Differential Histone H3 Lys-9 and Lys-27 Methylation Profiles on the X Chromosome (original) (raw)
Abstract
Histone H3 tail modifications are among the earliest chromatin changes in the X-chromosome inactivation process. In this study we investigated the relative profiles of two important repressive marks on the X chromosome: methylation of H3 lysine 9 (K9) and 27 (K27). We found that both H3K9 dimethylation and K27 trimethylation characterize the inactive X in somatic cells and that their relative kinetics of enrichment on the X chromosome as it undergoes inactivation are similar. However, dynamic changes of H3K9 and H3K27 methylation on the inactivating X chromosome compared to the rest of the genome are distinct, suggesting that these two modifications play complementary and perhaps nonredundant roles in the establishment and/or maintenance of X inactivation. Furthermore, we show that a hotspot of H3K9 dimethylation 5′ to Xist also displays high levels of H3 tri-meK27. However, analysis of this region in G9a mutant embryonic stem cells shows that these two methyl marks are dependent on different histone methyltransferases.
X-chromosome inactivation is one of the most striking examples of epigenetic regulation since it acts at the level of an entire chromosome and involves the differential treatment of two identical chromosome homologues within the same nucleus (14). This process takes place during early embryonic development in the mouse, where imprinted inactivation of the paternal X is observed initially (32). This status is maintained in extraembryonic lineages but is reversed in the inner cell mass (16, 19) where, subsequently, random X inactivation takes place. Random X inactivation also occurs in female embryonic stem (ES) cells when they are induced to differentiate in vitro (23). ES cells thus represent a useful cell culture model system for X-inactivation studies. X inactivation is under the control of the Xist gene, which generates a long, untranslated RNA that coats in cis the chromosome from which it is transcribed (3, 5). Prior to X inactivation in undifferentiated ES cells, Xist is expressed at low levels from every X chromosome. The initiation of X inactivation involves the stabilization and accumulation of the Xist transcript on what will become the inactive X (Xi) and the silencing of Xist expression from the other allele, so that in female somatic cells, only the Xi will express Xist (20, 26). Upon stabilization, Xist RNA coats the X chromosome, rapidly inducing inactivation (within just one or two cell cycles) (35). The mechanism for _Xist_-mediated silencing is unknown, although it is likely to involve recruitment of silencing factors and the induction of a series of epigenetic changes to the chromosome. Such changes include a shift to late replication timing (33); hypoacetylation of histones H2A, H2B, H3, and H4 (2, 11); incorporation of the histone variant macro H2A (6); and DNA (CpG) methylation (18). More recently, histone H3 methylation of lysines 9 (K9) and 27 (K27) was also described as a characteristic of the Xi, as was hypomethylation of H3 lysine 4 (K4) (1, 9, 17, 22, 27). These changes are likely to be involved in transcriptional shutdown and/or in the clonal stability and heritability of the inactive state.
In a previous study, involving chromatin immunoprecipitation (ChIP) and immunofluorescence analysis, we showed that a domain 5′ to Xist displays very high constitutive levels of H3K9 dimethylation, even before the global enrichment of the inactive X chromosome for this mark (9). We proposed that this constitutive H3K9 methylation hotspot might act as a nucleation center that might participate in the formation and propagation of a ribonucleoprotein complex when Xist RNA becomes stabilized and thus in the transcriptional shutdown of the chromosome. More recently, it was reported that trimethylated H3 K27 also becomes globally enriched on the X chromosome (22, 27). However, the enrichment at the 5′ Xist hotspot was not investigated in these studies.
Although the enzyme responsible for the X chromosome H3K9 dimethylation has yet to be determined, H3K27 trimethylation on this chromosome is thought to be mediated by Enx1 in conjunction with Eed, both being members of the mouse polycomb group complex. Eed was originally described as affecting the maintenance of imprinted X inactivation (15, 34). More recently, Eed and Enx1 were shown to bind the paternal X chromosome early on during imprinted X inactivation (16, 19). Xist RNA-dependent binding of Eed and Enx1 to the X chromosome was also reported to occur during random X inactivation (22, 27). Intriguingly, the enrichment for Eed/Enx1 and for trimethylated H3K27 on the Xi appear to be transient and can be lost in cells differentiated over long time periods (22). Furthermore, a mutant Xist cDNA transgene that is unable to trigger silencing in male ES cells (in contrast to a full-length Xist cDNA transgene that is capable of inducing X inactivation) but can coat the X chromosome still induces the recruitment of Enx1-Eed and H3K27 trimethylation (22). This suggests that, during random X inactivation at least, the initial enrichment on the future inactive X for trimethylated H3K27 mediated by the Enx1/Eed complex may not be sufficient to induce silencing or to maintain it and an additional activity, normally recruited by wild-type Xist RNA might be required. Such an activity could for instance mediate H3K9 dimethylation.
