFP or PTPLB-GFP, and GFP was monitored by immunofluorescence microscopy. (C) The _Bap31_-null and _Bap31_-null/BAP31 cell lines were transfected with vector pcDNA3.1 or a vector expressing GFP or PTPLB-GFP. After 36 h, the cells were fixed in 1% paraformaldehyde in phosphate-buffered saline for 30 min and analyzed by flow cytometry. The data were gated (R1) to exclude emissions obtained in the vector control cells.
DICUSSION
Protein-protein interaction plays a major role in the regulation of many biological processes, and elucidating these interactions can be instructive for proteins that have no known function or sequence homology with other proteins in the data banks (11). One-third of the predicted open reading frames in S. cerevisiae are still classified as proteins with unknown functions (44), and the published functions for many proteins may represent only part of their full activity spectrum. In the past decade, traditional two-hybrid techniques have identified a plethora of interactions between soluble proteins that are important in different signal transduction pathways (50). However, novel associations between membrane proteins have not been well studied because of inherent technical limitations.
Here we have provided details for exploiting the split-ubiquitin system as a screening tool and identified the BAP3-interacting protein PTPLB, a novel member of the PTPL family. Of note, although PTPL family members contain PTP signatures, we were unable to provide PTPLB with PTPase activity in detergent extracts by converting proline 101 to arginine and thus reconstituting the PTP catalytic motif (I/V)HCXXGXXR(S/T). As documented in the literature, receptor-type PTPs normally contain two tandem PTP domains disposed to the cytosol, and both domains have the PTP catalytic motif; however, only domain 1 has PTP activity, whereas domain 2 lacks PTP activity but has a regulatory function (23). Whether PTPLB has an analogous regulatory function remains to be determined. Such functions might include coordinating ER dynamics or ER transitions during the cell cycle.
The fact that the rapid turnover of PTPLB appears to come under the control of BAP31 is consistent with the predicted function of BAP31 as a chaperone or quality control factor for the biogenesis of integral membrane proteins at the ER (28, 38, 40). By reconstituting BAP31 into mouse cells with the Bap31 gene deleted, we could show that BAP31 is required to maintain even detectable levels of PTPLB. One possible scenario is therefore that BAP31 is required for the proper folding or assembly of the four-spanning PTPLB protein in the lipid bilayer of the ER. PTPLB molecules that do not assemble properly, on the other hand, are eliminated by the ER-associated degradation pathway. It is possible that other PTPL family members represent ER-associated degradation substrates, since they share high homologies and there have been no reports describing the protein expression of any members. The cystic fibrosis transmembrane conductance regulator was the first ER-associated degradation substrate identified, in part through the use of inhibitors of ubiquitin-dependent degradation by the proteasome (22, 55), and ER-associated degradation has since been extended to many other proteins (14, 16, 42, 57).
It will be important to determine if BAP31 has a general or restricted role in the turnover of membrane proteins at the ER. PTPLB was stabilized by placing an extended Myc tag at the N terminus, probably indicating that, in contrast to the cystic fibrosis transmembrane conductance regulator, rapid elimination of the protein is associated with ubiquitination of a free amine at the N terminus. Interestingly, we found that the BAP31 bait also picked up multiple hits for Sec61β in the split-ubiquitin screen (unpublished data). The Sec61 complex constitutes the core of both the forward translocation and retrotranslocation machinery for protein transport across the ER membrane. It consists of three subunits, the α subunit (Sec61α), which spans the membrane 10 times, and the β and γ subunits (Sec61β and Sec61γ, respectively), each of which spans the membrane once (33, 43). Since Sec61-dependent retrotranslocation of unfolded ER membrane proteins is a critical step in their delivery to the proteasome (14, 16, 19), an association of BAP31 with this complex might facilitate its contribution to the ER-associated degradation pathway.
.
