A Dominant Negative Mutant of Cyclin-Dependent Kinase A Reduces Endoreduplication but Not Cell Size or Gene Expression in Maize Endosperm (original) (raw)
Abstract
Cells in maize (Zea mays) endosperm undergo multiple cycles of endoreduplication, with some attaining DNA contents as high as 96C and 192C. Genome amplification begins around 10 d after pollination, coincident with cell enlargement and the onset of starch and storage protein accumulation. Although the role of endoreduplication is unclear, it is thought to provide a mechanism that increases cell size and enhances gene expression. To investigate this process, we reduced endoreduplication in transgenic maize endosperm by ectopically expressing a gene encoding a dominant negative mutant form of cyclin-dependent kinase A. This gene was regulated by the 27-kD γ-zein promoter, which restricted synthesis of the defective enzyme to the endoreduplication rather than the mitotic phase of endosperm development. Overexpression of a wild-type cyclin-dependent kinase A increased enzyme activity but had no effect on endoreduplication. By contrast, ectopic expression of the defective enzyme lowered kinase activity and reduced by half the mean C-value and total DNA content of endosperm nuclei. The lower level of endoreduplication did not affect cell size and only slightly reduced starch and storage protein accumulation. There was little difference in the level of endosperm gene expression with high and low levels of endoreduplication, suggesting that this process may not enhance transcription of genes associated with starch and storage protein synthesis.
INTRODUCTION
Endoreduplication is a form of polyploidization that occurs in somatic cells of many eukaryotes (Brodsky and Uryvaeva, 1977). It is the result of a cell cycle in which part or all of the genome is replicated without subsequent sister chromatid separation, nuclear division, and cytokinesis (Edgar and Orr-Weaver, 2001). In animals, endoreduplication occurs in a variety of cells and tissues, where it is thought to be related to differentiation, cell size, and the level of gene expression (White, 1973; Zimmet and Ravid, 2000). In plants, endoreduplication occurs in cells of vegetative tissues (Traas et al., 1998; Cebolla et al., 1999) as well those related to flowering (Kudo and Kimura, 2002), embryogenesis (Nagl, 1974; Lemontey et al., 2000), endosperm development (Kowles and Phillips, 1985), and fruit formation (Joubès et al., 1999). Despite its widespread occurrence in plants, the function of endoreduplication and mechanism(s) by which it occurs remain poorly understood. Based on its temporal relationship with cell expansion and increased metabolic activity, endoreduplication is generally thought to provide a mechanism for increasing cell size and gene transcription (Sugimoto-Shirasu and Roberts, 2003). Notably, the degree to which endoreduplication occurs in seed storage tissues has been correlated with the accumulation of starch and storage proteins as well as yield (Kowles et al., 1992; Cavallini et al., 1995; Lemontey et al., 2000).
Maize (Zea mays) endosperm provides a useful model system to investigate the mechanism of endoreduplication and study its physiological consequences in a seed storage tissue. Endosperm tissue is a major portion of the maize kernel and is the fusion product of two polar nuclei and one sperm nucleus during fertilization. In the initial phase of development, the triploid endosperm nucleus undergoes synchronous divisions without cytokinesis, forming a syncitium composed of several hundred nuclei (Kiesselbach, 1949). Cell walls form around the nuclei, so that by 4 d after pollination (DAP) the endosperm is completely cellularized. A rapid period of growth follows that is marked by numerous cell divisions and the differentiation of several cell types: aleurone, basal endosperm transfer cells, embryo-surrounding cells, and starchy endosperm (Olsen, 2001). This mitotic phase of development is essentially complete by 10 to 12 DAP, after which cell divisions are primarily restricted to peripheral regions of the endosperm. At this time, cells in the central starchy endosperm cease to divide and start to engage in multiple cycles of DNA endoreduplication (Kowles and Phillips, 1985; Dilkes et al., 2002). Coincidentally, these cells become enlarged and begin to accumulate starch and storage proteins (Lopes and Larkins, 1993). Accumulation of storage metabolites continues until 40 to 45 DAP, but beginning around 20 DAP, the large central starchy endosperm cells begin to undergo programmed cell death (Young et al., 1997); this process progresses from the center of the endosperm toward the crown and then the base, finally affecting the entire tissue except the aleurone cells (Young and Gallie, 2000).
Cell cycle regulation is well characterized in yeast and animals, and many details of the molecular mechanisms regulating this process are known. Progression through its different phases (G1, S, G2, and M) is controlled by a class of heterodimeric protein kinases composed of a catalytic subunit called a cyclin-dependent kinase (CDK) and a regulatory subunit, cyclin (Morgan, 1997). Many classes of CDKs have been identified in animals, and functional homologs of the yeast p34cdc2/cdc28 were widely demonstrated to be involved in central cell cycle functions (Morgan, 1997). These enzymes have a highly conserved amino acid sequence, including a PSTAIRE motif involved in cyclin binding. Among the substrates of these enzymes are proteins involved in CDK regulation, DNA replication, actin polarization, spindle assembly, and mitosis (Ubersax et al., 2003).
Cell cycle regulation in plants is less well characterized than in yeast and model animal systems, but during the past decade, many important genes have been identified, and we have begun to gain some understanding of their function and regulation (Vandepole et al., 2002; DeWitte and Murray, 2003). These studies have shown that plant and animal cell cycle proteins are highly conserved, as are some aspects of their regulation. In plants, p34cdc2/cdc28 homologs, referred to as type A CDKs (CDKA), contain the conserved PSTAIRE motif (Joubès et al., 2000), and they appear to control cell cycle progression much in the way it occurs in yeast and animal cells.
The endoreduplication cell cycle is a variant of the standard mitotic cell cycle in which there are recurrent S- and G-phases without M-phase (Edgar and Orr-Weaver, 2001). The mechanisms that enable cells to sequentially replicate chromosomes without progressing through M-phase have recently begun to be investigated, and they involve modulating the levels of CDK activity. Low levels of CDK activity during the G1-phase permit the formation of prereplication complexes (ORC proteins, Cdc6, Cdt1/Dup1, and MCM proteins) at origins of replication, licensing DNA for replication while inhibiting M-phase induction. Cdc6 and MCM proteins dissociate from these complexes during S-phase, when levels of CDK activity are high. A subsequent reduction of S-phase CDK activity, resulting from cyclin destruction and accumulation of CDK inhibitors, allows origins of replication to become relicensed for another round of DNA replication (Edgar and Orr-Weaver, 2001; Larkins et al., 2001). Based on the conservation of cell cycle regulation in plants and animals, it is likely that the mechanisms associated with endoreduplication are also conserved between plants and animals. There have been relatively few studies that address the molecular details of endoreduplication in plants, but several examples support this hypothesis (Grafi and Larkins, 1995; Grafi et al., 1996; Cebolla et al., 1999; Sun et al., 1999a, 1999b).
Dominant negative mutations of CDKs have been useful for revealing specific functions of different classes of these enzymes in human (Vandenheuvel and Harlow, 1993), yeast (Fleig and Nurse, 1991), and plant cells (Hemerly et al., 1995, 2000). In this study, we examined the function of maize Zeama;CDKA;1 (CDKA) in the endoreduplication cell cycle by expressing a gene encoding a dominant negative mutant form of the enzyme in the endosperm of transgenic maize. This gene (cdc2ZmA) was previously described by Colasanti et al. (1991), although studies to describe specific cell cycle functions and regulation of the enzyme it encodes have not been reported. A mutant form of the enzyme was created by converting Asp146 to Asn146. This modification does not affect interaction with a cyclin, but it blocks ATP binding, resulting in the loss of kinase activity. By overexpressing this mutated gene in endosperm with the 27-kD γ-zein promoter, the endogenous wild-type CDKA was diluted, leading to lower CDKA activity. Because the 27-kD γ-zein promoter is activated from 10 to 12 DAP, only the endoreduplication phase of endosperm development was affected. Using this approach, we were able to investigate the effect of endoreduplication on gene expression and examine the consequence of reduced endoreduplication on the growth and development of endosperm cells.
RESULTS
Construction of Transgenic Maize Plants Overexpressing Endosperm-Specific CDKA-WT and CDKA-DN Genes
To create a dominant negative mutant of the CDKA expressed in maize endosperm, we obtained a cDNA clone corresponding to the maize p34cdc2 (cdc2ZmA) described by Colasanti et al. (1991). An oligonucleotide encoding the hemagglutinin (HA) epitope was inserted in a _Hin_dIII site at the 5′ end of the mRNA coding sequence to distinguish the protein encoded by this gene (CDKA-WT) from the endogenous CDKA. A mutation in the coding sequence of CDKA-WT that converted Asp146 to Asn146 was made by site-directed mutagenesis, creating the CDKA-DN gene. As a consequence of this mutation, the ATP-dependent phosphorylation activity of the CDK was lost, and a dominant negative mutation was created (Fleig and Nurse, 1991; Vandenheuvel and Harlow, 1993). These sequences were cloned into the pHP3630 expression vector, which directs high levels of endosperm-specific expression via the 27-kD γ-zein promoter (Russell and Fromm, 1997). The CDKA-WT and CDKA-DN constructs were introduced into maize embryos along with a selectable marker gene (moPAT) by particle bombardment; herbicide-resistant calli containing the recombinant genes were cultured on selective media, and plants were regenerated (Gordon-Kamm et al., 2002). Several transformation events were recovered for the CDKA-WT transgene, but only two events could be regenerated for the CDKA-DN construct. Preliminary analysis by flow cytometry and immunoblotting revealed similar phenotypes for both CDKA-DN events. Therefore, we selected the apparently highest expressing transgenic event of each construct for detailed biochemical analysis. T1 plants were backcrossed to the B73 inbred, and the resulting (BC2 and BC3) seeds were used for flow cytometric and biochemical analyses.
