Fibroblast growth factor-223 binds directly to the survival of motoneuron protein and is associated with small nuclear RNAs (original) (raw)
Abstract
The SMN (survival of motoneuron) protein is mutated in patients with the neurodegenerative disease spinal muscular atrophy. We have shown previously that a high-molecular-mass isoform of FGF (fibroblast growth factor) 2 (FGF-223) is in a complex with SMN [Claus, Döring, Gringel, Müller-Ostermeyer, Fuhlrott, Kraft and Grothe (2003) J. Biol. Chem. 278, 479–485]. FGF-2 is a neurotrophic factor for motoneurons, and is known not only as a classical extracellular growth factor, but also as a nuclear protein. In the present study, we demonstrate that SMN binds to the arginine-rich N-terminus of FGF-223. In turn, FGF-223 interacts with amino acid residues 1–90 of the human SMN protein. This sequence displays nucleic-acid-binding capacity and overlaps partially with known binding sites for Gemin2/SIP1 (SMN-interacting protein 1) and p53. Finally, as a functional consequence of FGF-223 binding to SMN, FGF-223 is in a complex with the small nuclear RNAs U2 and U4. Since SMN functions as an assembly factor for snRNPs (small nuclear ribonucleoprotein particles), these results suggest binding of FGF-223 to snRNPs.
Keywords: fibroblast growth factor 2 (FGF-2), growth factor, small nuclear ribonucleoprotein (snRNP), spinal muscular atrophy, survival of motoneuron protein (SMN protein)
Abbreviations: AC buffer, affinity chromatography buffer; DsRed, Discosoma red fluorescent protein; FGF, fibroblast growth factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; GST, glutathione S-transferase; HMGB, high-mobility-group B; IP buffer, immunoprecipitation buffer; LSm, Sm-like protein; PRMT, protein arginine methyltransferase; RBB, RNA-binding buffer; SIP1, survival-of-motoneuron-interacting protein 1; SMA, spinal muscular atrophy; SMN, survival of motoneuron; snoRNP, small nucleolar ribonucleoprotein; snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein
INTRODUCTION
SMA (spinal muscular atrophy) is an autosomal-recessive disease that results in the degeneration of motoneurons of the spinal cord. SMA is associated with deletion and mutation, respectively, of the SMN1 (survival of motoneuron 1) gene [1]. The extent of the disease is related to the existing amount of functional SMN protein produced by the second gene SMN2 [2,3]. Children with type I SMA (Werdnig–Hofmann disease) demonstrate symptoms already at 6 months of age, and most of these children die before they reach the age of 2 years. SMA is the most frequent genetic cause of death in infants and affects one in 6000–10000 live births [4]. Several studies have shown that the SMN protein plays an important role as an assembly factor for ribosomal complexes [snoRNPs (small nucleolar ribonucleoproteins)], small nuclear RNA complexes [snRNPs (small nuclear ribonucleoproteins)] and possibly in transcription [5,6].
Mutations of the Tudor domain, the central interaction domain of SMN, prevent the binding of snRNP core factors (Sm proteins), so that assembly of snRNPs is impaired [7]. Deletion of the 27 N-terminal amino acid residues of SMN leads to inhibition of snRNP assembly, splicing and transcription [8,9]. Regarding the intracellular distribution, SMN is present mainly in the cytoplasm, where it is seen as part of the complex with spliceosomal Sm proteins [10,11], Gemin2 [previously known as SIP1 (SMN-interacting protein 1)] [11] and Gemin3–7 [12–17]. In the nucleus, SMN is found in the nuclear bodies called gems (gemini of Cajal bodies) and Cajal bodies, where it interacts with coilin [18].
The Sm proteins B/B', D1 and D3, and LSm4 (Sm-like protein 4) contain RG (arginine/glycine)-enriched domains in the C-termini that are responsible for interaction with SMN [19,20]. Furthermore, symmetrical dimethylation of arginine residues seems to be important for efficient binding of several interacting proteins [20,21]. However, other interacting proteins such as the snoRNP GAR1 and fibrillarin are asymmetrically dimethylated at arginine residues, and are able to bind to SMN [9,22,23]. In addition, the methylation state of GAR1 is not important for SMN binding [23].