In the present study, we have sought to compare the profiles of H3K9 dimethylation and H3K27 trimethylation on the X chromosome in somatic cells, as well as in ES cells, before and during the onset of X inactivation. The H3K9 and H3K27 residues are surrounded by very similar amino acid sequences that can lead to cross-reaction between epitopes by using some antibodies and has led to much confusion in the field concerning the specificities of various anti-H3-methyl K9 antibodies. It is therefore important to note that the anti-H3 dimethyl K9 used by Heard et al. (9) and in the present study shows no cross-reaction with methylated H3K27. We have reanalyzed the hotspot of H3K9 dimethylation 5′ to Xist in ES cells and report that the hotspot extends beyond the limits initially described and shows a bipartite nature. Although this region also shows the presence of high levels of trimethylated H3K27, the relative enrichment of this region for H3K27 methylation compared to the rest of the X chromosome is less than that found for H3K9 dimethylation. The distribution of H3K9 dimethylation in ES cells therefore appears more restricted than that of H3K27 methylation, with a marked specific enrichment of the region 5′ to Xist. Analysis of G9a mutant ES cells reveals a specific loss of H3K9 dimethylation in this region without affecting H3K27 trimethylation. Different histone methyltransferases therefore mediate these two marks in the hotspot region in undifferentiated ES cells and possibly during X inactivation as well. Indeed, although the kinetics of chromosomewide H3K9 and K27 methylation during the onset of X inactivation appear similar in wild-type female ES cells, we show that the distribution of these two histone tail modifications in female somatic cells differs markedly. Whereas in such cells H3K27 methylation is highly specific for the inactive X chromosome, H3K9 dimethylation appears widely distributed throughout the genome, with the enrichment of promoter regions of X-linked genes being the only inactive X-specific association. The distinct profiles of H3K9 and H3K27 methylation on the inactivating X compared to the rest of the genome both before and during X inactivation suggest that these two modifications are not simply redundant but rather play complementary roles in the establishment and/or maintenance of X inactivation.
MATERIALS AND METHODS
Cell culture.
Female mouse embryonic fibroblasts, prepared from 13.5-day-old embryos were cultured in Dulbecco modified Eagle medium (DMEM) with GlutMAX (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) (10). Both the wild-type and the mutant G9a male ES cell lines have been described previously (31, 36). For X-inactivation kinetics studies, the female ES cell line LF2 (a gift from Austin Smith) grown on gelatin-coated flasks or plates was used. ES cells were maintained in an undifferentiated state in DMEM with GlutMAX, 15% fetal calf serum (Gibco), 10−4 mM 2-mercaptoethanol (Sigma), and 1,000 U of leukemia inhibitory factor (LIF) (Chemicon)/ml. Differentiation of ES cells can be induced by removing LIF and by using 100 nM all-trans-retinoic acid (Sigma) in DMEM supplemented with 10% fetal bovine serum and 10−4 mM 2-mercaptoethanol (28). The differentiation medium was changed daily. All cells were grown at 37°C in 8% CO2.
Antibody specificity.
In a previous report (9), we used both a highly sensitive serum from a first bleed of a rabbit (provided by C. David Allis), and a commercially available H3 di-meK9 antibody (Upstate Biotechnology, Lake Placid, N.Y.) with similar results. Although the H3 di-meK9 “first bleed” serum was similar in its specificity to the affinity-purified commercially available antibody (J. Rice and C. D. Allis, unpublished data), the efficiency of this serum for both immunofluorescence and ChIP experiments was superior. All of the ChIP data presented here were generated by using the commercially available H3 di-meK9 antibody (see below for batch numbers) from Upstate Biotechnology since the first bleed serum was no longer available. Dot blot tests and peptide competitions for the Upstate antibody shows that it is specific for H3 di-meK9 (see Fig. S1 and S2 in the supplemental material). Importantly, this antibody shows no cross-reaction with H3 di- or trimethylation of K27.
ChIP.
Immunoprecipitations of DNA associated with methylated histone H3 were performed as described previously (9). Briefly, cells were cross-linked with 1% formaldehyde for 15 min at room temperature. Isolated nuclei were sonicated to an average length of 300 to 1,000 bp. We used 5 μl of dimethylated H3 K9 antibody (batch numbers 23424, 27271, and 23032; Upstate Biotechnology) and 1 μl of di- or trimethylated H3K27 antibody (7B11) (25).
Quantitative analysis by real-time PCR.
Immunoprecipitated DNA and input DNA were analyzed by real-time PCR with SYBR Green Universal Mix and an ABI Prism 7700 (Perkin-Elmer Applied Biosystems). Each PCR was run in triplicate to control for PCR variation. To standardize each experiment, the results are presented as a percentage of immunoprecipitation, calculated by dividing the average value of the immunoprecipitation by the average value of the corresponding input, with both values being first normalized by the dilution factor. Each experiment was repeated on independent chromatin preparations two to three times.
Immunofluorescence and RNA FISH analysis.
Immunofluorescence combined with RNA fluorescence in situ hybridization (FISH) was performed as described previously (4). We used rabbit polyclonal H3 di-meK9 antibody at 1/300 (Upstate Biotechnology; “first bleed serum”), mouse monoclonal H3 di/tri-meK27 antibody at 1/100 (7B11), and rabbit polyclonal H3 di-meK27 antibody at 1/300 (25). A Leica DMR fluorescence microscope with a Cool SNAP fx camera (Photometrics) and Metamorph software (Roper) was used for image acquisition.
RESULTS AND DISCUSSION
H3K9 and H3K27 methylation profiles in undifferentiated female ES cells.
Our previous analysis of H3K9 dimethylation in undifferentiated ES cells revealed the existence of a hotspot of H3K9 methylation in the vicinity of the Xist gene (9). This was initially defined as a 100- to 150-kb region, extending upstream of a region ∼7 kb 5′ to Xist. In order to characterize this hotspot further, we extended our systematic ChIP analysis in undifferentiated female ES cells to additional regions of the X chromosome by using an antibody highly specific for dimethylated H3K9 (Upstate Biotechnology; see Materials and Methods and Fig. S1 and S2 in the supplemental material). The level of H3 dimethyl K9 enrichment at specific sequences was determined by quantitative real-time PCR with primer pairs across the regions of interest.