CDKA-DN Protein and RNA Accumulate at High Levels during Endosperm Development
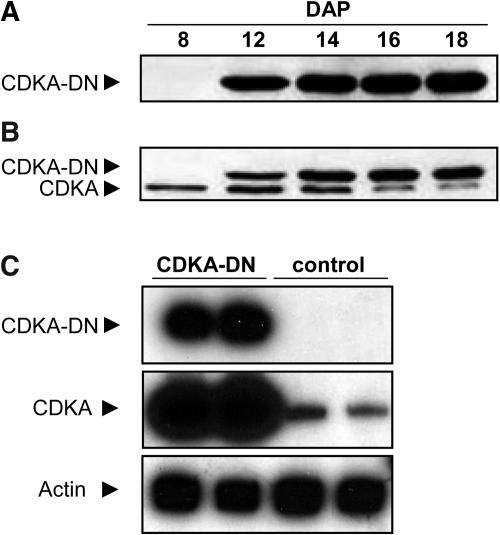
The expression levels of the CDKA-WT and CDKA-DN transgenes were assessed in developing endosperm by immunoblotting and RNA transcript analysis. Figure 1 shows the developmental accumulation of protein and RNA transcripts encoded by the CDKA-DN gene in endosperm between 12 and 18 DAP. Based on immunodetection with a monoclonal HA- antibody, the recombinant protein was present by 12 DAP (Figure 1A), after which its concentration appeared to remain relatively constant. Immunoblotting with a polyclonal anti-CDKA antibody detected both the endogenous CDKA and the recombinant CDKA-DN protein. The transgenic protein had a slightly slower electrophoretic mobility, which was most likely a consequence of the HA-epitope tag. Based on the intensity of immunostaining, it appeared that the ratio of the recombinant to the endogenous CDK increased from 1:1 to >5:1 as the endosperm developed. However, this difference in protein concentration was not reflected at the RNA level. RT-PCR analysis of RNA from endosperm of independent transgenic and nontransgenic kernels revealed extremely large differences in steady state transcript levels (Figure 1C). Based on phosphoimaging analysis, the radioactivity measured for the CDKA-DN and endogenous CDKA RT-PCR products showed an ∼100-fold difference at 16 DAP, indicating that the 27-kD γ-zein promoter resulted in much higher levels of transcription than that of the endogenous CDKA gene. Similarly, high levels of expression were measured for the CDKA-WT transgene (data not shown).
Figure 1.
Detection of CDKA-DN Expression in Developing Transgenic Maize Endosperms.
Endosperm protein extracts (40 μg/lane) from 8- to 18-DAP kernels were separated by 12.5% SDS-PAGE and blotted onto nitrocellulose.
(A) Immunoblotting with monoclonal anti-HA antibody detected a 36-kD protein corresponding to CDKA-DN.
(B) Immunoblotting with polyclonal anti-CDKA antibody detected CDKA-DN and a 34-kD protein corresponding to the endogenous CDKA.
(C) CDKA transcripts were detected in 16-DAP endosperm by semiquantitative RT-PCR using gene-specific primers to amplify the transgene (CDKA-DN), both the endogenous and transgenic CDKA (CDKA + CDKA-DN), and actin, which was used as loading control. RT-PCR products from individual CDKA-DN endosperms are shown in the first two lanes (CDKA-DN), and those from nontransgenic controls are in the last two lanes (control).
Expression of CDKA-WT and CDKA-DN Alters Protein Kinase Activity in Developing Endosperm
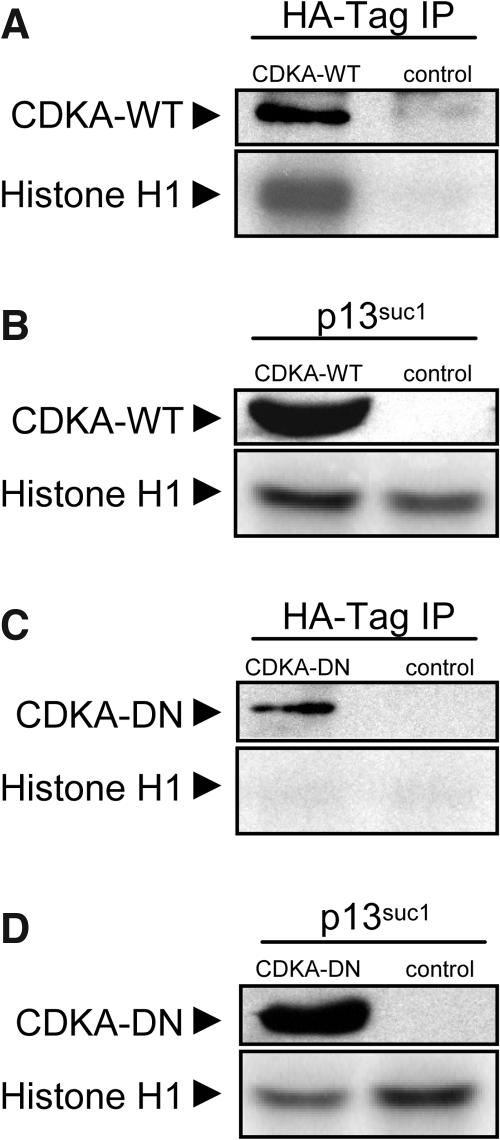
To determine whether CDKA-WT encoded a functional CDK and if expression of CDKA-DN reduced CDK activity in developing endosperm, HA-immunoprecipitated and p13suc1-adsorbed CDK activity in wild-type and transgenic kernels were assayed by histone H1 phosphorylation. Endosperms from 12 DAP kernels were genotyped based on expression of the HA-epitope tag (top panels of Figures 2A to 2D). Figure 2A shows that the HA-antibody immunoprecipitated histone H1 kinase activity from CDKA-WT endosperms but not from those of nontransgenic kernels. Consistent with this, there was more p13suc1-adsorbed histone H1 kinase activity from CDKA-WT than nontransgenic endosperm (Figure 2B). Based on the relative level of histone H1 phosphorylation, there was ∼50% more kinase activity in transgenic compared with control endosperms, although we did not systematically measure this difference. These results demonstrated that the protein encoded by the CDKA-WT gene is a functional CDK and that increased kinase activity was present in the endosperm.
Figure 2.
Assay of Histone H1 Phosphorylation by CDK Immunoprecipitates and p13suc1 Pulldowns from Nontransgenic Control, CDKA-WT, and CDKA-DN Transgenic Endosperms.
The presence or absence of transgenic HA-tagged CDKA in the assay is indicated in the top panels of (A) to (D); lanes corresponding to transgenic and control endosperms are designated.
(A) Histone H1 phosphorylation by HA-immunoprecipitate of 12-DAP CDKA-WT and nontransgenic control endosperms.
(B) Histone H1 phosphorylation by p13suc1-pulldown of 12-DAP CDKA-WT and nontransgenic control endosperms.
(C) Histone H1 phosphorylation by HA-immunoprecipitate of 12-DAP CDKA-DN and nontransgenic control endosperms.
(D) Histone H1 phosphorylation by p13suc1-pulldown of 12-DAP CDKA-DN and nontransgenic control endosperms.
Although immunoprecipitation with HA-antibody of CDKA-DN endosperm extracts recovered the epitope-tagged protein, the enzyme had no detectable histone H1 kinase activity (Figure 2C). When extracts from endosperms expressing the mutant CDK were incubated with p13suc1 beads, measurable levels of histone H1 kinase activity were detected, but they were about half of those recovered from the nontransgenic endosperms (Figure 2D). Consequently, it appeared that expression of the CDKA-DN gene markedly reduced the level of CDK activity in developing endosperm.
High Levels of CDKA-DN Expression Reduce Endoreduplication during Endosperm Development
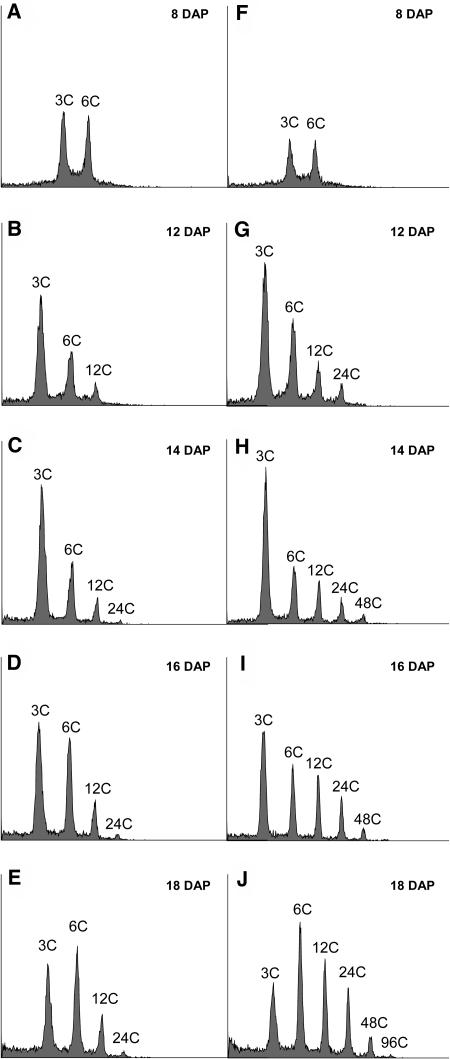
To determine the effects of CDKA-WT and CDKA-DN expression on endoreduplication, developing endosperms from at least 20 kernels were analyzed by flow cytometry and genotyped by HA-immunoblotting. Whereas overexpression of the CDKA-WT gene had no influence on the degree of endoreduplication in developing endosperm (data not shown), endosperms expressing CDKA-DN showed clear evidence of reduced endoreduplication by 12 DAP. Figure 3 shows representative flow cytometric profiles of nuclei from 8 to 18 DAP endosperms obtained from a BC2 ear segregating for the CDKA-DN transgene, and Figure 4 presents an analysis of these data. At 8 DAP, during the period of mitotic growth and development, 3C and 6C nuclei were present in similar proportions in the transgenic (Figure 3A) and control (Figure 3F) endosperms. However, by 12 DAP, which coincided with accumulation of detectable levels of CDKA-DN protein in transgenic endosperm (Figure 1), there was evidence of reduced endoreduplication in the mutant. At this early stage of development, 3C, 6C, 12C, and 24C nuclei were detectable in the nontransgenic control (Figure 3G), whereas only 3C, 6C, and a few 12C nuclei were detected in the CDKA-DN mutant. Between 14 and 18 DAP, endoreduplication progressed in the nontransgenic control endosperm (Figures 3H to 3J), as evidenced by the reduction in the number of 3C nuclei, the increased proportion of 6C, 12C, and 24C nuclei, and the appearance of 48C and 96C nuclei. By comparison, endoreduplication in endosperms containing the CDKA-DN protein was markedly reduced (Figures 3C to 3E): there was an increase in the number of 6C and 12C nuclei, but only a few 24C nuclei formed, and there was no evidence of nuclei with C-values larger than this.
Figure 3.
Flow Cytometric Analysis of Nuclei from Developing Transgenic CDKA-DN and Nontransgenic Control Endosperms.
Nuclei were isolated in PARTEC buffer and analyzed by flow cytometry as described in Methods.
(A) to (E) Endosperms from 8- to 18-DAP CDKA-DN kernels.
(F) to (J) Endosperms from 8- to 18-DAP nontransgenic control kernels. C-value is indicated for each nuclear peak.
