DNA/RNA Helicase Gene Mutations in a Form of Juvenile Amyotrophic Lateral Sclerosis (ALS4) (original) (raw)
Abstract
Juvenile amyotrophic lateral sclerosis (ALS4) is a rare autosomal dominant form of juvenile amyotrophic lateral sclerosis (ALS) characterized by distal muscle weakness and atrophy, normal sensation, and pyramidal signs. Individuals affected with ALS4 usually have an onset of symptoms at age <25 years, a slow rate of progression, and a normal life span. The ALS4 locus maps to a 1.7-Mb interval on chromosome 9q34 flanked by D9S64 and D9S1198. To identify the molecular basis of ALS4, we tested 19 genes within the ALS4 interval and detected missense mutations (T3I, L389S, and R2136H) in the Senataxin gene (SETX). The SETX gene encodes a novel 302.8-kD protein. Although its function remains unknown, SETX contains a DNA/RNA helicase domain with strong homology to human RENT1 and IGHMBP2, two genes encoding proteins known to have roles in RNA processing. These observations of ALS4 suggest that mutations in SETX may cause neuronal degeneration through dysfunction of the helicase activity or other steps in RNA processing.
Introduction
Amyotrophic lateral sclerosis (ALS), also known as “Lou Gehrig’s disease,” denotes a heterogeneous group of severe, progressive neurological disorders associated with degeneration of motor neurons in the cerebral cortex, brain stem, and spinal cord (Morrison and Harding 1994). Approximately 90% of ALS cases are sporadic, and ∼10% are familial (FALS) (Strong et al. 1991). FALS is clinically and genetically heterogeneous (Majoor-Krakauer et al. 2003). To date, causal mutations have been found for only two forms of ALS. ALS1 is an adult-onset, fatal, autosomal dominant (AD) disorder associated with mutations in the Cu/Zn superoxide dismutase (SOD1) gene on chromosome 21q21 (Rosen et al. 1993). ALS2 is a juvenile-onset, slowly progressive, autosomal recessive disorder that maps to chromosome 2q33 and is associated with mutations in the alsin gene, a putative GTPase regulator (Hadano et al. 2001; Yang et al. 2001). The mechanisms through which mutations in either SOD1 or alsin lead to motor neuron degeneration are unknown.
ALS4 (MIM 602433 [also known as “distal hereditary motor neuronopathy” with pyramidal features or dHMN]) is a rare, childhood- or adolescent-onset, AD form of ALS that is characterized by slow disease progression, limb weakness, severe muscle wasting, and pyramidal signs associated with degeneration of motor neurons in the brain and spinal cord. The phenotype of ALS4 includes a long duration of disease, absence of overt sensory abnormalities, and the sparing of bulbar and respiratory muscles (Chance et al. 1998; Rabin et al. 1999; De Jonghe et al. 2002).
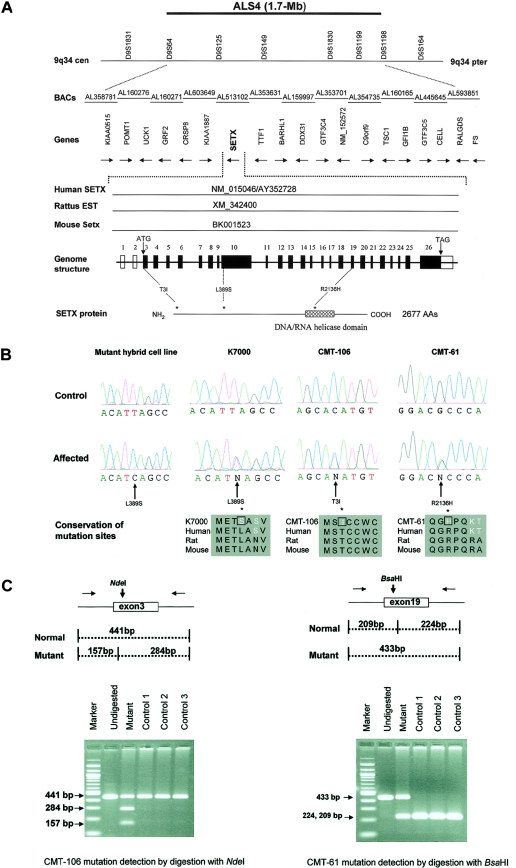
We mapped a gene for ALS4 to chromosome 9q34 in a large Maryland family (K7000) in which 49 family members were affected (Chance et al. 1998; Rabin et al. 1999; Blair et al. 2000). Analysis in an additional three families (CMT-61, CMT-106, and F-54) with a similar phenotype supported linkage to the same region in 9q34 (De Jonghe et al. 2002). To identify a critical gene for this disorder, we searched for mutations in 19 candidate genes mapping within the ALS4 interval on chromosome 9q34 (fig. 1_a_). Our analysis detected missense mutations in the Senataxin gene (SETX) or KIAA0625 (HUGE Protein Database), a gene that encodes a novel DNA/RNA helicase in three unrelated families with ALS4.