Acknowledgments
We thank S. Heesen and I. Staglar for the gift of the Nub- and Cub-containing plasmids and S. Heesen and M. Aebi for hospitality to A.H. and guidance in using the split-ubiquitin system. The BAP31-null cell lines described in reference 7 were derived from ES cells originally provided by S. Kuppig and M. Reth.
This work was supported by operating grants from the Canadian Institutes of Health Research (G.C.S., J.P., and A.H.) and the National Cancer Institute of Canada (G.C.S.).
REFERENCES
- 1.Adachi, T., W. W. Schamel, K. M. Kim, T. Watanabe, B. Becker, P. J. Nielsen, and M. Reth. 1996. The specificity of association of the IgD molecule with the accessory proteins BAP31/BAP29 lies in the IgD transmembrane sequence. EMBO J. 15**:**1534-1541. [PMC free article] [PubMed] [Google Scholar]
- 2.Annaert, W. G., B. Becker, U. Kistner, M. Reth, and R. Jahn. 1997. Export of cellubrevin from the endoplasmic reticulum is controlled by BAP31. J. Cell Biol. 139**:**1397-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronheim, A., D. Engelberg, N. Li, N. Alawi, J. Schlessinger, and M. Karin. 1994. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78**:**949-961. [DOI] [PubMed] [Google Scholar]
- 4.Aronheim A., E. Zandi, H. Hennemann, S. J. Elledge, M. and Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17**:**3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronheim, A. 2001. Protein recruitment systems for the analysis of protein-protein interactions. Methods 24**:**29-34. [DOI] [PubMed] [Google Scholar]
- 6.Bellec, Y., Y. Harrar, C. Butaeye, S. Darnet, C. Bellini, and J. D. Faure. 2002. Pasticcino2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis. Plant J. 32**:**713-722. [DOI] [PubMed] [Google Scholar]
- 7.Breckenridge, D. G., M. Nguyen, S. Kupping, M. Reth, and G. C. Shore. 2002. The procaspase-8 isoform, procaspase-8L, recruited to the BAP31 complex at the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 99**:**4331-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breckenridge, D. G., M. Stojanovic, R. C. Marcellus, and G. C. Shore. 2003. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 160**:**1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17**:**5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducret, A., M. Nguyen, D. G. Breckenridge, and G. C. Shore. 2003. The resident endoplasmic reticulum protein, BAP31, associates with gamma-actin and myosin B heavy chain. Eur. J. Biochem. 270**:**342-349. [DOI] [PubMed] [Google Scholar]
- 11.Dunham, I., N. Shimizu, B. A. Roe, et al. 1999. The DNA sequence of human chromosome 22. Nature 402**:**489-495. [DOI] [PubMed] [Google Scholar]
- 12.Dunnwald, M., A. Varshavsky, and N. Johnsson. 1999. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell 10**:**329-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faure, J. D., P. Vittorioso, V. Santoni, V. Fraisier, E. Prinsen, I. Barlier, H. Van Onckelen, M. Caboche, and C. Bellini. 1998. The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development 125**:**909-918. [DOI] [PubMed] [Google Scholar]
- 14.Fiebiger, E., C. Story, H. L. Ploegh, and D. Tortorella. 2002. Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein. EMBO J. 21**:**1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature (London) 340**:**245-246. [DOI] [PubMed] [Google Scholar]
- 16.Friedlander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2**:**379-384. [DOI] [PubMed] [Google Scholar]
- 17.Haberer, G., S. Erschadi, and R. A. Torres-Ruiz. 2002. The Arabidopsis gene PEPINO/PASTICCINO2 is required for proliferation control of meristematic and non-meristematic cells and encodes a putative anti-phosphatase. Dev. Genes Evol. 212**:**542-550. [DOI] [PubMed] [Google Scholar]
- 18.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267**:**1018-1021. [DOI] [PubMed] [Google Scholar]
- 19.Hegde, R. S., S. Voigt, T. A. Rapoport, and V. R. Lingappa. 1998. TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell 92**:**621-631. [DOI] [PubMed] [Google Scholar]