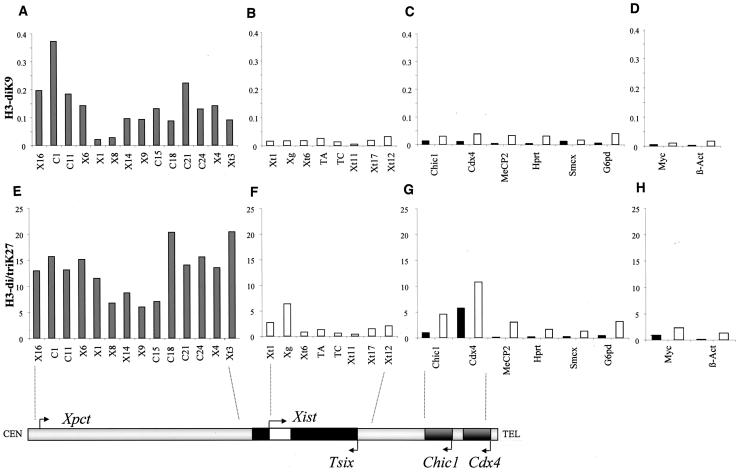
ChIP analysis revealed, in confirmation of our earlier results, that the ∼120-kb region upstream of Xist (between positions Xt3 and X14) was highly enriched for histone H3 methylated at K9, with maximum levels of enrichment ca. 11-fold greater than those seen at the Xist P1 promoter (position Xt1, Fig. 1A and B). This domain is flanked, on its centromere-proximal side, by a region (positions X8 and X1) showing basal, very low levels of H3K9 methylation that led to the postulate that this region was the upstream boundary of the hotspot. However, as we have extended our analysis further upstream, levels of H3K9 methylation were found to increase again and to reach values similar to or even higher than those seen in the proximal part of the hotspot (i.e., closest to Xist). The hotspot therefore spans a large domain extending over 340 kb 5′ to Xist and seems to have a bipartite structure, organized around a 30-kb central domain showing low H3K9 dimethylation and high H3K4 dimethylation (see below). When we extended our analysis to the Xist gene and its 3′ region, we observed only very low levels of H3K9 dimethylation (Fig. 1B), similar to those found for other X-linked genes (Fig. 1C), whether within the X inactivation center region (Chic1 and Cdx4) or elsewhere on the X chromosome (MeCP2, Hprt, Smcx, and G6pd) and for autosomal genes (Fig. 1D). The hotspot of H3K9 dimethylation we describe in the region 5′ to Xist appears therefore to be the only domain of the X chromosome to display such high levels of H3K9 methylation.
FIG. 1.
ChIP analysis of H3 lysines 9 and 27 methylation in female ES cells. Sonicated chromatin was immunoprecipitated using antibodies specific for dimethylated H3K9 (A to D) and di/trimethylated H3K27 (E to H). The immunoprecipitated DNA was analyzed by quantitative real-time PCR with primers distributed across a 350-kb region 5′ to Xist (A and E), across the Xist/Tsix region (B and F), in promoters (▪) and exons (□) of X-linked genes (C and G), and of autosomal genes (D and H). The graphs represent the percentage of immunoprecipitation obtained for each position tested. The differences in scale obtained by using the anti-dimethyl K9 and the anti-di/trimethyl K27 most likely correspond to differences in both the degree of enrichment for the modified histones, as well as in the efficiency of each antibody to immunoprecipitate chromatin. A partial map of the X inactivation center (Xic) is represented below the graphs to schematize the relative positions of the Xic primers used. The map ends after Chic1 and Cdx4, since the other genes tested map outside the Xic.
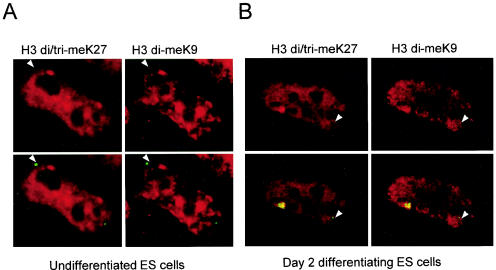
Since H3K27 trimethylation is, like H3K9 dimethylation, an early mark of the X chromosome as it inactivates (22, 27), we wondered whether such a modification would also be present in the region 5′ to Xist. We used an antibody recognizing trimethylated H3K27 which, although cross-reacting with dimethylated H3K27, shows no specificity for H3K9 di- or trimethylation (see below and Fig. S2 in the supplemental material). Systematic ChIP revealed an enrichment for H3K27 methylation with this antibody in the region 5′ to Xist (Fig. 1E). However, the overall H3K27 methylation profile in this domain and on the X chromosome in general differs from that of H3K9 dimethylation. First, the central region of the bipartite hotspot, which shows low H3K9 methylation (positions X1 and X8) does not appear to be similarly depleted for H3K27 methylation compared to neighboring sequences. Second, in other regions of Xic, notably within Xist (positions Xt1 and Xg, Fig. 1F) and Cdx4 (Fig. 1G), H3K27 methylation reaches levels that are comparable to those of the hotspot. This broader distribution of H3K27 methylation across the Xic region probably explains why immunofluorescence detection of the hotspot was only possible with the anti-H3 di-meK9 antibody and never with anti-H3 di/tri-meK27 (Fig. 2) or H3 di-meK27 antibodies (data not shown). In summary, prior to X inactivation the 340-kb region 5′ of Xist is characterized by high levels of H3K9 dimethylation and H3 K27 di/trimethylation. H3K9 dimethylation appears to be restricted to this hotspot region, whereas methylated H3K27 seems to be more widely distributed within the Xic and elsewhere on the X chromosome.
FIG. 2.