Figure 1.
Identification of the ALS4 gene. a, ALS4 gene region on 9q34. Markers D9S64 and D9S1198 define a 1.7-Mb interval containing 19 genes. Sequencing of 340 exons and flanking regions in probands with ALS4 revealed mutations in SETX or KIAA0625, a gene encoding a novel member of the RNA/DNA helicase family. Corresponding human, mouse, and rat cDNA sequences are shown. The SETX gene consists of 26 exons. An asterisk (*) indicates the locations of mutations in three unrelated pedigrees with ALS4. b, Mutations in SETX in pedigrees with ALS4 (K7000, CMT-106, and CMT-61). As shown, mutations are heterozygous, except for the mutant hybrid cell line (prepared from an affected individual from K7000). Protein alignment showed that mutations of human SETX are well conserved with rat and murine orthologs. c, Restriction pattern of the _Nde_I digest of the 441-bp exon 3 fragment, with the c.8C→T mutation seen in CMT-106 giving rise to two bands of 284 bp and 157 bp. _Bsa_HI exon 19 restriction digestion pattern of a 284-bp fragment and 157-bp fragment, with the c.6407G→A mutation seen in CMT-61, resulting in an undigested band of 433 bp.
Material and Methods
Families with ALS4
We obtained family data and blood samples, under a protocol of informed consent approved by the institutional review board of the University of Washington, Seattle, and the other relevant approved review boards for each medical institution, according to the Declaration of Helsinki.
Pedigrees K7000, CMT-61, CMT-106, and F-54 are from the United States, Belgium, Austria, and England, respectively. Detailed clinical, genetic, and electrophysiological features of individuals affected with ALS4 from these four unrelated pedigrees have been described elsewhere (Rabin et al. 1999; De Jonghe et al. 2002). Affected individuals have a slowly progressive motor neuronopathy with evidence of upper and lower motor-neuron dysfunction. There is predominantly distal muscle weakness and atrophy associated with pyramidal signs, including brisk deep-tendon reflexes and a positive Babinski maneuver. The mean ages at onset for each pedigree are 17 years (K7000), <6 years (CMT-61), 8 years (CMT-106), and 21 years (F-54). In pedigree K7000, ∼10% of affected persons had minimal sensory impairment, usually limited to a slight elevation of vibratory threshold in middle-aged or elderly patients. Otherwise, affected persons have no overt clinical signs of sensory-nerve impairment. There is wide variation of phenotypic expression, ranging from some individuals with only mild gait abnormalities and brisk reflexes to those who may be wheelchair-bound with no functional hand use by the 5th or 6th decade of life.
Whereas the phenotype in pedigree F-54 includes distal weakness and amyotrophy with normal sensation, affected individuals in this family are somewhat different from those seen in the other three pedigrees. All patients in F-54 presented with foot deformities (pes cavus) during early childhood, and some patients had been diagnosed as having spastic paraplegia with amyotrophy (Silver disease). This pedigree was reported elsewhere as having LOD scores supporting linkage to the ALS4 region (De Jonghe et al. 2002); however, we performed analysis with additional affected persons, and linkage to the ALS4 region was no longer found (table 1). Furthermore, haplotypes constructed with markers from the ALS4 interval do not segregate with the disease in this pedigree (data not shown).
Table 1.