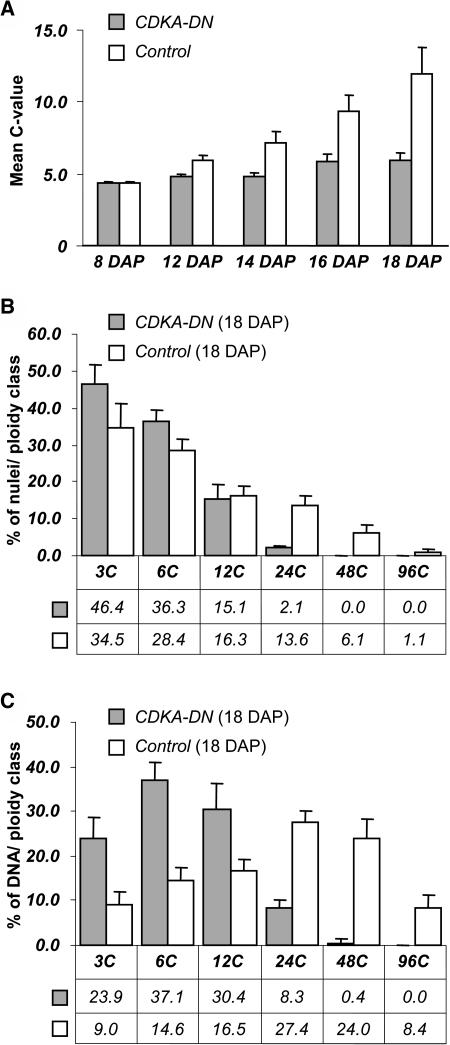
Figure 4.
Analysis of Flow Cytometric Data Describing Nuclear C-Values for Developing CDKA-DN and Nontransgenic Control Endosperms.
(A) The mean C-value was calculated for each stage of endosperm development shown in Figure 3.
(B) Calculation of the percent of nuclei in each C-value class in CDKA-DN and nontransgenic control endosperm at 18 DAP.
(C) Calculation of the proportion of total DNA contributed by nuclei in each C-value class in CDKA-DN and nontransgenic control endosperm at 18 DAP. The error bars show the standard deviation of the measurements.
The effect of reduced endoreduplication in the CDKA-DN endosperm on the mean C-value and the distribution of nuclei in various C-value classes is shown in Figure 4. At 8 DAP, endosperms in the transgenic and nontransgenic kernels both had a mean C-value of 5.0, but by 18 DAP it was 5.9 in the CDKA-DN mutant versus 11.9C in the nontransgenic control (Figure 4A). This twofold reduction in mean C-value in the CDKA-DN mutant was primarily a consequence of the reduction of highly endoreduplicated nuclei. Whereas 37% of the nuclei in the nontransgenic control were between 12C and 96C, these classes represented 17% of the nuclei in the CDKA-DN mutant (Figure 4B). Coincidentally, there was a 20% increase in the relative number of 3C and 6C nuclei in the CDKA-DN mutant. Figure 4C shows that whereas endoreduplicated nuclei with C-values of 12C or greater accounted for 76% of the DNA in control nuclei, they accounted for only 39% of the DNA in the CDKA-DN mutant.
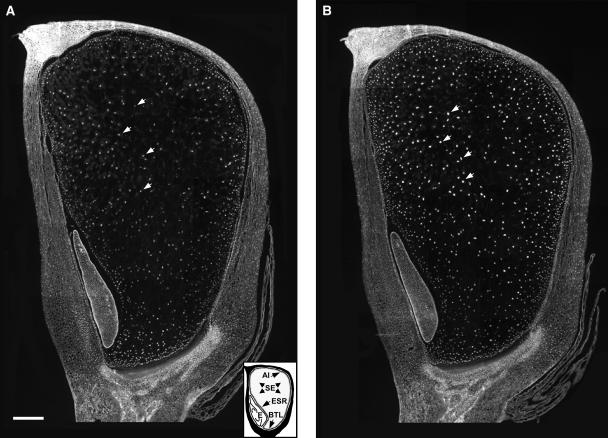
To investigate the spatial effects of CDKA-DN expression on endoreduplication, endosperms from 14 DAP transgenic and nontransgenic kernels were examined after staining with 4′,6-diamidino-2-phenylindole (DAPI). Figure 5 shows median longitudinal sections of CDKA-DN and nontransgenic control kernels. Normally, endoreduplication commences around 10 DAP in the center of the endosperm and progresses centripetally toward the aleurone (Kowles and Phillips, 1988; Vilhar et al., 2002). As evidenced by their larger sizes, nuclei in cells of the central endosperm region of both the CDKA-DN (Figure 5A) and nontransgenic control (Figure 5B) had higher C-values than those in the aleurone and subaleurone cell layers or the 2C nuclei of the embryo and pericarp. Nuclei near the basal transfer region also appeared to be more highly endoreduplicated than those in the starchy endosperm subaleurone region. Expression of the CDKA-DN gene appeared to affect endoreduplication in nuclei throughout most of the endosperm; in particular, nuclei in the very large cells in the center of the endosperm were reduced in size (cf. region defined by arrows in Figures 5A and 5B). These effects reflected the temporal and spatial expression of the 27-kD γ-zein promoter (Woo et al., 2001) and are consistent with the reduction in high C-value nuclei (e.g., 24C, 48C, and 96C) measured by flow cytometry (Figure 3).
Figure 5.
DAPI-Stained Longitudinal Sections of 14-DAP CDKA-DN and Nontransgenic Control Endosperms.
Paraffin-embedded kernels were sectioned near the site of silk attachment and stained with DAPI as described in Methods. CDKA-DN (A) and nontransgenic (B) control kernels are shown. Arrows identify nuclei of contrasting size in comparable cells of the central starchy endosperm region. The inset in (A) illustrates the location of the aleurone (Al), starchy endosperm (SE), embryo surrounding region (ESR), basal transfer layer (BTL), and embryo (E). Bar = 0.5 mm.
The long-term effects of CDKA-DN gene expression on endoreduplication were assessed by measuring the DNA content of 35-DAP control and CDKA-DN endosperms. At this stage of development, the CDKA-DN endosperm contained half as much DNA (1.20 versus 2.51 μg per 50 mg of flour endosperm) as the nontransgenic control.
CDKA-DN Endosperms Manifest Normal Cell Sizes
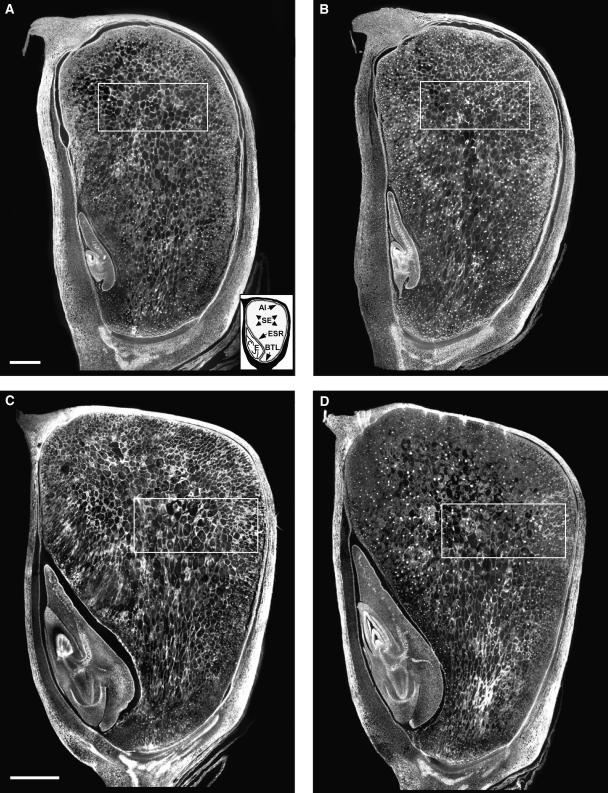
A structural comparison was made with CDKA-DN and nontransgenic control endosperm to assess the consequences of reduced endoreduplication on cell number and size. Figure 6 shows representative kernels of CDKA-DN endosperms at 13 (Figure 6A) and 19 DAP (Figure 6C), and comparable nontransgenic controls are shown in Figures 6B and 6D, respectively. The morphology of the transgenic and nontransgenic kernels was comparable at both stages of development: the basal transfer layer, aleurone, embryo-surrounding region, and starchy endosperm had similar sizes and organization. Indeed, no striking differences in the number and sizes of cells were observed among any of the cell types in transgenic and nontransgenic endosperms. We measured the average cross-sectional area of cells in the center of the starchy endosperm because they showed the greatest reduction in endoreduplication based on nuclear size. A fixed area was determined in the middle of the endosperm (Figures 6A and 6C), and the number of cells in this region was counted in six different CDKA-DN and nontransgenic endosperms. In this region, the average cell cross-sectional area at 13 DAP was 5792 ± 1103 μm2 and 6954 ± 712 μm2 for the CDKA-DN and nontransgenic control endosperms (cf. Figures 6A and 6B), respectively. At 19 DAP, the sizes were 14,784 ± 2265 μm2 and 13,040 ± 889 μm2 for the CDKA-DN and nontransgenic control endosperms (cf. Figures 6C and 6D), respectively. Statistical analysis (Student's t test) of these measurements suggested that, in accordance with our visual observations, there were no significant differences in the sizes of starchy endosperm cells in the CDKA-DN and nontransgenic control endosperms.
Figure 6.
Comparison of Cell Sizes in CDKA-DN and Nontransgenic Control Endosperms.
Comparison of cell sizes in CDKA-DN ([A] and [C]) and nontransgenic control ([B] and [D]) endosperms at 13 ([A] and [B]) and 19 DAP ([C] and [D]). Paraffin-embedded kernel sections were double stained with Calcofluor-white and DAPI; the procedure for identification of the region selected for cell size measurements is described in Methods. The inset in (A) identifies the location of the aleurone (Al), starchy endosperm (SE), embryo surrounding region (ESR), basal transfer layer (BTL), and embryo (E). Bar = 0.5 mm for (A) and (B); bar = 1.0 mm for (C) and (D).