- 20.**Hubsman, M., G. Yudkovsky, and A. Aronheim.2001. A novel approach for the identification of protein-protein interaction with integral membrane proteins. Nucleic Acids Res. 29:**4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isakoff, S. J., T. Cardozo, J. Andreev, Z. Li, K. M. Ferguson, R. Abagyan, M. A. Lemmon, A. Aronheim, and E. Y. Skolnik. 1998. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17**:**5374-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, T. J., M. A. Loo, S. Pind, D. B. Williams, A. L. Goldberg, and J. R. Riordan. 1995. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83**:**129-135. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, K. G. and D. Van Vactor. 2003. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 83**:**1-24. [DOI] [PubMed] [Google Scholar]
- 24.Johnsson, N., and A. Varshavsky. 1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA 91**:**10340-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joung, J. K. 2001. Identifying and modifying protein-DNA and protein-protein interactions using a bacterial two-hybrid selection system. J. Cell Biochem. Suppl. 37**:**53-57. [DOI] [PubMed] [Google Scholar]
- 26.Kim, K. M., T. Adachi, P. J. Nielsen, M. Terashima, M. C. Lamers, G. Kohler, and M. Reth. 1994. Two new proteins preferentially associated with membrane immunoglobulin D. EMBO J. 13**:**3793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klopotowski, T., and A. Wiater. 1965. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch. Biochem. Biophys. 112**:**562-566. [DOI] [PubMed] [Google Scholar]
- 28.Lambert, G., B. Becker, R. Schreiber, A. Boucherot, M. Reth, and K. Kunzelmann. 2001. Control of cystic fibrosis transmembrane conductance regulator expression by BAP31. J. Biol. Chem. 276**:**20340-20345. [DOI] [PubMed] [Google Scholar]
- 29.Levy, F., N. Johnsson, T. Rumenapf, and A. Varshavsky. 1996. Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. USA 93**:**4907-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, D., O. Gonzalez, L. L. Bachinski, and R. Roberts. 2000. Human protein tyrosine phosphatase-like gene: expression profile, genomic structure, and mutation analysis in families with ARVD. Gene 256**:**237-243. [DOI] [PubMed] [Google Scholar]
- 31.Licitra, E. J., and J. O. Liu. 1996. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Natl. Acad. Sci. USA 93**:**12817-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massaad, M. J., and A. Herscovics. 2001. Interaction of the endoplasmic reticulum alpha 1,2-mannosidase Mns1p with Rer1p using the split-ubiquitin system. J. Cell Sci. 114**:**4629-4635. [DOI] [PubMed] [Google Scholar]
- 33.Matlack, K. E., W. Mothes, and T. A. Rapoport. 1998. Protein translocation: tunnel vision. Cell 92**:**381-390. [DOI] [PubMed] [Google Scholar]
- 34.Ng, F. W., M. Nguyen, T. Kwan, E. Branton, P. D. W. Nicholson, J. A. Cromlish, and G. C. Shore. 1997. p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum. J. Cell Biol. 139**:**327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng, F. W., and G. C. Shore. 1998. Bcl-XL cooperatively associates with the Bap31 complex in the endoplasmic reticulum, dependent on procaspase-8 and Ced-4 adaptor. J. Biol. Chem. 273**:**3140-3143. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, M., D. G. Breckenridge, A. Ducret, and G. C. Shore. 2000. Caspase-resistant BAP31 inhibits _fas_-mediated apoptotic membrane fragmentation and release of cytochrome c from mitochondria. Mol. Cell. Biol. 20**:**6731-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi, F., C. A. Charlton, and H. M. Blau. 1997. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc. Natl. Acad. Sci. USA 94**:**8405-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schamel, W. W. A., S. Kupping, B. Becker, K. Gimborn, H. Hauri, and M. Reth. 2003. A high-molecular-weight complex of membrane proteins BAP29/BAP31 is involved in the retention of membrane-bound IgD in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 100**:**9861-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengupta, D. J., B. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93**:**8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiliotis, E. T., M. Osorio, M. C. Zuniga, and M. Edidin. 2000. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity 13**:**841-851. [DOI] [PubMed] [Google Scholar]