In an effort to analyse the function of nuclear FGF (fibroblast growth factor)-2, we have shown recently that SMN is able to bind specifically in a common complex to a high-molecular-mass iso-form of FGF-2, known as FGF-223 [24]. The N-terminus of FGF-223 is arginine-rich with RGR (Arg-Gly-Arg) motifs, which are known to be methylated [25,26]. The FGF-2 protein is expressed in different isoforms, which are regulated translationally by a common mRNA with alternative start codons. The 18 kDa FGF-2 (FGF-218) is translated from an AUG start codon, whereas translation of the higher-molecular-mass isoforms FGF-221 and FGF-223 (21 kDa and 23 kDa) is initiated at CUG start codons [27]. Apart from their role as extracellular signalling molecules, all FGF-2 isoforms are localized differentially to the nuclei of cells [24,28–30]. FGF-223 is co-localized with SMN in nuclear gems [24]. In motoneurons, FGF-2 functions, among other proteins, as a neurotrophic factor [31,32], indicating the importance of the SMN–FGF-2 complex for the survival or degeneration of these cells.
In the present study, we extend our analyses of the SMN– FGF-223 complex [24] by mapping the domains responsible for this protein–protein interaction. Our data show that the interaction is not mediated by a RNA bridge, but is a direct interaction between two distinct domains of each protein: SMN binds to the N-terminus of FGF-223, which contains several RGR motifs. FGF-223 binds reciprocally to a N-terminal domain of SMN which was demonstrated previously to be important for nucleic acid binding and protein–protein interactions. Moreover, FGF-223 is associated with snRNPs via contacts with SMN, since we could demonstrate co-immunoprecipitation of FGF-223 with snRNAs (small nuclear RNAs).
MATERIALS AND METHODS
Plasmid constructs and subcloning
Full-length (amino acid residues 1–294) human SMN was amplified by PCR from clone IMAGp998P0910174Q2 (RZPD, Berlin) in pCMV-SPORT6 with specific primers. Deletion mutants were produced by PCR with appropriate primers comprising 5′ and 3′ ends of the respective coding sequences and introducing _Eco_RI and _Sal_I restriction sites for subsequent cloning. All PCR products were cloned into pGEM-T, sequenced and subcloned into pET-41a (Novagen) allowing in-frame ligation with GST (glutathione S-transferase) as a tag in the N-terminal position. Recombinant full-length SMN and deletion mutants in pET-41a were transformed into Escherichia coli BL21(DE3) for protein expression.
Cell culture and transfection
Immortalized rat Schwann cells [33] were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with a 10% final concentration of foetal calf serum, 2 mM glutamine and 100 units/ml penicillin/streptomycin at 37 °C in humidified 5% CO2/95% air incubator. Transfections were performed using Effectene (Qiagen) or Metafectene (Biontex).
Immunoprecipitations
Immunoprecipitations were carried out as described previously [20]. For the analysis of RNase resistance of the protein complexes, extracts in RIPA buffer [20] were incubated overnight with anti-SMN and anti-FGF-2 antibodies, respectively, followed by incubation with Protein-G– and Protein-A–agarose respectively for 1.5 h and treatment with RNase A for 30 min. For the analysis of association of FGF-223 with snRNAs, immunoprecipitations were performed as described with IP buffer [immunoprecipitation buffer: 20 mM Tris/HCl, pH 7.5, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1% Triton X-100, 1% desoxycholate, 1×Complete™ protease inhibitors (Roche)], followed by a RNA extraction with TRIzol® reagent (Invitrogen) and a second RNA-purification step with RNeasy columns (Qiagen). The snRNAs were reverse-transcribed as described previously [34], with specific primers for U2, U4 and U6 snRNAs respectively, which were also employed as primers in PCR. Input material was analysed by isolation of total RNA directly from a fraction of the lysates. To demonstrate that no genomic DNA was co-purified, and to test for the specificity of the immunoprecipitation, primers for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) have been employed for reverse transcription PCR.
In vitro detection of protein–protein interactions
Overnight cultures (2.5 ml) with 30 μg/ml kanamycin were used to inoculate 25 ml of 2×LB (Luria–Bertani) medium. Cultures were induced with 1 mM IPTG (isopropyl β-D-thiogalactoside) for 6 h at 37 °C. Lysates from previously frozen pellets were incubated with glutathione–agarose beads (Novagen) for 30 min at room temperature (21 °C) and were subsequently washed once with WB (wash buffer: 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl and 2.7 mM KCl, pH 7.3), twice with supplemented WB [WB with 500 mM NaCl and 0.1% (v/v) Tween 20] and once with WB. Amounts of purified proteins on beads were determined by SDS/PAGE with Coomassie Blue staining and BCA (bicinchoninic acid) protein assay (Pierce). Equal amounts of proteins were used for pull-down assays in WB. Recombinant FGF-223 was expressed and purified as described in [35].