Simultaneous analysis of the H3K9 and H3K27 methylation enrichment in the vicinity of the Xist gene in ES cells. Representative nuclei of undifferentiated or differentiating ES cells hybridized with Xist RNA (green) and double stained with antibodies to H3 di-meK9, detected by using goat anti-rabbit Alexa 568 secondary antibody (Molecular Probes) (pseudocolored red, left), and H3 tri-meK27, detected by using goat anti-mouse Alexa 680 secondary antibody (Molecular Probes) (pseudocolored red, right). Arrowheads indicate the position of the unstable Xist/Tsix primary transcript and an accompanying hotspot of H3 di-meK9 but a lack of such a hotspot of H3 tri-meK27 in both undifferentiated and differentiating ES cells. The arrow indicates the accumulation of Xist RNA accompanied by enrichment of both H3 di-meK9 and H3 tri-meK27 in differentiating ES cells. It should be noted that the peptide competitions shown in Fig. S2 in the supplemental material are also applicable to the data shown here.
We have previously suggested that the incapacity of large single copy _Xist_-containing yeast artificial chromosome (YAC) transgenes to induce X inactivation, which correlates with their inability to retain or spread H3K9 methylation, was due to the fact that they lacked part of this hotspot (9). Since these transgenes only contain 100 kb of sequence upstream of Xist, the present ChIP analysis, which revealed that the hotspot covers at least a 340-kb region, reinforces this hypothesis. The juxtaposition of two or more copies of a “partial” hotspot region present in the YACs used could reconstitute a functional domain and explain the capacity of transgenes in multicopy arrays to function as ectopic _Xic_s. Interestingly, unlike multicopy transgenes, single-copy transgenes cannot recruit Eed/Enx1 either in differentiating ES cells (E. Heard, unpublished results). The exact role of this region and the proteins that bind to it are currently under investigation.
Analysis of H3K9 and K27 methylation within the hotspot 5′ to Xist in G9a mutant ES cells.
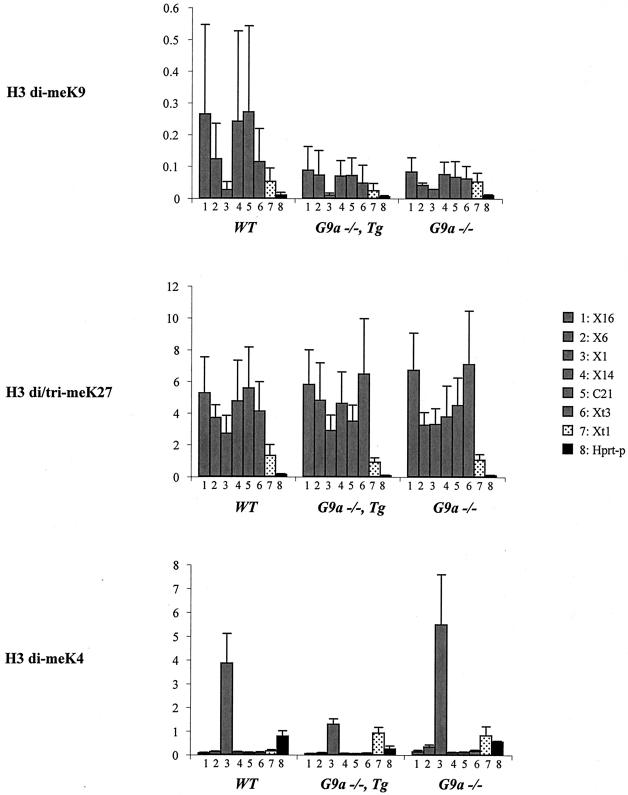
To gain insight into whether the H3K9 and H3K27 methylation marks are mechanistically linked, we evaluated the profiles of these modifications within the region 5′ to Xist in ES cells mutant for the G9a histone methyltransferase. The G9a enzyme is able to methylate H3K9 and, to a lesser extent H3K27, at least in vitro (30). Although in vivo studies suggest that G9a is only responsible for H3K9 dimethylation preferentially in euchromatic regions (21, 24, 31), it remains possible that this enzyme might show different activities in specific regions, such as the hotspot 5′ to Xist. Levels of H3 di-meK9, H3 tri-meK27 and, as a control, H3 di-meK4 across the hotspot region were examined by using ChIP in G9a mutant ES cells. H3K4 dimethylation is generally thought to show an opposite pattern to H3K9 methylation, as illustrated in the central domain of the hotspot region, which displays low H3K9 and high H3K4 methylation levels (position X1, Fig. 3). Given that the hotspot is present in both male and female ES cells (9; unpublished results), we exploited three available male ES cell lines: a double-null G9a mutant (G9a−/−) (31), a G9a double-null carrying a G9a transgene (G9a−/− Tg) (36), and the parental line (WT). In the G9a−/− line, the high levels of H3K9 dimethylation within the hotspot region decreased substantially. The low H3K9 methylation levels in other regions of the X chromosome, such as the promoters of Xist (Xt1) and Hprt (Hprt-p), did not appear to be affected (Fig. 3). Loss of high levels of H3K9 dimethylation at the hotspot and throughout the nucleus was also observed by immunofluorescence (Fig. S3). In contrast, H3K27 methylation levels were highly similar at all positions tested by ChIP (Fig. 3) and more globally by immunofluorescence (Fig. S3 in the supplemental material) between WT and G9a−/− lines. Our ChIP results demonstrate that G9a is responsible for the presence of high levels of H3K9 dimethylation but not H3K27 trimethylation in the hotspot region. Further, the loss of H3K9 dimethylation in G9a mutant cells does not affect the maintenance of H3K27 methylation in this region. The fact that levels of H3K4 dimethylation were either unchanged or slightly raised at some positions in the G9a−/− mutant compared to WT further underscores the significance of the drop in H3K9 dimethylation we observed in G9a mutants. Interestingly, the addition of a functional G9a transgene (G9a−/− Tg), although restoring global levels of H3 di-meK9 in the nucleus (Fig. S3 in the supplemental material), did not restore H3 di-meK9 enrichment in the hotspot region (Fig. 3). Thus, although G9a is clearly responsible for maintaining the high level of H3K9 methylation in the hotspot, it may not be involved in its establishment. Alternatively, early developmental factors responsible for targeting G9a to the hotspot may no longer be present in ES cells.