LOD Scores (Z) in Pedigree F-54
| LOD at θ= | |||||||
|---|---|---|---|---|---|---|---|
| Marker | .0 | .01 | .05 | .10 | .2 | .3 | .4 |
| D9S1847 | −3.53 | −2.34 | −.97 | −.41 | .01 | .10 | .05 |
| D9S2126 | −4.51 | −2.07 | −.69 | −.16 | .21 | .24 | .13 |
| D9S1199 | −4.51 | −2.76 | −1.27 | −.62 | −.08 | .07 | .05 |
Electrophysiological studies, including nerve-conduction velocity (NCV) and electromyography (EMG), of patients with ALS4 documented the presence of a chronic motor neuronopathy with partial denervation-reinnervation, in a graded pattern of distal muscles being more severely affected than proximal muscles (Rabin et al. 1999; De Jonghe et al. 2002). EMG testing disclosed reduced amplitudes of compound muscle-action potentials, positive sharp waves, and fibrillations. Sural and median sensory nerve–action potential amplitudes were normal. Motor and sensory NCV testing showed normal values or minimal slowing.
Pathological Studies
Postmortem examinations of the brain and spinal cord have been undertaken in two individuals (aged 75 and 88 years) from pedigree K7000 (Rabin et al. 1999). In both individuals, there were atrophic spinal cords with marked loss of anterior horn cells and degeneration of corticospinal tracts. Despite an absence of significant sensory impairment on clinical examinations, in both individuals, there was a definite loss of neurons in the dorsal-root ganglia and degeneration of the posterior columns. Axonal spheroids were present in the gray matter of the spinal cord, the dorsal-root entry zones, and the peripheral nerves. Motor and sensory roots, as well as peripheral nerves, showed significant axonal loss.
Generation of Somatic Cell Hybrids
This procedure was performed as described elsewhere (Yan et al. 2000; Marra et al. 2001). In brief, lymphoblasts from the K7000 proband were electrofused to mouse E2 cell lines. Unfused mouse E2 clones and unfused human lymphocyte clones were negatively selected using HAT and geneticin, respectively. Fused colonies appearing after 2 wk were selected and expanded for an additional 2 wk prior to DNA analysis. Chromosome 9 polymorphic markers were used to isolate two hybrid clones, one containing the affected human monochromosome 9 and the second containing the normal human monochromosome 9. The ALS4-affected hybrid was selected by haplotype analysis with known disease-associated polymorphic markers (data not shown).
Mutation Detection
We performed PCR amplification and DNA sequencing of 340 exons derived from 19 candidate genes within the ALS4 disease interval. We compared DNA sequences obtained from affected patients and normal individuals, using primer pairs designed from Internet-accessible sources (see the “Electronic-Database Information” section). One hundred DNA samples taken from a racially mixed population were used as controls.
Northern-Blot Analysis
We performed northern-blot analysis of human 12-lane multiple tissue blot (Clontech) with an SETX (KIAA0625) IMAGE clone insert (number 4136196) labeled with dCTP-[α-32P]. Standard conditions for hybridization and washing were employed, with the ExpressHyb buffer, according to the manufacturer’s specifications (Clontech).
Results
Mutation Analysis of a Monochromosome 9 Hybrid Cell Line
We searched for deletions, insertions, or other rearrangements in the ALS4 region by PCR-based STS/EST content mapping in the affected chromosome 9 cell line. We did not detect evidence for such deletions/insertions or rearrangements following the amplification of all 340 exon fragments (data not shown). We next searched for disease-associated DNA sequence alterations in these same exons and their flanking regions, derived from 19 candidate genes for ALS4 (fig. 1_a_). In the ALS4-affected monochromosome 9 hybrid line (pedigree K7000), we found a single-nucleotide change, a c.1166T→C transversion in exon 10 of the SETX gene (GenBank accession number AY362728), predicted to cause an L389S substitution.