- 41.Stagljar, I., C. Korostensky, N. Johnsson, and S. T. Heesen. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95**:**5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki, T., and A. Varshavsky. 1999. Degradation signals in the lysine-asparagine sequence space. EMBO J. 18**:**6017-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai, B., and T. A. Rapoport. 2002. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J. Cell Biol. 159**:**207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uetz, P., L. Giot, G. Cagney, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403**:**623-627. [DOI] [PubMed] [Google Scholar]
- 45.Uwanogho, D. A., Z. Hardcastle, P. Balogh, G. Mirza, K. L. Thornburg, J. Ragoussis, and P. T. Sharpe. 1999. Molecular cloning, chromosomal mapping, and developmental expression of a novel protein tyrosine phosphatase-like gene. Genomics 15**:**406-416. [DOI] [PubMed] [Google Scholar]
- 46.VanSlyke, J. K., and L. S. Musil. 2002. Dislocation and degradation from the ER are regulated by cytosolic stress. J. Cell Biol. 157**:**381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varshavsky, A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93**:**12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidal, M., R. K. Brachmann, A. Fattaey, E. Harlow, and J. D. Boeke. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93**:**10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vittorioso, P., R. Cowling, J. D. Faure, M. Caboche, and C. Bellini. 1998. Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol. Cell. Biol. 18**:**3034-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallach, D., M. P. Boldin, A. V. Kovalenko, N. L. Malinin, I. L. Mett, and J. H. Camonis. 1998. The yeast two-hybrid screening technique and its use in the study of protein-protein interactions in apoptosis. Curr. Opin. Immunol. 10**:**131-136. [DOI] [PubMed] [Google Scholar]
- 51.Wang, B., K. Kishihara, D. Zhang, H. Hara, and K. Nomoto. 1997. Molecular cloning and characterization of a novel human receptor protein tyrosine phosphatase gene, hPTP-J: down-regulation of gene expression by PMA and calcium ionophore in Jurkat T lymphoma cells. Biochem. Biophys. Res. Commun. 231**:**77-81. [DOI] [PubMed] [Google Scholar]
- 52.Wang, B., K. Kishihara, D. Zhang, T. Sakamoto, and K. Nomoto. 1999. Transcriptional regulation of a receptor protein tyrosine phosphatase gene hPTP-J by PKC-mediated signaling pathways in Jurkat and Molt-4 T lymphoma cells. Biochim. Biophys. Acta 1450**:**331-340. [DOI] [PubMed] [Google Scholar]
- 53.Wang, B., S. Lemay, S. Tsai, and A. Veillette. 2001. SH2 domain-mediated interaction of inhibitory protein tyrosine kinase Csk with protein tyrosine phosphatase-HSCF. Mol. Cell. Biol. 21**:**1077-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, B., M. Nguyen, D. G. Breckenridge, M. Stojanovic, P. A. Clemons, S. Kuppig, and G. C. Shore. 2003. Uncleaved BAP31 in association with A4 protein at the endoplasmic reticulum is an inhibitor of Fas-initiated release of cytochrome c from mitochondria. J. Biol. Chem. 278**:**14461-14468. [DOI] [PubMed] [Google Scholar]
- 55.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83**:**121-127. [DOI] [PubMed] [Google Scholar]
- 56.Wittke, S., N. Lewke, S. Muller, and N. Johnsson. 1999. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell 10**:**2519-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yedidia, Y., L. Horonchik, S. Tzaban, A. Yanai, and A. Taraboulos. 2001. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 20**:**5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, J., and S. Lautar. 1996. A yeast three-hybrid method to clone ternary protein complex components. Anal. Biochem. 242**:**68-72. [DOI] [PubMed] [Google Scholar]