FIG. 3.
Analysis of H3K9, H3K27, and H3K4 methylation profiles in the region 5′ to Xist in G9a mutant ES cell lines. Three male cell lines were used in the present study, a double-null G9a (G9a−/−) mutant (31), a double-null G9a carrying a G9a transgene (G9a−/− Tg) (36), and the parental line (WT). The results of three experiments carried out on independent chromatin preparations are shown. Three different batches of H3 di-meK9 antibody with variable ChIP efficiencies were used. The low efficiency of some of the batches (batches 27271 and 23032) compared to others (batch 23424) explains the high standard deviations seen in the WT cell line. The same batches of H3 di/tri-meK27 (7B11) and H3 di-meK4 (24747) were used throughout. Six positions within the hotspot of H3K9 methylation were analyzed (░⃞), as well as the promoters of Xist (Xt1) and Hprt (Hprt-p).
Comparative kinetics of accumulation of dimethylated H3K9 and trimethylated H3K27 during the onset of X inactivation.
The early enrichment (soon after Xist RNA coating) for H3 di-meK9 (9, 17) and H3 tri-meK27 (22, 27) on the X chromosome have been reported independently but have not thus far been investigated in the same cells. Such comparative analysis could provide insight into whether these marks are simultaneously or independently deposited during X inactivation. In order to address this issue, we performed dual immunofluorescence staining with the rabbit polyclonal H3 di-meK9 and mouse monoclonal H3 di/tri-meK27 antibodies, together with Xist RNA FISH, on differentiating female ES cells from day 0 to day 9 (Fig. 4). Since the use of an anti-H3 di-meK27 antibody shows that H3K27 dimethylation is excluded from the inactive X-chromosome somatic cells (see Fig. 6) and in differentiating ES cells, this indicates that the anti-H3 di/tri-meK27 antibody only detects H3K27 trimethylation enrichment on the inactive X chromosome, a finding consistent with a previous report (22). We found that for both H3K9 and K27 methylation, enrichment on the Xist RNA-coated X chromosome occurs very early, from day 1 onward (Fig. 4). H3K9 dimethylation first appears on the X chromosome at the same time point as H3 tri-meK27. In no case did we find cells that were enriched in H3 di-meK9 that were not enriched in H3 tri-meK27, although the inverse was true. Thus, H3K27 trimethylation may precede H3K9 dimethylation. On the other hand, the proportion of Xist RNA-coated X chromosomes showing H3K9 methylation was lower than for H3K27 methylation at all stages, and we verified that this was not due to a cell cycle effect (J. Chaumeil and E. Heard, data not shown). Thus, the detection of the H3K9 dimethyl mark appears to be less robust than that of the H3K27 trimethylation mark. It is therefore equally possible that these two marks occur simultaneously during X inactivation. In agreement with this, by using ChIP we found no significant difference in the kinetics of appearance of these marks at the promoters of X-linked genes during female ES cell differentiation (data not shown). At later stages of differentiation (from day 7), we also observed a reduction in H3 tri-meK27 levels on the Xist RNA-coated X chromosome, as reported by Plath et al. (22). There is a similar decrease in the proportion of cells with an Xi enriched in H3 di-meK9 (in our previous study we did not look beyond day 7). However, this reduced number of positive cells may be due to the cellular heterogeneity inherent in ES cell differentiation and the appearance of certain cell types, with a potentially higher degree of differentiation, in which these marks are no longer present. Consistent with this, we found that there is a substantial degree of heterogeneity between cells at both the nuclear and the X chromosomal levels of H3K27 and K9 methylation in fibroblasts from adult (8-week-old) mice compared to embryonic fibroblasts at 13.5 days postcoitus (dpc) (see Fig. 6 and discussion below).
FIG. 4.
Kinetics of histone H3K27 and H3K9 methylation on the Xist RNA-coated X chromosome during the differentiation of female ES cells. (A) Representative immunofluorescence for histone H3 trimethyl K27 (H3 di/tri-meK27, columns 2 and 3) and dimethyl K9 (H3 di-meK9, columns 4 and 5) combined with Xist RNA FISH. Immunodetections with goat anti-mouse Alexa 680- and goat anti-rabbit Alexa 568-conjugated secondary antibodies (in red, columns 2 and 4), were combined with Xist RNA FISH (Spectrum green-labeled probe, green, columns 3 and 5). The first row shows a typical example of a nucleus with an enrichment only for H3 di/tri-meK27 on the Xist RNA domain (column 3, arrowhead; yellow indicates the overlap of green and red signals). The second row shows an example of enrichment for both H3 di/tri-meK27 (arrowheads) and di-meK9 (arrows) on the Xist RNA domain (columns 3 and 5, yellow). DNA is stained with DAPI (blue, column 1). The bar represents 5 μm in each case. (B) Relative kinetics of H3K9 and H3K27 methylation on the X chromosome undergoing inactivation in female ES cells during differentiation (9 days of differentiation of female ES cells). Between 50 and 200 cells were counted for each time point.