Pull-down assays with recombinant FGF-223 were performed in IP buffer supplemented with 0.5% (w/v) dried milk, and incubated for 25 min on an overhead shaker followed by three washing steps with IP buffer and PBS. Pull-down assays with Schwann cell extract were performed by blocking of the beads with 3% (w/v) dried milk in supplemented IP buffer [with a final concentration of 1 M NaCl and 0.1% (v/v) Nonidet P40], followed by incubation with lysate of FGF-223–DsRed (Discosoma red fluorescent protein)-overexpressing Schwann cells [20]. After 1 h at room temperature, the beads were washed twice with supplemented IP buffer and three times with PBS. Beads from both experimental procedures were finally incubated with 1× Laemmli buffer at 95 °C for 5 min, and samples were analysed on 12.5% polyacrylamide gels. Proteins were blotted on to ECL® (enhanced chemiluminescence) nitrocellulose membranes (Amersham Biosciences) and analysed with monoclonal anti-FGF-2 antibody (Transduction Laboratories) in combination with horseradish-peroxidase-conjugated anti-mouse secondary antibody and ECL® reagents (Amersham Biosciences). For far-Western blot assays, equal amounts of recombinant FGF-2 isoforms were blotted after electrophoresis on to an ECL® nitrocellulose membrane, blocked with 5% milk powder in AC buffer {affinity chromatography buffer: 10% (v/v) glycerol, 100 mM NaCl, 20 mM Tris/HCl, pH 7.6, 0.5 mM EDTA and 0.1% (v/v) Tween 20; [36]} and probed with SMN from a coupled in vitro transcription–translation reaction (TnT T7 System, Promega) for 12 h at 4 °C with human SMN in pCMV-SPORT6 as a template. After four washing steps with AC buffer, the standard Western blot protocol [20] was followed with anti-SMN (Transduction Laboratories) as primary antibody.
RNA-binding assay
The U2 snRNA template was obtained by PCR amplification using the following oligonucleotide pairs: PC157-U2_F, 5′-ATCGCTTCTCGGCCTTTTGGCTAAGA-3′ and PC158-U2_R, 5′-GGGTGCACCGTTCCGGAGGTACTGCAATA-3′. A total of 27 rounds of amplification were performed using KlenThemTaq Platinum polymerase (GeneCraft). The obtained amplicon was subsequently cloned into the pGEM-T vector (Promega) resulting in clone pGEM-T-U2snRNA and sequenced. U2 snRNA was transcribed in vitro and labelled with [α-33P]rUTP using T7 RNA polymerase (Stratagene). For RNA binding, 10 μg of GST–SMN fusion proteins were incubated with approx. 25 ng of radiolabelled U2 snRNA. The binding reaction was carried out for 10 min in 400 μl of RBB [RNA-binding buffer: 150 mM NaCl, 20 mM Tris/HCl, pH 7.5, 1 mM dithiothreitol, 0.2% (v/v) Nonidet P40] on ice while inverting occasionally. As an non-specific competitor, 25 ng of poly(dG-dC)·poly(dG-dC) (Amersham Biosciences) or tRNA was used. Bound fractions were washed five times in 800 μl of pre-chilled RBB, pelleted, and the RNA was released and purified from the fusion proteins by using TRIzol® and subsequently visualized by dot blotting.
RESULTS
SMN binds to the arginine-rich N-terminus of FGF-223 in vitro, but not to other FGF-2 isoforms
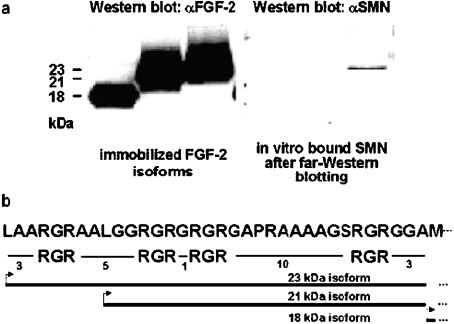
Immunoprecipitations have revealed an interaction between SMN and FGF-223, but not with the shorter FGF-218 which lacks an arginine-rich N-terminal sequence [24]. However, a possible binding of SMN to the FGF-221 isoform with fewer RGR motifs (Figure 1b) was not analysed. Binding of SMN to the 18, 21 and 23 kDa isofoms of FGF-2 was therefore tested on a far-Western blot. For this purpose, recombinant FGF-2 isoforms (18, 21 and 23 kDa) were immobilized on a nitrocellulose membrane by Western blotting. These isoforms differ in the length of their respective N-termini, with FGF-223 containing the longest N-terminal extension in front of the common FGF-218 core sequence (Figure 1b). An analysis of the sequences displays a high portion of arginine residues in the N-termini of FGF-221 and FGF- 223, which are organized in RGR motifs, and are surrounded by hydrophobic amino acid residues (Figure 1b). The immobilized FGF-2 isoforms were incubated with in vitro translated SMN. After washing, the blots were subsequently probed with an anti-SMN antibody. SMN bound to the immobilized high-molecular-mass FGF-223 isoform, but not to FGF-218 or FGF-221 isoforms respectively (Figure 1a). This demonstrates SMN binding to the arginine-rich N-terminus of FGF-223. Additionally, a critical sequence length, number of arginine residues or conformation seems to be required for efficient binding, since the FGF-221 isoform was not bound by SMN.