Reduced Levels of Endoreduplication in CDKA-DN Endosperm Have Little Effect on Starch and Storage Protein Accumulation
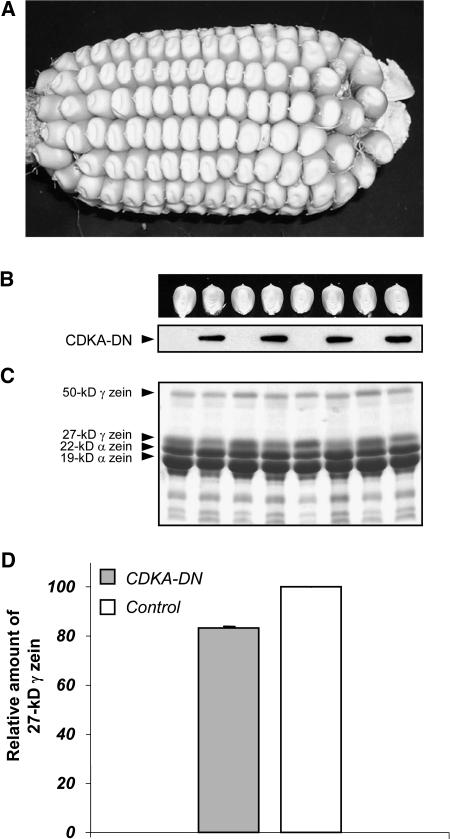
To determine if the lower level of endoreduplication in CDKA-DN endosperm affected starch and storage protein synthesis, we measured the average dry weight and zein content of mature transgenic and nontransgenic BC2 kernels. Figure 7A illustrates that there was no apparent difference in kernel size, and it was necessary to distinguish the genotypes based on expression of the HA-epitope in the endosperm (Figure 7B). The average dry weight of the CDKA-DN kernels (n = 36) was 303 ± 28.3 mg, whereas that of the nontransgenic control (n = 36) was 324 ± 30.0 mg, suggesting there was a slight difference (6.5%) in starch accumulation. To compare storage protein synthesis, the zein prolamin proteins were separated by SDS-PAGE and stained with Coomassie blue. Extracts from equal amounts of CDKA-DN and nontransgenic endosperm flour revealed little difference in the accumulation of the highly abundant 22- and 19-kD α-zeins nor the less abundant 50-kD γ-zein (Figure 7C). However, there appeared to be reduced accumulation of the 27-kD γ-zein in CDKA-DN endosperm. This was measured more accurately by an ELISA with antiserum against the 27-kD γ-zein protein (Wallace et al., 1990). The analysis showed an ∼17% reduction in the concentration of the 27-kD γ-zein in the transgenic kernels (Figure 7D).
Figure 7.
Accumulation of Zein Storage Proteins in Mature CDKA-DN and Nontransgenic Control Endosperms.
(A) A BC2 ear segregating for CDKA-DN and nontransgenic kernels.
(B) Identification of CDKA-DN and nontransgenic kernels based on immunodetection of the HA-epitope tag. After photographing the kernels, endosperm protein was extracted and separated by 12.5% SDS-PAGE; immunoblotting was the same as described in Figure 1.
(C) Analysis of zein storage proteins in transgenic CDKA-DN and control kernels shown in (B). After SDS-PAGE, the gel was stained with Coomassie blue.
(D) ELISA of 27-kD γ-zein in CDKA-DN and control endosperms shown in (B). Zein proteins were extracted and immobilized for ELISA analysis as described in Methods.
Gene Expression Assays Show Little Difference between the Steady State RNA Levels or Gene Transcription in CDKA-DN and Nontransgenic Control Endosperms
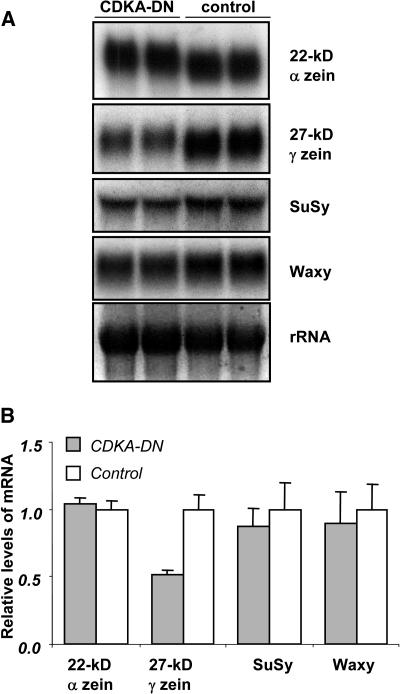
Because the lower level of endoreduplication in CDKA-DN endosperm appeared to have only minor effects on starch and storage protein accumulation in mature endosperm, RNA transcripts of genes involved in the synthesis of these products were examined. Figure 8 shows representative RNA gel blots of RNAs from 20-DAP endosperm of CDKA-DN and nontransgenic kernels. The level of 22-kD α-zein RNAs was relatively indistinguishable between the two genotypes. As suggested by the 27-kD γ-zein protein level, its RNA transcripts were reduced in CDKA-DN endosperm. There also appeared to be a slight reduction in RNAs encoding sucrose synthase and granule-bound starch synthase1 (waxy), but these differences were not significant. Thus, the reduced level of endoreduplication in CDKA-DN endosperm appeared to have relatively modest effects on the level of the RNA transcripts responsible for starch and storage protein synthesis.
Figure 8.
RNA Gel Blot Analysis of RNA Transcripts in 20-DAP CDKA-DN and Nontransgenic Control Endosperms.
Ten micrograms of total RNA was separated electrophoretically in 1.5% agarose and blotted to nylon membranes as described in Methods. RNA samples from CDKA-DN (first two lanes) and nontransgenic control endosperms (last two lanes) are designated. After hybridization, the nylon filters were exposed to a PhosphorImager screen (A), and the radioactivity was measured by PhosphorImager analysis. The mean radioactive values for three independent blotting experiments are shown in (B).
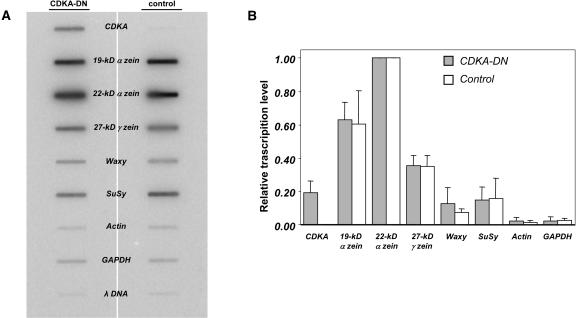
It is possible that starch and storage protein accumulation in maize endosperm are not under stringent transcriptional control. For example, starch synthesis could largely be a consequence of enzyme levels, and zein storage protein accumulation could reflect relatively stable steady state RNA levels. Therefore, as another method to assess the effect of endoreduplication on gene expression, nuclear run-on transcription assays were done using 20-DAP transgenic and nontransgenic endosperm. These assays were based on using equal amounts (fresh weight) of CDKA-DN and nontransgenic endosperm; hence, each reaction should have contained approximately equal numbers of nuclei. Nuclei were isolated by Percoll gradient centrifugation and added to in vitro transcription reactions containing 32P-CTP to label the RNA. Upon completion of the reaction, the radioactive RNA was isolated and incubated with nylon filters containing immobilized DNA probes. Figure 9A shows representative phosphoimages comparing the hybridization of nuclear run-on transcripts from CDKA-DN endosperm (on the left) and nontransgenic control nuclei (on the right). The first slot, which contained DNA corresponding to the CDKA sequence (Colasanti et al., 1991), showed much higher levels of radioactivity for the CDKA-DN nuclei compared with the nontransgenic control, which was not detectable by this assay. Notably, the hybridization intensity of the CDKA transcripts from CDKA-DN nuclei was comparable to that of the 27-kD γ-zein in the transgenic and nontransgenic control nuclear run-on transcription assays (Figure 9A). The synthesis of RNA transcripts for the 19- and 22-kD α-zeins and 27-kD γ-zeins was similar between the two samples, as were transcripts corresponding to starch biosynthetic enzymes, granule-bound starch synthase (waxy), and sucrose synthase. Likewise, there were comparable levels of actin and glyceraldehyde phosphate dehydrogenase transcripts produced by CDKA-DN and nontransgenic control nuclei.
Figure 9.
Levels of Nuclear Transcripts in CDKA-DN and Nontransgenic Control Endosperms.
(A) Illustration of an autoradiograph obtained after hybridization of RNAs obtained by in vitro run-on transcription with CDKA-DN (left) and nontransgenic control nuclei (right) 20 DAP. λ DNA was used as a background control; slot blots corresponding to 19- and 22-kD α-zeins and 27-kD γ-zeins, waxy, sucrose synthase (SuSy), glyceraldehyde phosphate dehydrogenase (GAPDH), and actin are indicated.
(B) Mean values of hybridization intensities calculated for three independent run-on transcription assays with 18-DAP CDKA-DN and nontransgenic control endosperm nuclei. The error bars indicate the standard deviation of the measurements.
Figure 9B shows the relative level of nuclear transcripts for each of these genes compared with that of the 22-kD α-zeins based on three independent assays. As suggested by the data in Figure 9A, no significant differences could be detected in the production of nuclear transcripts for any of these genes, except CDKA. Based on this analysis, a 50% reduction in the mean C-value of endosperm nuclei had relatively little effect on the production of nuclear transcripts for these genes.
DISCUSSION
A Reduction in CDKA Activity Inhibits the Progression of Endoreduplication Cell Cycles
Since the original cloning of the maize CDKA (Colasanti et al., 1991), there have been no reports demonstrating its activity in mitotic cell cycle regulation or endoreduplication, although immunolocalization of the protein implicated its involvement in cell division, based on its association with microtubules of the pre-prophase band and the phragmoplast (Colasanti et al., 1993). Our results suggest this enzyme is involved in progression of the endoreduplication cell cycle, most likely by functioning during the G1/S transition. This hypothesis is based on the similarity in peak symmetry of the flow cytometric histograms of nuclei from CDKA-DN and nontransgenic control endosperms. These data suggest that although there was a delay in the progression of S-phase in the CDKA-DN mutant, once initiated, the process of DNA replication was similar to that in the nontransgenic control.
Overexpression of CDKA-WT had no measurable effect on endoreduplication, but it led to an increase in p13suc1-associated histone H1 phosphorylation activity, indicating that the enzyme was active and associated with a cyclin. Immunoblotting experiments have been done to identify potential cyclin partners that might be associated with this enzyme, but thus far this approach has not been successful. That there was no apparent cell cycle phenotype associated with overexpression of this enzyme implies that CDKA is not rate limiting, at least for endoreduplication. This observation is consistent with that of Hemerly et al. (1995), who found that constitutive expression of Arabidopsis thaliana wild-type CDKA in tobacco (Nicotiana tabacum) cells produced no cell cycle phenotype. By contrast, expression of CDKA-DN substantially reduced p13suc1-associated kinase activity in the endosperm, and this was associated with a reduction in the number of endoreduplication cycles such that the final mean C-value was half that of the nontransgenic control endosperm (Figure 4A).