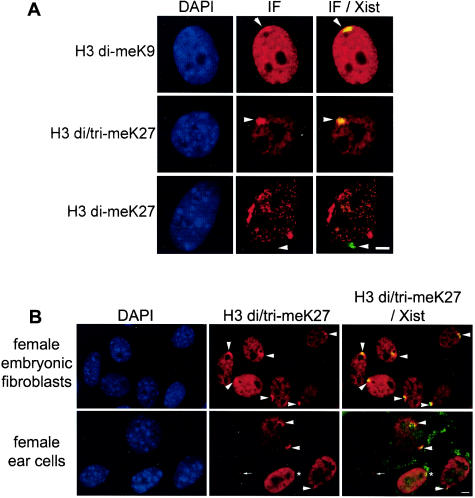
FIG. 6.
Histone H3 dimethylation at K9 and di- and trimethylation at K27 on the inactive X chromosome in female fibroblasts. (A) Representative immunofluorescence for histone H3 modifications in mouse embryonic (13.5 dpc) fibroblasts. The histone H3 modifications (column 2), H3 di-meK9 (row 1), H3 di/tri-meK27 (row 2), and H3 di-meK27 (row 3) are shown combined with Xist RNA FISH data (column 3). Green coloration indicates the absence of the modification on the Xist RNA-coated X chromosome; yellow coloration (overlap of green and red signals) shows the enrichment of the modification on the Xist domain (arrowheads). DNA is stained with DAPI (blue, column 1). (B) Representative immunofluorescence for histone H3 modifications in ear fibroblasts from an 8-week-old female. Note the absence of H3K27 methylation enrichment on the Xist RNA-coated X chromosome (asterisk) and the variable nuclear levels of H3K27 methylation in some nuclei. The bar represents 5 μm in each case.
In conclusion, we found that, although H3 tri-meK27 may precede H3 di-meK9, both of these marks show similar early kinetics of global enrichment on the X chromosome undergoing random inactivation, suggesting that the deposition of these two modifications may be linked during random X inactivation. This could imply that the same enzyme is responsible for both H3K27 and H3K9 methylation on the X chromosome. Indeed, the histone methyltransferase responsible for the H3K27 methylation mark on the X chromosome, Enx1/Ezh2, also has a methyltransferase activity toward the K9 residue of H3 (7, 13). Alternatively, H3K9 methylation on the X chromosome could be attributable to a distinct histone methyltransferase, such as G9a, which could act either synergistically or independently from Enx1. Given our finding that in the hotspot region methylation of H3K9 (and not of H3K27) is dependent on G9a, this histone methyltransferase may also be responsible for X-chromosome-wide H3K9 dimethylation induced during X inactivation.
H3K9 and H3K27 methylation profiles in female somatic cells.
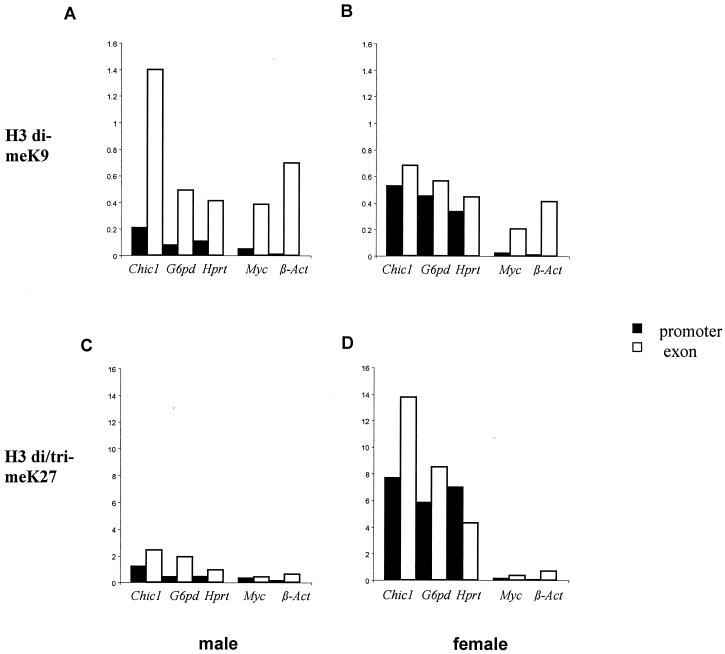
We have previously reported that the promoters of X-linked genes were enriched for dimethylated H3K9 on the inactive X chromosome (9). Analysis of two additional X-linked genes (Chic1 and Hprt) confirmed this observation, since the enrichment could be detected only in female and not in male embryonic fibroblasts (Fig. 5). H3K9 methylation of promoters is therefore associated with transcriptional silencing of the inactive X chromosome. In contrast to the promoter regions, H3K9 dimethylation in exonic regions was high in both males and females, suggesting that it is associated with both the active and the inactive X chromosomes (Fig. 5). Furthermore, H3K9 dimethylation was elevated in the exonic regions of autosomal genes (Myc and β-actin). This demonstrates that the presence of dimethylated H3K9 within exons is not specific to genes on the inactive X chromosome and does not necessarily impair transcription.
FIG. 5.
ChIP analysis of H3 lysines 9 and 27 methylation in 13.5 dpc mouse embryonic fibroblasts. The levels of H3K9 dimethylation (A and B) and H3K27 di/trimethylation (C and D) within several X-linked (Chic1, G6pd, and Hprt) and autosomal (Myc and β-actin) genes were analyzed in both male (A and C) and female (B and D) fibroblasts. Two positions were analyzed for each gene: one in the promoter (▪) and one in an exon (□). Promoter-specific primers were designed to span the transcription initiation site.