Identification of SETX Mutations in Three Pedigrees with ALS4
Following the identification of an SETX disease-associated mutation in the somatic cell hybrid, all 24 coding exons of this gene were sequenced in probands from each of the four pedigrees mapping to 9q34. All affected individuals from pedigree K7000 were heterozygous for the L389S substitution, whereas unaffected members had a homozygous normal pattern. The L389S mutation was not detected in 100 unrelated control individuals. In pedigree CMT-61, affected individuals were found to carry a heterozygous c.6407G→A transition in exon 19, leading to an R2136H substitution. This mutation eliminated a Bsa_HI restriction site, allowing us to scan a panel of 100 unrelated control individuals without detecting this mutation. In pedigree CMT-106, we detected a heterozygous c.8C→T transversion in exon 3 (exons 1 and 2 are noncoding) leading to a T3I substitution. This mutation generated an Nde_I restriction site that was not present in unaffected family members, nor was it found in 100 unrelated control individuals (fig. 1_b_ and 1_c_; table 2). No mutation was detected in pedigree F-54 as a result of our initial examination of the coding exons or exon/intron boundaries of SETX. We performed a more extensive search by sequencing additional genomic DNA, including 1 kb of 5′ UTR, the two noncoding exons (1 and 2), 2.8 kb of the 3′ UTR, and the canonical polyadenylation signal. However no disease-associated mutation of SETX was identified in this pedigree. The SETX mutations segregate with the disease phenotype in three families, consistent with the AD mode of inheritance and supporting the hypothesis that mutations in the SETX gene are associated with ALS4.
Table 2.
Mutations of SETX and Clinical Data for Patients with ALS4
| Family | No. ofAffected/UnaffectedIndividuals | Geographic Origin | HeterozygousMutationa | Amino Acid Substitution | Average Ageat Onset(years) |
|---|---|---|---|---|---|
| K7000 | 55/65 | United States | c.1166T→C | L389S | 17 |
| CMT-61 | 5/3 | Belgium | c.6407G→A | R2136H | <6 |
| CMT-106 | 7/10 | Austria | c.8C→T | T3I | 8 |
Structure and Deduced Protein Sequence of SETX
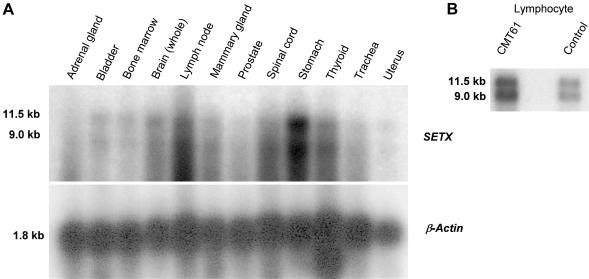
The SETX gene encodes a 302.8-kD protein consisting of 2,677 amino acids (fig. 1_a_). Northern-blot analysis identified two prominent transcripts of 11.5 kb and 9.0 kb in all tissues examined, including brain and spinal cord (fig. A1_a_ and A1_b_ [online only]). Both bands detected by northern blot were observed with each of several different SETX probes that were used to target both 5′ and 3′ regions of the transcript (data not shown). We suggest that a canonical polyadenylation signal 3,174 bp beyond the stop codon accounts for the stronger 11.5-kb band and that a less efficient cryptic polyadenylation signal be used to generate the weaker 9.0-kb band.
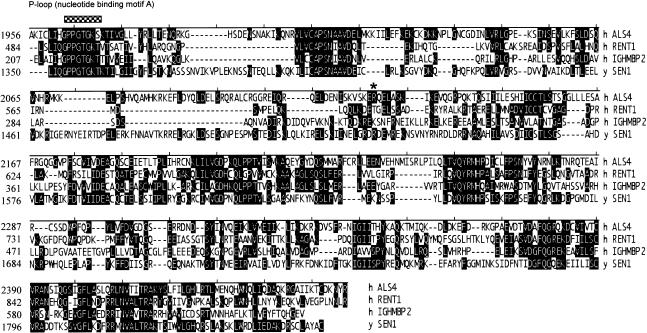
The SETX protein sequence was analyzed with the domain prediction programs InterproScan (European Bioinformatics Institute), SMART, and BLASTP. Each program predicted a superfamily I DNA/RNA helicase domain at residues 1931–2456. InterproScan predicted an ATP/GTP–binding site motif A (P-loop) at residues 1963–1970 (fig. 2) that has been shown to be essential for DNA unwinding in other systems (Molnar et al. 1997). A bipartite nuclear localization signal was also predicted by InterproScan and by the peptide signal–sorting program P-sort (residues 2070–2087). It is interesting that the majority of this large protein appears novel, since no program or homology search could identify additional domains or structural features within residues 1–1930.