Regulation by the 27-kD γ-Zein Promoter Restricted the Effects of the CDKA-DN Mutant to the Endoreduplication Phase of Endosperm Development
The 27-kD γ-zein promoter was very useful for overexpressing the wild-type and CDKA-DN mutant genes because it is not active during the early, mitotic stages of endosperm development and it is highly and uniformly expressed throughout the starchy endosperm by 10 DAP (Russell and Fromm, 1997; Woo et al., 2001). Consequently, the physiological effects of reduced CDKA activity were restricted to the period of endoreduplication. The semiquantitative RT-PCR measurements we made (Figure 1C) showed that the steady state level of RNA transcripts produced by the chimeric CDKA transgene was ∼100-fold greater than that produced by the endogenous CDKA gene. But despite the much higher level of CDKA-DN gene transcription, there appeared to be only an approximately fivefold higher level of CDKA-DN protein by 18 DAP, suggesting that some form of posttranscriptional and/or posttranslational regulation led to lower than expected levels of the mutant protein.
Although the 27-kD γ-zein promoter is thought to be endosperm specific, based on the low recovery of transgenic maize plants expressing CDKA-DN, there appeared to be some degree of leaky expression in embryogenic calli. Hemerly et al. (1995) reported somewhat similar results when creating transgenic plants expressing a CDKA-DN mutant gene under control of the 35S promoter of Cauliflower mosaic virus. Multiple transformation events expressing an Arabidopsis wild-type CDKA sequence were regenerated into plants. However, they did not recover any Arabidopsis plants, and only two transgenic tobacco plants expressing CDKA-DN were regenerated. Consequently, it appears even low expression levels of the dominant negative mutant CDKA are deleterious to cell division and plant growth and development.
Based on the slight reduction in level of the 27-kD γ-zein RNA and protein in the CDKA-DN mutant, it appeared this transgenic event led to a small degree of posttranscriptional gene silencing. The recombinant gene construct used to regulate expression of CDKA-DN contained the noncoding regions of the 27-kD γ-zein mRNA, and it is possible that either the 5′ or 3′ noncoding sequence of the transcript was responsible for the reduction in 27-kD γ-zein synthesis (Brummell et al., 2003).
Expression of CDKA-DN Reduced Endoreduplication throughout the Endosperm but Had No Effect on Cell Size
Based on flow cytometric (Figure 3) and microscopic analyses (Figures 5 and 6), the reduction in CDKA activity resulting from expression of CDKA-DN affected endoreduplication throughout much of the starchy endosperm. The most obvious effect was in the central cells, which normally attain the highest C-values. Endocyles in these cells were not completely blocked, however, because a few 12C and 24C nuclei could be detected by flow cytometry at 14 DAP (Figure 3C). However, these nuclei failed to undergo additional cycles of endoreduplication because there was relatively little change in their numbers or C-values between 14 and 18 DAP, and no 48C and 96C nuclei were detected by flow cytometry. The central starchy endosperm cells begin to undergo programmed cell death around 20 DAP, so additional cycles of endoreduplication would not be expected beyond this stage of development (Young and Gallie, 2000). Expression of CDKA-DN had less of an impact on endoreduplication in the peripheral regions of the starchy endosperm in the CDKA-DN mutant, where the cells typically attain ploidy levels of only 3C and 6C (Vilhar et al., 2002). Nevertheless, endoreduplication appeared to have been affected in this region because there was a slower rate of conversion of 3C to 6C nuclei in the CDKA-DN mutant than the nontransgenic control (cf. Figures 3D and 3E and Figures 3I and 3J). Because the DNA content of mature CDKA-DN endosperm was half that of the nontransgenic control, similar to the difference measured at 18 DAP, it appeared there was no further DNA synthesis after mid-development nor a significant difference in DNA degradation as a consequence of programmed cell death.
In a recent study of the W22 inbred at 16 DAP, Vilhar et al. (2002) showed that whereas only 20% of the endosperm cells had nuclei of 12C and higher, these accounted for 80% of the endosperm volume. Cells with 3C and 6C nuclei were more numerous (80%) but occurred primarily at the periphery of the endosperm, where they accounted for ∼20% of the volume. Our flow cytometeric data (Figure 3; data not shown) suggested that, at a comparable stage of development, ∼14% of the nuclei in the nontransgenic control endosperm were 24C to 96C, but these high endoreduplication classes accounted for only 3% of the nuclei in CDKA-DN mutant endosperm. Approximately 70% of the nuclei in the nontransgenic control were 3C and 6C, whereas these two classes accounted for 84% of the nuclei in the CDKA-DN mutant. These results suggest that a small number of nuclei in the center of the CDKA-DN starchy endosperm had two and rarely three cycles of endoreduplication, but the process was arrested at one or no cycles in most cells. Consequently, whereas the mean C-value for nuclei in the CDKA-DN and nontransgenic control endosperms differed twofold, this value was most likely even greater for cells that contributed to most of the endosperm volume in the CDKA-DN mutant.
The reduced level of endoreduplication in the CDKA-DN endosperm affected the size of nuclei, particularly those in the central starchy endosperm cells (Figure 5), but it had little or no effect on cell size (Figure 6). Systematic measurement of the sizes of central endosperm cells in the CDKA-DN mutant and nontransgenic control at 13 and 19 DAP revealed fundamentally no difference in their average diameters and, hence, volumes. We did not attempt to measure the sizes of cells in the peripheral and basal regions of the endosperm because they are generally small and undergo only one or two cycles of endoreduplication. Consequently, it would have been difficult to evaluate the relationship between changes in endoreduplication and cell size in these areas. If there is a linkage between cell size and endoreduplication, it was effectively uncoupled by the low levels of CDKA activity in the CDKA-DN mutant. This result brings into question the relationship between nuclear size and cell volume, which has often been attributed to endoreduplication (Traas et al., 1998; Lemontey et al., 2000; Kudo and Kimura, 2002; Sugimoto-Shirasu and Roberts, 2003). However, it should be noted that this relationship has been questioned by others (Gendreau et al., 1998; Joubès and Chevalier, 2000), and Kowles et al. (1990) proposed that endoreduplication and cell size might not be tightly coupled in maize endosperm. This hypothesis was supported by the variation in cell volume and degree of endoreduplication observed in wild-type and miniature1 endosperm (Vilhar et al., 2002).
Reduced Levels of Endoreduplication Had Little Effect on Gene Expression in Maize Endosperm
A second function generally ascribed to endoreduplication is an increase in the level of gene transcription. Several previous reports provided indirect evidence supporting this relationship (Kowles et al., 1992; Engelen-Eigles et al., 2000; Lemontey et al., 2000), but these studies were largely correlative and endoreduplication could not be separated from pleiotropic effects associated with physiological changes or environmental effects. Surprisingly, we found only a modest (6.5%) reduction in average seed weight for the CDKA-DN mutant compared with the nontransgenic control. Because 90% of the kernel dry weight reflects starch accumulation, this implies that gene expression related to starch synthesis was only modestly affected by the reduction in endoreduplication. Likewise, with the exception of 27-kD γ-zein protein accumulation, which could have been affected by posttranscriptional gene silencing, there was relatively little difference in zein storage protein synthesis in CDKA-DN and nontransgenic control endosperm (Figure 7). The minor differences in starch and storage protein accumulation in CDKA-DN and nontransgenic control kernels were reflected in the steady state levels of the corresponding RNAs in developing endosperm (Figure 8).
Because accumulation of these storage metabolites might reflect relatively long-lived mRNAs and stable enzymes, we also measured the relative levels of nuclear transcripts of the corresponding genes. For this analysis, we compared nuclei from equal fresh weight samples of 20 DAP endosperm tissue of homozygous CDKA-DN and nontransgenic control kernels. We assume these reactions contained approximately equal numbers of nuclei because there was no difference in endosperm cell size at this stage of development (Figures 6C and 6D). When the DNA content of nuclear samples used in these reactions was measured, those from CDKA-DN endosperm contained half the amount found in the nontransgenic control (data not shown), consistent with the mean C-values measured by flow cytometry. Based on this rather sensitive measure of gene expression, no difference in the level of transcription in populations of nuclei that averaged ∼6C for the CDKA-DN mutant and 12C for the nontransgenic control was detected (Figure 9). This result suggests that gene transcription per nucleus was nearly the same, regardless of the C-value. Alternatively, the contribution of nuclear transcripts from highly endoreduplicated nuclei (12C and greater) in the nontransgenic control was not detectable by this assay.
These data imply that endoreduplication is inconsequential for the increased levels of gene expression required to support starch and storage protein synthesis. This hypothesis is also supported by results from previous experiments in which zein RNAs were detected by in situ hybridization (Woo et al., 2001). Between 10 and 15 DAP, the concentration of 27-kD γ-zein RNA, as well as other types of zein RNAs, were fairly uniform throughout the starchy endosperm, in spite of the fact that the central cells are more highly endoreduplicated. Although these experiments were not designed to quantitatively measure RNA concentration, they were done in probe excess and differences in the concentration of specific zein transcripts (e.g., 22-kD α-zeins) were detected in some cells of the starchy endosperm.
There are, nevertheless, extenuating factors that could influence these conclusions. We measured steady state levels of RNAs and nuclear transcription in 20 DAP endosperm (Figures 8 and 9), a period that typically coincides with the peak of RNA, DNA, and protein synthesis (Young and Gallie, 2000). Consequently, we expect these measurements reflected the peak of gene expression in developing endosperm. However, coincident with this period of development, the central endosperm cells, particularly those with the highest level of endoreduplication, begin to undergo programmed cell death (Young and Gallie, 2000). Unfortunately, little is known about transcription and translation in the central cells of the starchy endosperm at 18 to 20 DAP. Viability staining indicated that these cells begin to lose membrane integrity around this time (Young et al., 1997), and in situ hybridization suggested that zein RNAs are not retained within them (Woo et al., 2001). Endosperm extracts showed an increase in nuclease activity beginning around 24 DAP (Young et al., 1997), and by 28 DAP nucleosomal fragments could be detected (Young and Gallie, 2000). However, our flow cytometric analysis of 20-DAP endosperm nuclei provided no evidence of chromatin breakdown; the nuclei were intact and showed no evidence of fragility. We did not detect nuclei larger than 96C by flow cytometry analysis of 20-DAP endosperm extracts, but this is typical (Kowles et al., 1990; Dilkes et al., 2002). There are not many of these nuclei (Vilhar et al., 2002), and their large size and fragility makes them subject to damage by starch (Dilkes et al., 2002).