Methylation of H3K27 was also examined by using immuno-FISH and ChIP in female embryonic fibroblasts. The inactive X chromosome stains strongly with the anti-di/tri-meH3K27 antibody in 100% of these cells (Fig. 4 and 6 ), whereas the overall levels of signal in the nucleus appear to be lower than those detected in ES cells. The exclusion of the H3 dimethyl K27 mark from the inactive X chromosome seen in 100% of cells (Fig. 6) suggested that the specific enrichment detected by the H3 di/tri-meK27 antibody on the inactive X chromosome was for the H3K27 trimethyl mark only.
ChIP analysis confirmed the high enrichment for methylated H3K27 on the inactive X (Fig. 5C and D). In contrast to H3K9 methylation, the inactive X-specific H3K27 methylation concerns both promoters and exons of X-linked genes, as well an intergenic region tested in the vicinity of the G6pd gene (data not shown). Importantly, all of the other regions tested, whether on the active X chromosome (see male fibroblasts, Fig. 5C) or on the autosomes, display low levels of H3K27 methylation that are similar to or even lower than those of ES cells (compare the percentages of IP for Chic1, G6pd, Hprt, Myc, and β-actin in fibroblasts (Fig. 5C) and ES cells (Fig. 1G). This is in agreement with the reduced nuclear staining in fibroblasts compared to ES cells observed by immuno-FISH (see above and Fig. 6). In somatic cells therefore, H3K27 methylation appears to be highly specific for the inactive X chromosome. In contrast, comparison of both immunofluorescence (Fig. 2 and 6) and ChIP (Fig. 1A to D and 5) data between ES cells and fibroblasts suggests that there is a global, genomewide elevation of dimethylated H3K9 upon differentiation. We observed, for instance, in promoter regions of active X-linked genes, a 10- to 20-fold increase in the percentage of immunoprecipitation of methylated H3K9 between ES cells and male fibroblasts (Fig. 1C and 5A). The inactive X chromosome in female fibroblasts is characterized by a further specific increase of H3K9 methylation levels in promoter regions (Fig. 5A) and possibly other intergenic regions (data not shown).
In conclusion, the chromatin of the inactive X is characterized by the presence of methylated H3K9 and H3K27. The unique combination of H3 di-meK9 and H3 tri-meK27 on the inactive X chromosome implies that these two methyl marks could be required together for the establishment and the maintenance of the inactive state. In this context, two hypotheses can be envisioned. In the first, the roles of H3K9 and H3K27 methylation on the X chromosome would be redundant, and their combined presence could reinforce the efficiency of the silencing and contribute to the “locking in” of the inactive state, together with other epigenetic marks such as DNA methylation. In an alternative model, these two modifications could be complementary, which implies that methylating H3K9 alone or H3K27 alone would be insufficient to ensure X inactivation. Several observations support the latter hypothesis. First, the incapacity of a mutant Xist cDNA transgene to induce silencing in male ES cells, despite its ability to recruit Eed/Enx1 and H3K27 methylation, suggests that H3K27 methylation may not be sufficient to trigger X inactivation (22), although it should be noted that another Xist mutant, as reported by Silva et al., was unable to induce either silencing or Enx1 recruitment (27). Second, we observe that H3K9 and K27 methylation profiles on the X chromosome, and in the genome in general, evolve very differently between ES and somatic cells. H3K9 methylation, for example, is more restricted in ES cells than in somatic cells, whereas the opposite is true for H3K27 methylation. These findings strongly suggest that these two modifications have distinct functions across the genome during cellular differentiation.
In the case of X inactivation, the combined enrichment for methylated H3K9 and K27 at promoters of X-linked genes could be required to target specific proteins or complexes to the inactive X chromosome. This would be consistent with the notion of a histone code, whereby combinations of modifications are used to establish specific states of activity within the genome (29). The silencing induced by these two histone tail modifications could be mediated by distinct and specific chromodomain-containing proteins that are able to discriminate between both methyl-lysine marks (8). Alternatively, inactivation of the X chromosome could involve X-chromosome-specific silencing proteins with domains that would specifically target the combination of H3K9 and H3K27 methylation marks. The latter possibility appears all the more likely as neither HP1 (9) nor members of Polycomb group family (15, 27) have thus far been found to be enriched on the inactive X chromosome during ES cell differentiation. Finally, the combined presence of H3K9 and H3K27 methylation might also be necessary to recruit additional activities, such as DNA methylation, that contribute to the stabilization of the inactive state. DNA methylation has been shown to be present at the promoters of X-linked genes in somatic cells and to be a late event in the X-inactivation process during ES cell differentiation (12). In this context, it will be interesting to examine H3K9 and H3K27 methylation at the promoters of marsupial X-linked genes where DNA methylation is not a feature of the inactive X chromosome.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank M. Guggiari and S. Pichard for technical assistance; members of the UMR 218, particularly G. Almouzni and C. Maison, for discussions and advice; and Yoichi Shinkai for the generous gift of G9a mutant ES cells.
This study was supported by the CNRS, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Medicale, a Program Incitatif et Collaboratif from the Curie Institute to E.H., and an ACI Biologie Moleculaire et Structurale to P.A. J.C. is funded by a Ph.D. grant from the Ministère de la Recherche et de la Technologie.