Figure 2.
Alignment of the helicase domain regions of the human SETX, IGHMBP2, RENT1, and yeast SEN1 proteins. Identities of the SETX helicase domain with that of IGHMBP2, RENT1, and yeast SEN1 are 42%, 46%, and 42%, respectively. SETX has an ATP/GTP–binding site motif A (P-loop), indicated by a bar. An asterisk (*) indicates the position of the mutation in CMT-61.
As mentioned above, the C-terminal end of the SETX protein contains a superfamily I helicase domain from residues 1931–2456. This helicase domain has high homology with the rat orthologue (at 85% identity), and the mouse orthologue (at 90% identity). By BLASTP analysis, the SETX helicase domain has 46% identity with the regulator of nonsense transcripts-1 protein (RENT1). Human RENT1 is thought to be part of a distinct subset within the superfamily that includes the yeast-splicing endonuclease 1 gene (Sen1). The helicase domain of SETX also shows strong identity to a second human protein, the immunoglobulin μ-binding protein 2 (IGHMBP2), at 42% identity (fig. 2).
Discussion
We previously mapped the gene for ALS4 to chromosome 9q34 in a large Maryland kindred, K7000 (Chance et al. 1998; Blair et al. 2000). Genetic analysis of additional families with similar phenotypic features yielded combined LOD scores suggesting linkage with multiple markers within the ALS4 interval (De Jonghe et al. 2002). In this article, we focused our analysis on these four pedigrees and searched for causal mutations within a 1.7-Mb interval encompassing 19 known candidate genes (fig. 1_a_). Mutations in SETX, a gene encoding a novel DNA/RNA helicase, were found in three pedigrees. In a fourth pedigree (F-54), we did not find any disease-associated mutations. Although initial studies suggested linkage to the ALS4 interval (De Jonghe et al. 2002), subsequent analysis including additional affected individuals confirmed that the gene in pedigree F-54 is unlinked to 9q34.
The clinical and genetic nosology of inherited disorders affecting motor systems is complex and evolving. Frequently, the overlap of phenotypic features seen within forms of familial motor neuronopathies, hereditary spastic paraplegias, and hereditary peripheral polyneuropathies (e.g., Charcot-Marie-Tooth [CMT] neuropathy) may complicate their classification. Individuals in pedigree K7000 were first reported as having a variant of CMT, despite the presence of upper motor-neuron dysfunction and lack of sensory impairment (Myrianthopoulos et al. 1964). Pedigrees CMT-61 and CMT-106 were also initially diagnosed to have variant forms of CMT; however, genetic analysis suggested linkage to chromosome 9q34, and they were reported to have a form of dHMN or ALS4. Furthermore, linkage to chromosome 9q34 in the ALS4 interval was found in an Italian pedigree with spastic paraplegia designated “SPG19” (Valente et al. 2002). Regardless of the diagnostic terminology for these pedigrees, the results of our analysis show that an important gene for motor-neuron function maps to chromosome 9q34 and is associated with mutations in SETX. It will be crucial to test other kindreds having motor-neuron disorders with similar phenotypes for mutations in SETX.
It is interesting that it has been reported that putative loss-of-function mutations (nonsense) in the SETX gene lead to an unrelated disorder, ataxia-oculomotor apraxia type 2 (AOA2 [MIM 606002]) (Moreira et al. 2004). Ataxia-oculomotor apraxia (AOA) is a genetically heterogeneous group of autosomal recessive disorders characterized by cerebellar ataxia/atrophy, oculomotor apraxia, early loss of reflexes, late peripheral neuropathy, slow progression leading to severe motor handicap and the absence of telangiectasias, and immunodeficiency. One form, AOA1, maps to chromosome 9q21 and is caused by mutations in the aprataxin gene (Date et al. 2001; Moreira et al. 2001). Another form, AOA2, maps to chromosome 9q34 within the same interval to which the ALS4 gene maps. Moreira et al. (2004) reported 15 different mutations of SETX in patients with AOA2, and 10 of these mutations were predicted to lead to premature termination of the SETX protein. Heterozygous carriers of mutations in SETX associated with AOA2 have a normal phenotype. Although there are clear differences in the phenotypes of ALS4 and AOA2 and different modes of inheritance (AD and recessive, respectively), they have several features in common. Both disorders lead to development of a peripheral motor neuropathy (albeit with a later onset in subjects with AOA2), and reduced evoked amplitudes have been documented in AOA2 (Watanabe et al. 1998) and in patients in pedigree K7000 (Rabin et al. 1999). Disease progression is slow, leading to severe motor handicap, whereas longevity is unaffected in both disorders. Given the types of SETX mutations seen thus far in ALS4 and AOA2, it is tempting to speculate that a dominant-acting, partial loss of function or a toxic gain of function could lead to development of primarily upper and lower motor-neuron degeneration, as seen in patients with ALS4, whereas the recessive, putative loss-of-function mutations could lead to a more widespread disorder, as seen in AOA2 (Watanabe et al. 1998; Moreira et al. 2004).