We observed no evidence indicating that the highly endoreduplicated nuclei (e.g., 24C, 48C, and 96C) in cells of the nontransgenic control endosperm were damaged or transcriptionally inactive in the run-on assay. Numerically, these accounted for ∼20% of the nuclei, and they were only 2 to 3% of the nuclei in endosperm extracts from the CDKA-DN mutant. Although the number of these nuclei is small, they accounted for more than half of the genomes represented in the assay (Figure 4C). If we conclude that the highly endoreduplicated nuclei are transcriptionally inactive, the inability to detect differences in level of RNAs in run-on transcription assays with control and CDKA-DN nuclei at 20 DAP could be explained on the basis that the number of 3C and 6C nuclei in these assays differed by only 20%. Resolution of questions relating to the transcriptional activity of endoreduplicated nuclei will require that we investigate the structure of DNA and chromatin in nuclei with markedly different C-values and measure the transcriptional activity of nuclei in specific C-value classes. Alternatively, methods to more effectively reduce or eliminate endoreduplication cycles will need to be found.
Endoreduplication in the endosperm and cotyledons of developing seeds is widely believed to provide a mechanism for increasing cell size and gene expression. However, as reviewed by Kowles et al. (1986), it could also occur as a mechanism to provide a store of phosphate and nucleotides for the developing/germinating embryo. Unfortunately, little research has been published in which this process was investigated (see Wildon et al., 1971). It is well documented that endosperm contains nucleases and that the DNA in the central cells of the starchy endosperm starts to become degraded around 20 DAP (Young and Gallie, 2000). Though we cannot yet rule out a role in gene transcription, we must seriously consider the hypothesis that endoreduplication in maize endosperm functions primarily to provide a store of nucleotides during embryogenesis and/or germination.
METHODS
Construction of Recombinant Wild-Type Zeama;CDKA;1 (CDKA-WT) and Zeama;CDKA;1D146N (CDKA-DN) Genes and Creation of Transgenic Maize Plants
A maize CDKA cDNA (Colasanti et al., 1991) was mutated in vitro as described by Hemerly et al. (1995), such that the Asp at position 146 was replaced with Asn. Conversion of Asp146 (GAC) to Asn146 (AAC) was performed with a Transformer site-directed mutagenesis kit (Clontech, Palo Alto, CA) using the mutagenic primer, 5′-GCACTGAAGCTTGCAaACTTTGGTTTAGCCAG-3′; the nucleotide sequence alteration was confirmed by DNA sequencing. For both the wild-type and mutant CDKA sequences, an oligonucleotide encoding the HA-epitope (nine amino acids) was inserted into a _Hin_dIII site at the 5′ end of the mRNA coding sequence, seven amino acids after the first Met residue. The CDKA-WT and CDKA-DN sequences were then cloned into pHP3630, an expression vector that contains the maize 27-kD γ-zein promoter and 5′ noncoding sequence (Lopes et al., 1995), a multiple cloning site, and the 27-kD γ-zein 3′ noncoding sequence and transcriptional terminator. The CDKA-WT and CDKA-DN constructs were introduced into scutellar cells of the maize Hi-II genotype along with a selectable marker plasmid that contained a maize-optimized phosphinothricin acetyl transferase (moPAT) gene and moGFP (moPAT∼moGFP). Particle-mediated transformation, selection of herbicide resistant calli, and regeneration of fertile plants were essentially as described by Gordon-Kamm et al. (2002). Initial genotyping was done with endosperms of T1 kernels; transgenic plants were backcrossed using pollen from a B73 inbred line. With exception of the nuclear run-on transcription assays, which were done with kernels from homozygous BC3-S2 ears, the data reported were obtained with developing or mature kernels from segregating BC2 or BC3 ears.
Immunodetection of Endogenous and Transgenic CDKA
Dissected endosperms were ground in three volumes of NETT buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 20 mM EDTA, 0.5% Triton X-100, 5 mM NaF, 1 mM Na orthovanadate, with freshly added 1 mM phenylmethylsulfonyl fluoride, and 1 mM DTT) and protease inhibitor cocktail (10 μM benzamidine, 1 μg/mL of phenanthroline, 10 μg/mL of aprotinin, 10 μg/mL of leupeptin, 10 μg/mL of pepstatin A, and 10 mM sodium bisulfate dissolved in ethanol). The extract was cleared by centrifugation at 14,000_g_ in an Eppendorf microfuge at 4°C (Eppendorf, Hamburg, Germany), and the protein concentration of the supernatant was determined by Bradford assay (Bio-Rad, Hercules, CA). Forty micrograms of protein from each sample was separated by 12.5% SDS-PAGE and blotted onto nitrocellulose using a wet transfer apparatus (Bio-Rad mini trans-blot cell) at 200 Vh. The membrane was blocked with TTBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) containing 1.5% nonfat milk for 1 h, followed by overnight incubation on a rotating plate with a 1:10,000 dilution of mouse monoclonal anti-HA epitope tag (12AC5; Roche, Basel, Switzerland) or rabbit polyclonal anti-ZmCDKA antibody (our unpublished data). After three washes with TTBS, the membranes were incubated 2 h with the appropriate secondary antiserum, either anti-mouse IgG conjugated with horse radish peroxidase (Roche) or anti-rabbit IgG conjugated with horse radish peroxidase (Sigma, St. Louis, MO). Filters were washed in TTBS three times and incubated with chemiluminescent substrate (SuperSignal West Pico; Pierce, Rockford, IL) for 5 min. The membranes were exposed to x-ray film for 10, 30, and 60 s, which was developed using a Konica developer (Konica QX13A plus; Tokyo, Japan).
CDKA Immunoprecipitation, Pull-Down, and Histone H1 Phosphorylation Assays
For immunoprecipation of HA-tagged CDKA, three endosperm halves from 12 DAP transgenic kernels expressing the epitope-tagged CDKA were ground in 600 μL (approximately three volumes) of NETT buffer, cleared as for immunoblotting, and the supernatant (∼4.2 mg of protein) was preincubated with 40 μL of a slurry of Protein-A agarose beads (Sigma) by rocking at 4°C for 1 h. After removal of the agarose beads with a quick spin in the microfuge, 2 μg of anti-HA monoclonal antibody was added and incubated for 1 h on the rocker at 4°C. Subsequently, 40 μL of Protein-A agarose beads was added and the mixture incubated for 2 h as previously described. The Protein-A agarose beads were collected by centrifugation and washed three times with NETT buffer and once with kinase buffer (50 mM Tris-HCl, pH 7.5, 20 mM EGTA, 10 mM MgCl2, 1 mM DTT, and 1 mM β-glycerol phosphate); 5-μL aliquots were used for immunoblotting as previously described.
p13suc1 pull-downs were performed similarly to the immunoprecipitations, except endosperm extract was preincubated with glutathione agarose beads (Sigma) (Grafi and Larkins, 1995). Forty microliters of a GST-p13suc1 slurry (∼5 μg of protein) bound to glutathione agarose beads was added to the cleared extract and incubated for 3 h at 4°C and washed as described for immunoprecipitations.
The kinase activity associated with immunoprecipitates or p13suc1-pulldowns was assayed by histone H1 phosphorylation. Agarose beads were incubated in 7 μL of kinase buffer, 1 μL of 2.5 μg/μL histone H1 (Sigma), 1 μL of 4 mM ATP, and 1 μL of [γ-32P]ATP (10 μCi/μL) (Amersham, Piscataway, NJ). After 30 min of incubation at room temperature, the reactions were stopped by adding 5 μL of 5× Laemmli sample buffer (Laemmli, 1970), heated in a boiling water bath for 5 min, and analyzed by 12.5% SDS-PAGE. Gels were vacuum dried and exposed to x-ray film overnight.
Flow Cytometric Analysis of Endosperm Nuclei
Kernels from the center of developing ears segregating 1:1 for the transgene were dissected to recover the embryo and the endosperm. In some experiments, the embryos were rescued by tissue culture and grown to mature plants (Dilkes et al., 2002). The endosperm was bisected by cutting the kernel in half at the silk attachment site. One half of the endosperm was frozen for subsequent genotype analysis based on detection of the HA-epitope tag by immunoblotting. The other endosperm half was analyzed by flow cytometry to assess the degree of nuclear endoreduplication. Nuclei were released by chopping with a single-edged razor blade in the presence of 0.8 mL of filtered ice cold PARTEC buffer (200 mM Tris-HCl, pH 7.5, 4 mM MgCl2, and 0.1% Triton X-100) (Dilkes et al., 2002). The homogenate was aspirated through two layers of cheesecloth with a plastic pipette, passed through a 100-μm nylon mesh placed in the tip of a 3-cc syringe, and collected in a 3-mL polystyrene tube (Sarstedt 55.484; Newton, NC). The Petri dish was washed with an additional 0.8 mL of PARTEC buffer, which was combined with the initial nuclear extract. Nuclei were stained with 40 μL of a 100 mg/mL solution of DAPI and analyzed with a PARTEC CCAII flow analyzer (PARTEC, Münster, Germany). For each sample, at least 10,000 nuclei were collected and analyzed using a logarithmic scale display. Each flow cytometric histogram was saved with PARTEC CA3 software and analyzed with WinMDI 2.8 software (available at http://facs.scripps.edu/software.html). Mean C-value was calculated according to the following equation: [(n3 × 3) + (n6 × 6) + (n12 × 12) + …]/total number of nuclei, where n3 = total number of nuclei in the 3C peak, etc.