Footnotes
REFERENCES
- 1.**Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis.2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:**73-76. [DOI] [PubMed] [Google Scholar]
- 2.**Boggs, B. A., B. Connors, R. E. Sobel, A. C. Chinault, and C. D. Allis.1996. Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma 105:**303-309. [DOI] [PubMed] [Google Scholar]
- 3.**Brown, C. J., B. D. Hendrich, J. L. Rupert, R. G. Lafrenière, Y. Xing, R. G. Lawrence, and H. F. Willard.1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71:**527-542. [DOI] [PubMed] [Google Scholar]
- 4.**Chaumeil, J., I. Okamoto, M. Guggiari, and E. Heard.2002. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet. Cell Genet. 99:**75-84. [DOI] [PubMed] [Google Scholar]
- 5.**Clemson, C. M., J. A. Mc Neil, H. F. Willard, and J. B. Lawrence.1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132:**259-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.**Costanzi, C., P. Stein, D. M. Worrad, R. M. Schultz, and J. R. Pehrson.2000. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development 127:**2283-2289. [DOI] [PubMed] [Google Scholar]
- 7.**Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta.2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:**85-96. [DOI] [PubMed] [Google Scholar]
- 8.**Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh.2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 bt Polycomb and HP1 chromodomains. Genes Dev. 17:**1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.**Heard, E., C. Rougeulle, D. Arnaud, P. Avner, C. D. Allis, and D. L. Spector.2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:**727-738. [DOI] [PubMed] [Google Scholar]
- 10.**Hogan, B., R. Beddington, F. Constantini, and E. Lacy.**1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.**Jeppesen, P., and B. M. Turner.1993. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74:**281-289. [DOI] [PubMed] [Google Scholar]
- 12.**Keohane, A. M., L. P. O'Neil, N. D. Belyaev, J. S. Lavender, and B. M. Turner.1996. X inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol. 180:**618-630. [DOI] [PubMed] [Google Scholar]
- 13.**Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg.2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:**2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.**Lyon, M. F.1961. Gene action in the X chromosome of the mouse (Mus musculus L.). Nature 190:**372-373. [DOI] [PubMed] [Google Scholar]
- 15.**Mak, W., J. Baxter, J. Silva, A. E. Newall, A. P. Otte, and N. Brockdorff.2002. Mitotically stable association of polycomb group proteins Eed and Enx1 with the inactive X chromosome in trophoblast stem cells. Curr. Biol. 12:**1016-1020. [DOI] [PubMed] [Google Scholar]
- 16.**Mak, W., T. B. Nesterova, M. de Napoles, R. Appanah, S. Yamanaka, A. P. Otte, and N. Brockdorff.2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303:**666-669. [DOI] [PubMed] [Google Scholar]
- 17.**Mermoud, J. E., B. Popova, A. H. Peters, T. Jenuwein, and N. Brockdorff.2002. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr. Biol. 12:**247-251. [DOI] [PubMed] [Google Scholar]
- 18.**Norris, D. P., N. H. Brockdorff, and S. Rastan.1991. Methylation status of CpG-rich bands on active and inactive mouse X chromosomes. Mamm. Genome 1:**78-83. [DOI] [PubMed] [Google Scholar]
- 19.**Okamoto, I., A. P. Otte, C. D. Allis, D. Reinberg, and E. Heard.2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303:**644-649. [DOI] [PubMed] [Google Scholar]
- 20.**Panning, B., J. Dausman, and R. Jaenisch.1997. X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90:**907-916. [DOI] [PubMed] [Google Scholar]
- 21.**Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein.2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:**1577-1589. [DOI] [PubMed] [Google Scholar]
- 22.**Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang.2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:**131-135. [DOI] [PubMed] [Google Scholar]
- 23.**Rastan, S., and E. J. Robertson.1985. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J. Embryol. Exp. Morph. 90:**379-388. [PubMed] [Google Scholar]
- 24.**Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz, D. F. Hunt, Y. Shinkai, and C. D. Allis.2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12:**1591-1598. [DOI] [PubMed] [Google Scholar]
- 25.**Sarma, K., K. Nishioka, and D. Reinberg.2004. Tips in analyzing antibodies directed against specific histone tail modifications. Methods Enzymol. 376:**255-269. [DOI] [PubMed] [Google Scholar]
- 26.**Sheardown, S. A., S. M. Duthie, C. M. Johnston, A. E. Newall, E. J. Formstone, R. M. Arkell, T. B. Nesterova, G. C. Alghisi, S. Rastan, and N. Brockdorff.1997. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91:**99-107. [DOI] [PubMed] [Google Scholar]
- 27.**Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff.2003. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complex. Dev. Cell 4:**481-495. [DOI] [PubMed] [Google Scholar]
- 28.**Smith, A. G.1991. Culture and differentiation of embryonic stem cells. J. Tiss. Cult. Methods 13:**89-94. [Google Scholar]
- 29.**Strahl, B. D., and D. C. Allis.2000. The language of covalent histone modifications. Nature 403:**41-45. [DOI] [PubMed] [Google Scholar]
- 30.**Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai.2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:**25309-25317. [DOI] [PubMed] [Google Scholar]
- 31.**Tachibana, M., K. Sugimoto, M. Nozaki, J. Ueda, T. Ohta, M. Ohki, M. Fukuda, N. Takeda, H. Niida, H. Kato, and Y. Shinkai.2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16:**1779-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.**Takagi, N., and M. Sasaki.1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256:**640-642. [DOI] [PubMed] [Google Scholar]
- 33.**Takagi, N., O. Sugawara, and M. Sasaki.1982. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma 85:**275-286. [DOI] [PubMed] [Google Scholar]
- 34.**Wang, J., J. Mager, Y. Chen, E. Schneider, J. C. Cross, A. Nagy, and T. Magnuson.2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28:**371-375. [DOI] [PubMed] [Google Scholar]
- 35.**Wutz, A., and R. Jaenisch.2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5:**695-705. [DOI] [PubMed] [Google Scholar]
- 36.**Xin, Z., M. Tachibana, M. Guggiari, E. Heard, Y. Shinkai, and J. Wagstaff.2003. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 278:**14996-15000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]