The mechanisms through which mutations in SETX lead to ALS4 or AOA2 are unknown; however, DNA/RNA helicases are known to be involved in DNA repair, replication, recombination and transcription, RNA processing, transcript stability, and translation initiation (Tanner and Linder 2001). Recent studies showed that several human diseases, including spinal muscular atrophy (SMA), are associated with defects in proteins or protein complexes that possess helicase activity (Campbell et al. 2000; Meister et al. 2000; Grohmann et al. 2001). Pellizzoni et al. (2001) identified RNA helicase A as a novel SMN-interacting protein and showed that this interaction is defective in some SMA mutants, suggesting a critical role for the SMN complex in several aspects of mRNA biogenesis. Although the SMN protein does not contain helicase motifs, it is part of a major cellular complex including DP103, a member of the DEAD box family of RNA helicase (Campbell et al. 2000). It is interesting that SMA with respiratory distress type 1 (SMARD1) results from mutations in the IGHMBP2 gene that do contain a helicase motif (Grohmann et al. 2001). These results suggest that DNA/RNA helicase dysfunction may play an important role in the development of lower motor-neuron diseases.
The helicase domain of SETX also showed strong homology with RENT1 (46% identity), which, like IGHMBP2, also plays an important role in producing mature mRNA. Recent findings showed that RENT1 is involved in nonsense-mediated RNA decay (NMD) (Sun et al. 1998; Mendell et al. 2002). NMD probably protects the organism from deleterious peptides that could be expressed from nonsense alleles. Studies also showed that inhibition of RENT1 expression abrogated NMD of nonsense T-cell receptor beta transcripts (Mendell et al. 2002).
Given that the helicase domains of the SETX and IGHMBP2 proteins show strong homology (42% identity) and that mutations in the genes encoding these proteins lead to selected motor-neuron degeneration, our observations of ALS4 suggest that DNA/RNA mutant helicases play an important role in both lower and upper motor-neuron diseases. Like the IGHMBP2 protein, SETX may function in mRNA biogenesis. Similarly, the strong homology that SETX shows with RENT1 (46% identity) suggests a role for SETX in nonsense-mediated RNA decay. We suggest that mutations in SETX may lead to the dysfunction of the helicase activity of this protein. It is conceivable that the abnormal SETX protein in ALS4 impairs the capacity of neurons to produce error-free mature mRNA, thus leading to neuronal degeneration.
Acknowledgments
The authors are grateful to the patients and family members for their cooperation in this study. This work was supported by National Institutes of Health grant NS42810 (The National Institute of Neurological Disorders and Stroke), the Muscular Dystrophy Association, and the Amyotrophic Lateral Sclerosis Association (United States); the Fund for Scientific Research, the University of Antwerp, the Medical Foundation Queen Elisabeth, the Association Belge contre les Maladies Neuro-Musculaires, and the Interuniversity Attraction Poles program P5/19 of the Belgian Federal Science Policy Office (Belgium); and the Austrian Science Fund (Austria). I.D. is supported by a Ph.D. fellowship of the Institute for Science and Technology, Belgium.
Appendix
Figure A1.