RNA Isolation and Blotting
To avoid carbohydrate contamination, RNA was isolated from 20-DAP endosperms by a combination of phenol/chloroform extraction and the TRIzol procedure (Invitrogen, Carlsbad, CA). Dissected endosperms from CDKA-DN and control nontransgenic seeds were ground in RNA extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM LiCl, 5 mM EDTA, pH 8.0, and 1% SDS in diethyl pyrocarbonate–treated water) (Prescott and Martin, 1987) and incubated on ice for 5 min with an equal volume of phenol-chlorophorm in a phase-lock tube (Eppendorf, Hamburg, Germany). The samples were spun at room temperature for 10 min (full speed) in a microfuge and the aqueous phase re-extracted with an equal volume of chloroform. The aqueous phase was transferred to a new tube and the RNA extracted with 1 mL of TRIzol according to the manufacturer's directions. The RNA was dissolved in water, and 10 μg of each sample was denatured and separated by electrophoresis in a 1.5% agarose gel containing 0.66 M formaldehyde and blotted onto nylon membrane (MSI, Westboro, MA). DNA probes for 22-kD α-zeins and 27-kD γ-zeins (Hunter et al., 2002), granule-bound starch synthase (waxy) (Shure et al., 1983), and sucrose synthase (Geiser et al., 1980; Werr et al., 1985) were PCR amplified with gene-specific primers. After analysis by agarose gel electrophoresis, the amplified products were purified using a NucleoSpin extraction kit (Clontech) and radioactively labeled using [α-32P]dCTP in a random priming kit (Invitrogen). After RNA transfer, gel blots were hybridized with 32P-labeled probes. Hybridizations were performed at 65°C in a buffer containing 1.5× SSPE (1× SSPE is 0.18 M NaCl, 0.1 M NaH2PO4, and 0.01 M EDTA, pH 7.7), 6% polyethylene glycol 8000, 1% SDS, 1% nonfat dried milk (Carnation), 0.1% (v/v) diethyl pyrocarbonate, and 30 μg/mL of freshly denatured salmon sperm DNA (Sabelli and Shewry, 1993). After hybridization, the blots were washed once with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M Na citrate, pH 7) containing 0.1% SDS at room temperature, twice with 1× SSC containing 0.1% SDS for 15 min at 65°C, and finally with 0.1× SSC containing 0.1% SDS for 15 min at 65°C. The filters were exposed to PhosphorImager screens for varying periods of time, and the radioactive signals were measured with a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Semiquantitative RT-PCR Analysis of RNA Transcripts
RNA was isolated as described above and treated with DNase as recommended by the manufacturer (Invitrogen). RT-PCR reactions were performed with 25 ng of total RNA using the Titan One Tube RT-PCR system (Roche, Hamburg, Germany) according to the manufacturer's protocol, with the exception that the reactions were supplemented with 0.7 μL of [α-32P]dATP (10 mCi/mL) (NEN, Boston, MA). Transgene-specific amplification was achieved with a HA-tag annealing primer (HATagF, 5′-TACCCCTACGACGTGCCCGACTACGCC-3′) and a downstream CDKA primer (CDC2R, 5′-TCACTGTACCACTTCAAGGTC-3′). Actin and the endogenous CDKA were amplified using the following primer combinations: ACT1F, 5′-ATTCAGGTGATGGTGTGAGCCACAC-3′; ACT1R, 5′-GCCACCGATCCAGACACTGTACTTCC-3′; CDC2-F, 5′-ATGGAGCAGTACGAGAAGGTGGAGAAG-3′; CDC2R, 5′-TCACTGTACCACTTCAAGGTC-3′. The annealing temperature was 65°C in all cases; twenty cycles were used for amplification of actin and CDKA-DN RNAs, and 25 cycles were used for the endogenous CDKA RNA. PCR products were separated by electrophoresis in 4% (w/v) nondenaturing acrylamide containing TBE, and the gel dried and exposed to x-ray film or PhosphorImager screen. Radioactive signals were measured with a Storm 860 PhosphorImager.
Analysis of Endosperm Proteins
Single degermed kernels from a mature 1:1 segregating BC3 ear were used to determine the relative levels of zein proteins. Zein and nonzein protein fractions were extracted as described by Wallace et al. (1990) and analyzed by 12.5% SDS-PAGE. The amount of 27-kD γ-zein was estimated by the ELISA assay described by Moro et al. (1995). The nonzein fraction was dissolved in 8 M urea and used to determine the genotype of the corresponding kernel.
Nuclear Run-On Transcription Assay
The level of nuclear transcripts in CDKA-DN and nontransgenic control endosperms at 20 DAP was estimated by in vitro transcription with isolated nuclei. Nuclei from 2.5 g of freshly dissected endosperms of each genotype were isolated by Percoll gradient centrifugation as previously described (Luthe and Quatrano, 1980; Or et al., 1994). Endosperms were chopped with a single edged razor blade in a Petri dish containing Honda buffer (0.44 M sucrose, 2.5% Ficoll, 5% Dextran-T40, 25 mM Tris-HCl, pH 8.5, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.5% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, and 2 mM spermidine) and freshly added EDTA-free protease inhibitor cocktail (Roche). The homogenate was filtered through two layers of cheesecloth, followed by filtration through a 100-μm (pore size) nylon sheet. Nuclei were pelleted by centrifugation at 6000_g_ in a Sorvall HB4 rotor for 5 min at 4°C, resuspended in 5 mL of Honda buffer (minus spermidine), and carefully layered on 5-mL layers of 40, 60, and 80% Percoll in 0.45 M sucrose, 40 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 0.5% Triton X-100; the bottom of the gradient had a cushion of 2.5 M sucrose. After centrifugation at 6000_g_ for 20 min at 4°C, the nuclei were removed from the Percoll-sucrose interface and washed twice with Honda buffer (minus spermidine) and another two times with nuclei buffer (50 mM Tris-HCl, pH 8.5, 5 mM MgCl2, 50% glycerol, and 10 mM 2-mercaptoethanol). The purified nuclei were resuspended in 100 μL of transcription buffer (50 mM Tris-HCl, pH 8.5, 5 mM MgCl2, 50% glycerol, and 10 mM 2-mercaptoethanol) and added to the run-on transcription assay. The in vitro transcription reaction and isolation of nuclear RNA were performed as described by Dorweiler et al. (2000). Total RNA products from each transcription reaction were used for filter hybridization. Nitrocellulose filters with slot blots of cDNAs corresponding to CDKA (Colasanti et al., 1991), 19- and 22-kD α-zeins and 27-kD γ-zeins (Hunter et al., 2002), granule-bound starch synthase (waxy) (Shure et al., 1983), sucrose synthase (Geiser et al., 1980; Werr et al., 1985), actin (Dorweiler et al., 2000), and glyceraldehyde phosphate dehydrogenase (Russell and Sachs, 1989) were prepared by immobilizing 100 ng of PCR-amplified, gene-specific fragments for each sequence. Hybridization and washing procedures were those described by Dorweiler et al. (2000). Radioactivity was measured with a Storm 860 PhosphorImager and analyzed with ImageQuaNT (Amersham) software. For comparison of transcription levels in CDKA-DN and control nuclei, the background hybridization to λ phage DNA was subtracted from each probe signal before normalizing to the radioactivity of the 22-kD α-zein. The 22-kD α-zein was selected as a basis of comparison because the radioactive signals were significantly above background and the radioactive measurement of control and transgenic samples were similar. Run-on transcription assays were repeated three times using three different ears of each genotype. Comparable kernel samples from these ears were analyzed by flow cytometry and found to contain reproducible C-values for endoreduplicated nuclei.
DNA Isolation and Quantification
To estimate the amount of DNA at late stages of endosperm development, 50 mg of flour from lyophilized 35-DAP CDKA-DN and control endosperm was added to 500 μL of Sarcosyl extraction buffer (Tris-HCl, pH 8.5, 100 mM NaCl, 200 mM EDTA, and 1% sarcosyl) and incubated at room temperature for 10 min. Subsequently, an equal volume of phenol-chloroform was added and mixed by vortexing. After transfer to a phase-lock tube, the samples were incubated at room temperature for 5 min, and then the aqueous and organic phases were separated by centrifugation in a microfuge at 14,000 rpm for 10 min. The aqueous phase was transferred to a new tube and incubated at 37°C with 5 μL of RNase A (10 mg/mL). The DNA was precipitated by adding 300 μL of isopropanol and incubating at −80°C for 30 min. After pelleting by centrifugation for 10 min at top speed in a microfuge, the DNA was washed with 70% ethanol and air dried, dissolved in 50 μL of TE (10 mM Tris-HCl and 1 mM EDTA, pH 8.0) and the concentration measured with PicoGreen (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. DNA samples of 1 μL were added to 100 μL of TE and 1 μL of PicoGreen in flat bottom 96-well plates. After 5 min, the plate was read with a PhosphorImager (Molecular Dynamics) using blue fluorescence scanning. The DNA concentration was determined by ImageQuant software (Amersham) using a standard curve of known λ phage DNA concentrations (20 to 300 ng/μL). To identify the transgenic endosperms, the remainder of the flour sample was used for immunoblotting with HA antibody as previously described.
Microscopic Examination of Developing Endosperm
Kernels of uniform size were isolated from the middle of developing ears and trimmed on both sides to form a 2- to 3-mm-thick longitudinal median section containing the embryo. These slices were immediately fixed in 3.7% paraformaldehyde and 0.2% picric acid in 50 mM potassium phosphate and 5 mM EGTA buffer at pH 6.8, which was prepared in diethylpyrocarbonate-treated, double distilled water. The reminder of the kernel tissues was frozen for subsequent genotyping based on HA-immunoblotting, as previously described. After fixation for 3 to 4 h at room temperature, the thick sections were washed in the same buffer for 1 to 2 h with three changes to fresh buffer and then processed through a standard paraffin embedding procedure that included dehydration in gradient ethanol, substitution with xylene, and gradual infiltration of paraffin. Paraffin-embedded tissues were cut into 15-μm-thick sections that were attached to Histogrip-coated (Zymed Laboratories, South San Francisco, CA) glass slides. After microtome sectioning, comparable thin sections were stained with DAPI to detect nuclei and Calcofluor-white to visualize cell walls. Fluorescent images of kernels were collected at 10× magnification using a fluorescence microscope coupled to a CCD camera, and serial images were stored and analyzed using Adobe Photoshop (San Jose, CA). To measure the average cell cross-sectional area (we assume the cells are roughly isodiametric), cells in the central region of a longitudinal section from four 13-DAP and three 19-DAP kernels were measured. To ensure that cells in comparable regions were measured in all the kernels that were examined, the central region was defined by placing a fixed area (1.37 mm2 at 13 DAP and 2.78 mm2 at 19 DAP) in the center of the endosperm, such that it was aligned vertically by the left border of pedicel's vasculature and horizontally at a fixed distance below the silk scar. The average cross-sectional area of central endosperm cells was calculated by determining the number of cells in this region divided by the total area.
Acknowledgments
We thank Giovanni Bosco and Hong Nguyen for providing valuable suggestions for the improvement of the manuscript. We also wish to acknowledge the intellectual and technical contributions of Brian P. Dilkes and Rudolf Jung, who helped with the development of this project. This research was supported by a grant from the Department of Energy (DE-FG02-96ER20242) to B.A.L. and a grant from the Binational Agricultural Research and Development Fund (IS-2726-96) to B.A.L. and G.G. R.A.D. was supported by a scholarship from the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico of Brazil.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Brian A. Larkins (larkins@ag.arizona.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.022178.
References
- Brodsky, W.Y., and Uryvaeva, I.V. (1977). Cell polyploidy: Its relation to tissue growth and function. Int. Rev. Cytol. 50**,** 275–332. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A., Balint-Kurti, P.J., Harpster, M.H., Palys, J.M., Oeller, P.W., and Gutterson, N. (2003). Inverted repeat of a heterologous 3′-untranslated region for high-efficiency, high-throughput gene silencing. Plant J. 33**,** 793–800. [DOI] [PubMed] [Google Scholar]
- Cavallini, A., Natali, L., Balconi, C., Rizzi, E., Motto, M., Cionini, G., and Damato, F. (1995). Chromosome endoreduplication in endosperm cells of two maize genotypes and their progenies. Protoplasma 189**,** 156–162. [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Olah, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18**,** 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, J., Cho, S.O., Wick, S., and Sundaresan, V. (1993). Localization of the functional p34cdc2 homolog of maize in root tip and stomatal complex cells: Association with predicted division sites. Plant Cell 5**,** 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, J., Tyers, M., and Sundaresan, V. (1991). Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc. Natl. Acad. Sci. USA 88**,** 3377–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., and Murray, J.A.H. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54**,** 235–264. [DOI] [PubMed] [Google Scholar]
- Dilkes, B.P., Dante, R.A., Coelho, C., and Larkins, B.A. (2002). Genetic analyses of endoreduplication in Zea mays endosperm: Evidence of sporophytic and zygotic maternal control. Genetics 160**,** 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler, J.E., Carey, C.C., Kubo, K.M., Hollick, J.B., Kermicle, J.L., and Chandler, V.L. (2000). Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12**,** 2101–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B.A., and Orr-Weaver, T.L. (2001). Endoreplication cell cycles: More for less. Cell 105**,** 297–306. [DOI] [PubMed] [Google Scholar]
- Engelen-Eigles, G., Jones, R.J., and Phillips, R.L. (2000). DNA endoreduplication in maize endosperm cells: The effect of exposure to short-term high temperature. Plant Cell Environ. 23**,** 657–663. [Google Scholar]
- Fleig, U.N., and Nurse, P. (1991). Expression of a dominant negative allele of cdc2 prevents activation of the endogenous p34cdc2 kinase. Mol. Gen. Genet. 226**,** 432–440. [DOI] [PubMed] [Google Scholar]
- Geiser, M., Doring, H.P., Wostemeyer, J., Behrens, U., Tillmann, E., and Starlinger, P. (1980). A cDNA clone from Zea mays endosperm sucrose synthetase mRNA. Nucleic Acids Res. 8**,** 6175–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau, E., Hofte, H., Grandjean, O., Brown, S., and Traas, J. (1998). Phytochrome controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl. Plant J. 13**,** 221–230. [DOI] [PubMed] [Google Scholar]
- Gordon-Kamm, W., et al. (2002). Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc. Natl. Acad. Sci. USA 99**,** 11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi, G., and Larkins, B.A. (1995). Endoreduplication in maize endosperm: Involvement of M-phase-promoting factor inhibition and induction of S-phase-related kinases. Science 269**,** 1262–1264. [DOI] [PubMed] [Google Scholar]
- Grafi, G., Burnett, R.J., Helentjaris, T., Larkins, B.A., DeCaprio, J.A., Sellers, W.R., and Kaelin, W.G. (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 93**,** 8962–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly, A., Engler Jde, A., Bergounioux, C., Van Montagu, M., Engler, G., Inze, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14**,** 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly, A.S., Ferreira, P.C., Van Montagu, M., Engler, G., and Inze, D. (2000). Cell division events are essential for embryo patterning and morphogenesis: Studies on dominant-negative cdc2aAt mutants of arabidopsis. Plant J. 23**,** 123–130. [DOI] [PubMed] [Google Scholar]
- Hunter, B.G., Beatty, M.K., Singletary, G.W., Hamaker, B.R., Dilkes, B.P., Larkins, B.A., and Jung, R. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14**,** 2591–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès, J., and Chevalier, C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43**,** 735–745. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Phan, T.H., Just, D., Rothan, C., Bergounioux, C., Raymond, P., and Chevalier, C. (1999). Molecular and biochemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit. Plant Physiol. 121**,** 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inze, D., Umeda, M., and Renaudin, J.P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43**,** 607–620. [DOI] [PubMed] [Google Scholar]
- Kiesselbach, T.A. (1949). The Structure and Reproduction of Corn, Research Bulletin 161. (Lincoln, NE: Agricultural Experimental Station, University of Nebraska).
- Kowles, R.V., McMullen, M.D., and Phillips, R.L. (1986). Gene expression in developing maize kernels. In Regulation of Carbon and Nitrogen Reduction and Utilization in Maize, J.C. Shannon, D.P. Knievel, and C.D. Boyer, eds (Rockville, MD: American Society of Plant Physiologists), pp. 189–206.
- Kowles, R.V., McMullen, M.D., Yerk, G., Phillips, R.L., Kraemer, S., and Srienc, F. (1992). Endosperm mitotic activity and endoreduplication in maize affected by defective kernel mutations. Genome 35**,** 68–77. [Google Scholar]
- Kowles, R.V., and Phillips, R.L. (1985). DNA amplification patterns in maize endosperm nuclei during kernel development. Proc. Natl. Acad. Sci. USA 82**,** 7010–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowles, R.V., and Phillips, R.L. (1988). Endosperm development in maize. Int. Rev. Cytol. 112**,** 97–136. [Google Scholar]
- Kowles, R.V., Srienc, F., and Phillips, R.L. (1990). Endoreduplication of nuclear DNA in the developing maize endosperm. Dev. Genet. 11**,** 125–132. [Google Scholar]
- Kudo, N., and Kimura, Y. (2002). Nuclear DNA endoreduplication during petal development in cabbage: Relationship between ploidy levels and cell size. J. Exp. Bot. 53**,** 1017–1023. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227**,** 680–685. [DOI] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52**,** 183–192. [PubMed] [Google Scholar]
- Lemontey, C., Mousset-Declas, C., Munier-Jolain, N., and Boutin, J.P. (2000). Maternal genotype influences pea seed size by controlling both mitotic activity during early embryogenesis and final endoreduplication level/cotyledon cell size in mature seed. J. Exp. Bot. 51**,** 167–175. [DOI] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5**,** 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.A., Takasaki, K., Bostwick, D.E., Helentjaris, T., and Larkins, B.A. (1995). Identification of two opaque2 modifier loci in quality protein maize. Mol. Gen. Genet. 247**,** 603–613. [DOI] [PubMed] [Google Scholar]
- Luthe, D.S., and Quatrano, R.S. (1980). Transcription in isolated wheat nuclei. 1. Isolation of nuclei and elimination of endogenous ribonuclease activity. Plant Physiol. 65**,** 305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13**,** 261–291. [DOI] [PubMed] [Google Scholar]
- Moro, G.L., Lopes, M.A., Habben, J.E., Hamaker, B.R., and Larkins, B.A. (1995). Phenotypic effects of opaque2 modifier genes in normal maize endosperm. Cereal Chem. 72**,** 94–99. [Google Scholar]
- Nagl, W. (1974). Phaseolus suspensor and its polytene chromosomes. Zeit. Pflanzenphysiol. 73**,** 1–44. [Google Scholar]
- Olsen, O.A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52**,** 233–267. [DOI] [PubMed] [Google Scholar]
- Or, E., Boyer, S.K., and Larkins, B.A. (1994). Opaque2 modifiers act post-transcriptionally and in a polar manner on γ-zein gene expression in maize endosperm. Plant Cell 5**,** 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, A., and Martin, C. (1987). A rapid method for the quantitative assessment of levels of specific mRNA in plants. Plant Mol. Biol. Rep. 4**,** 219–224. [Google Scholar]
- Russell, D.A., and Fromm, M.E. (1997). Tissue-specific expression in transgenic maize of four endosperm promoters from maize and rice. Transgenic Res. 6**,** 157–168. [DOI] [PubMed] [Google Scholar]
- Russell, D.A., and Sachs, M.M. (1989). Differential expression and sequence analysis of the maize glyceraldehyde-3-phosphate dehydrogenase gene family. Plant Cell 1**,** 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P., and Shewry, P. (1993). Nucleic acid blotting and hybridisation. In Methods in Plant Biochemistry, Vol. 10, J. Bryant, ed (London: Academic Press), pp. 79–99.
- Shure, M., Wessler, S., and Fedoroff, N. (1983). Molecular identification and isolation of the waxy locus in maize. Cell 35**,** 225–233. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6**,** 544–553. [DOI] [PubMed] [Google Scholar]
- Sun, Y.J., Dilkes, B.P., Zhang, C.S., Dante, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (1999. a). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96**,** 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Flannigan, B.A., and Setter, T.L. (1999. b). Regulation of endoreduplication in maize (Zea mays L.) endosperm. Isolation of a novel B1-type cyclin and its quantitative analysis. Plant Mol. Biol. 41**,** 245–258. [DOI] [PubMed] [Google Scholar]
- Traas, J., Hulskamp, M., Gendreau, E., and Hofte, H. (1998). Endoreduplication and development: Rule without dividing? Curr. Opin. Plant Biol. 1**,** 498–503. [DOI] [PubMed] [Google Scholar]
- Ubersax, J.A., Woodbury, E.L., Quang, P.N., Paraz, M., Blethrow, J.D., Shah, K., Shokat, K.M., and Morgan, D.O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425**,** 859–864. [DOI] [PubMed] [Google Scholar]
- Vandenheuvel, S., and Harlow, E. (1993). Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262**,** 2050–2054. [DOI] [PubMed] [Google Scholar]
- Vandepole, K., Raes, J., De Veyleder, L., Rouzé, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14**,** 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhar, B., Kladnik, A., Blejec, A., Chourey, P.S., and Dermastia, M. (2002). Cytometrical evidence that the loss of seed weight in the miniature1 seed mutant of maize is associated with reduced mitotic activity in the developing endosperm. Plant Physiol. 129**,** 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, J.C., Lopes, M.A., Paiva, E., and Larkins, B.A. (1990). New methods for extraction and quantitation of zeins reveal a high content of gamma-zein in modified opaque-2 maize. Plant Physiol. 92**,** 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werr, W., Frommer, W.B., Maas, C., and Starlinger, P. (1985). Structure of the sucrose synthase gene on chromosome 9 of Zea mays L. EMBO J. 4**,** 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M.J.D. (1973). Animal Cytology and Evolution. (Cambridge: Cambridge University Press).
- Wildon, D.C., Wong, D., and Bryant, J.A. (1971). Turnover of deoxyribonucleic acid in seedlings of Pisum sativum. Biochem. J. 124**,** 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, Y.M., Hu, D.W.N., Larkins, B.A., and Jung, R. (2001). Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13**,** 2297–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, T.E., and Gallie, D.R. (2000). Programmed cell death during endosperm development. Plant Mol. Biol. 44**,** 283–301. [DOI] [PubMed] [Google Scholar]
- Young, T.E., Gallie, D.R., and DeMason, D.A. (1997). Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol. 115**,** 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmet, J., and Ravid, K. (2000). Polyploidy: Occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp. Hematol. 28**,** 3–16. [DOI] [PubMed] [Google Scholar]