Human multiple-tissue northern-blot (Clontech) analysis. a, Northern-blot analysis of SETX mRNA. We performed northern-blot analysis on human 12-lane MTN blot (Clontech), with a SETX 3′ IMAGE clone insert (number 4136196) labeled with dCTP-[α-32P], and we identified two transcripts in all tissues at 11.5 kb and 9.0 kb. The lower panel represents the same blots hybridized with human β-actin. b, Northern-blot analysis of 10 μg of total RNA isolated from normal control lymphoblast cell lines. Probe derived from exon 10 of SETX was labeled with dCTP-[α-32P], which identified two transcripts of 11.5 kb and 9.0 kb.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for BLASTP)
- Ensembl Genome Browser, http://www.ensembl.org/Homo_sapiens/contigview?chr=9&vc_start=129656154&vc_end=131311785 (for BLAST searches)
- Entrez SNP, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp (for SNP database)
- European Bioinformatics Institute, http://www.ebi.ac.uk/interpro/scan.html (for InterproScan sequence search)
- Expressed Sequence Tags Database, http://www.ncbi.nlm.nih.gov/dbEST/index.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human genomic contig [accession number NT_035014], Human KIAA0625 [accession numbers NM_015046, BX538166, and BX537849) or SETX [accession number AY362728]; human RENT1 [accession number NM_002911], human IGHMBP2 [accession number NM_002180], yeast SEN-1 [accession number S53416], KIAA0625 rat EST [accession number XM_342400], and mouse Setx [accession number BK001523])
- HUGE Protein Database, http://www.kazusa.or.jp/huge/ (for KIAA0625/SETX)
- Inherited Peripheral Neuropathies Mutation Database, http://molgen-www.uia.ac.be/CMTMutations/ (for mutation database)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ALS1, ALS2, ALS4, AOA1, AOA2, SPG19, and CMT)
- Pfam, http://pfam.wustl.edu/index.html
- PSORT, http://psort.ims.u-tokyo.ac.jp/
- SMART, http://smart.embl-heidelberg.de/
- University of California–Santa Cruz (UCSC) Genome Bioinformatics, http://genome.cse.ucsc.edu/cgi-bin/hgTracks (for genome assembly and BLAST alignments on human July 2003 freeze)
References
- Blair IP, Bennett CL, Abel A, Rabin BA, Griffin JW, Fischbeck KH, Cornblath DR, Chance PF (2000) A gene for autosomal dominant juvenile amyotrophic lateral sclerosis (ALS4) localizes to a 500-kb interval on chromosome 9q34. Neurogenetics 3:1–6 [DOI] [PubMed] [Google Scholar]
- Campbell L, Hunter KM, Mohaghegh P, Tinsley JM, Brasch MA, Davies KE (2000) Direct interaction of SMN with dp103, a putative RNA helicase: a role for SMN in transcription regulation? Hum Mol Genet 9:1093–1100 10.1093/hmg/9.7.1093 [DOI] [PubMed] [Google Scholar]
- Chance PF, Rabin BA, Ryan SG, Ding Y, Scavina M, Crain B, Griffin JW, Cornblath DR (1998) Linkage of the gene for an autosomal dominant form of juvenile amyotrophic lateral sclerosis to chromosome 9q34. Am J Hum Genet 62:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S (2001) Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet 29:184–188 10.1038/ng1001-184 [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Auer-Grumbach M, Irobi J, Wagner K, Plecko B, Kennerson M, Zhu D, De Vriendt E, Van Gerwen V, Nicholson G, Hartung HP, Timmerman V (2002) Autosomal dominant juvenile amyotrophic lateral sclerosis and distal hereditary motor neuronopathy with pyramidal tract signs: synonyms for the same disorder? Brain 125:1320–1325 10.1093/brain/awf127 [DOI] [PubMed] [Google Scholar]
- Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C, Bertini E, Leonhardt-Horti H, Muntoni F, Ouvrier R, Pfeufer A, Rossi R, Van Maldergem L, Wilmshurst JM, Wienker TF, Sendtner M, Rudnik-Schoneborn S, Zerres K, Hubner C (2001) Mutations in the gene encoding immunoglobulin μ-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet 29:75–77 10.1038/ng703 [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 10.1038/ng1001-166 [DOI] [PubMed] [Google Scholar]
- Majoor-Krakauer D, Willems PJ, Hofman A (2003) Genetic epidemiology of amyotrophic lateral sclerosis. Clin Genet 63:83–101 [DOI] [PubMed] [Google Scholar]
- Marra G, D’Atri S, Yan H, Perrera C, Cannavo’ E, Vogelstein B, Jiricny J (2001) Phenotypic analysis of hMSH2 mutations in mouse cells carrying human chromosomes. Cancer Res 61:7719–7721 [PubMed] [Google Scholar]
- Meister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U (2000) Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal SM proteins. Hum Mol Genet 9:1977–1986 10.1093/hmg/9.13.1977 [DOI] [PubMed] [Google Scholar]
- Mendell JT, ap Rhys CM, Dietz HC (2002) Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298:419–422 10.1126/science.1074428 [DOI] [PubMed] [Google Scholar]
- Molnar GM, Crozat A, Kraeft SK, Dou QP, Chen LB, Pardee AB (1997) Association of the mammalian helicase MAH with the pre-mRNA splicing complex. PNAS 94:7831–7836 10.1073/pnas.94.15.7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M (2001) The gene mutated in ataxia-oculomotor apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet 29:189–193 10.1038/ng1001-189 [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Ber IL, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, Pandolfo M, Schulz JB, Pouget J, Calvas P, Shizuka-Ikeda M, Shoji M, Tanaka M, Izatt L, Shaw CE, M’Zahem A, Dunne E, Bomont P, Benhassine T, Bouslam N, Stevanin G, Brice A, Guimaraes J, Mendonca P, Barbot C, Coutinho P, Sequeiros J, Durr A, Warter JM, Koenig M (2004) Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 36:225–227 10.1038/ng1303 [DOI] [PubMed] [Google Scholar]
- Morrison KE, Harding AE (1994) Disorders of the motor neurone. Baillieres Clin Neurol 3:431–445 [PubMed] [Google Scholar]
- Myrianthopoulos NC, Lane MH, Silberberg DH, Vincent BL (1964) Nerve conduction and other studies in families with Charcot-Marie-Tooth disease. Brain 87:589–610 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G (2001) Functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol 152:75–85 10.1083/jcb.152.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin BA, Griffin JW, Crain BJ, Scavina M, Chance PF, Cornblath DR (1999) Autosomal dominant juvenile amyotrophic lateral sclerosis. Brain 122:1539–1550 10.1093/brain/122.8.1539 [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- Strong MJ, Hudson AJ, Alvord WG (1991) Familial amyotrophic lateral sclerosis, 1850–1989: a statistical analysis of the world literature. Can J Neurol Sci 18:45–58 [DOI] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE (1998) A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. PNAS 95:10009–10014 10.1073/pnas.95.17.10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8:251–262 10.1016/S1097-2765(01)00329-X [DOI] [PubMed] [Google Scholar]
- Valente EM, Brancati F, Ferraris A, Graham EA, Davis MB, Breteler MM, Gasser T, Bonifati V, Bentivoglio AR, De Michele G, Durr A, Cortelli P, Wassilowsky D, Harhangi BS, Rawal N, Caputo V, Filla A, Meco G, Oostra BA, Brice A, Albanese A, Dallapiccola B, Wood NW, European Consortium on Genetic Susceptibility in Parkinson’s Disease (2002) Novel locus for autosomal dominant pure hereditary spastic paraplegia (SPG19) maps to chromosome 9q33-q34. Ann Neurol 51:681–685 10.1002/ana.10204 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Sugai Y, Concannon P, Koenig M, Schmitt M, Sato M, Shizuka M, Mizushima K, Ikeda Y, Tomidokoro Y, Okamoto K, Shoji M (1998) Familial spinocerebellar ataxia with cerebellar atrophy, peripheral neuropathy, and elevated level of serum creatine kinase, γ-globulin, and α-fetoprotein. Ann Neurol 44:265–269 [DOI] [PubMed] [Google Scholar]
- Yan H, Papadopoulos N, Marra G, Perrera C, Jiricny J, Boland CR, Lynch HT, Chadwick RB, de la Chapelle A, Berg K, Eshleman JR, Yuan W, Markowitz S, Laken SJ, Lengauer C, Kinzler KW, Vogelstein B (2000) Conversion of diploidy to haploidy. Nature 403:723–724 10.1038/35001659 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 10.1038/ng1001-160 [DOI] [PubMed] [Google Scholar]