Figure 1. SMN binds to the arginine-rich N-terminus of FGF-223 in vitro, but not to other FGF-2 isoforms.
(a) Recombinantly expressed FGF-2 isoforms (18, 21 and 23 kDa) were immobilized on a nitrocellulose membrane and probed with in vitro translated SMN (far-Western blot). SMN binding was subsequently analysed using anti-SMN antibody and chemiluminescence detection. SMN bound to the FGF-223 isoform, but not to the smaller isoforms, demonstrating that the number or configuration of RGR-motifs is critical for efficient binding. (b) The sequence shows the N-termini of FGF-223 and FGF-221 with RGR motifs and hydrophobic spacer sequences. Numbers denote length of spacers as numbers of amino acid residues. Thick lines with arrows, length of the N-termini 21 and 23 kDa isoforms of FGF-2 respectively with translation starts at CUG codons (leucine). FGF-218 starts with a methionine residue from an AUG codon.
The SMN–FGF-223 complex is RNase-resistant
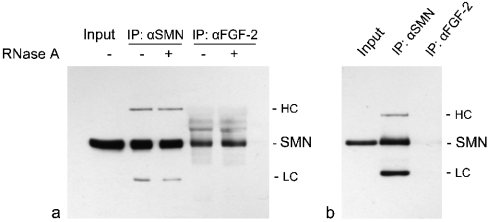
In a recent study, we have shown by co-immunoprecipitation analyses that SMN is in a complex together with the high-molecular-mass isoform of FGF-2 [24]. To test whether this observation is based on RNA as a bridging molecule, we performed co-immunoprecipitations with subsequent digestion of the immunoprecipitated material with RNase A. Lysates of immortalized Schwann cells overexpressing DsRed-tagged FGF-223 [24] were incubated with anti-FGF-2 and anti-SMN antibodies respectively, and the immunoprecipitated and Protein-A–agarose-immobilized protein complexes were treated extensively with RNase A. After gel electrophoresis, Western blots were probed with anti-SMN antibody. The binding of SMN to FGF-223 was not disrupted upon treatment with RNase A (Figure 2a). No difference could be observed between immunoprecipitations of treated and non-treated SMN–FGF-2 complexes, indicating a protein–protein interaction between these two molecules without RNA as a bridging molecule. As a control for the specificity of the anti-FGF-2 antibody, control immunoprecipitations were performed in extracts from rat Schwann cells overexpressing the FGF-218 isoform (Figure 2b). As shown previously, this isoform is not able to interact with SMN [24]. An immunoprecipitation with anti-FGF-2 antibody followed by detection with anti-SMN on a Western blot showed only a very weak band (Figure 2b, lane 3), probably due to the presence of small amounts of endogenous FGF-223. However, considering only this assay, the possibility cannot be excluded that a bridging RNA molecule could be protected by the complex against digestion with RNase A.
Figure 2. The SMN–FGF-223 complex is RNase-resistant.
(a) Co-immunoprecipitations with an anti-FGF-2 polyclonal antibody of lysates from Schwann cells overexpressing FGF-223 revealed no change of SMN binding to FGF-223 upon extensive digestion with RNase A. Immunoprecipitation (IP) with anti-SMN antibody is a control. Detection antibody for the Western blot was anti-SMN. (b) Control for the specificity of the employed anti-FGF-2 polyclonal antibody. Lysates from FGF-218-overexpressing Schwann cells were subjected to immunoprecipitations with anti-FGF-2. Detection with anti-SMN revealed only a very weak band due to the presence of endogenous FGF-223. HC, heavy chain; LC, light chain of IP antibodies.
FGF-223 binds directly to SMN between amino acid residues 1–90
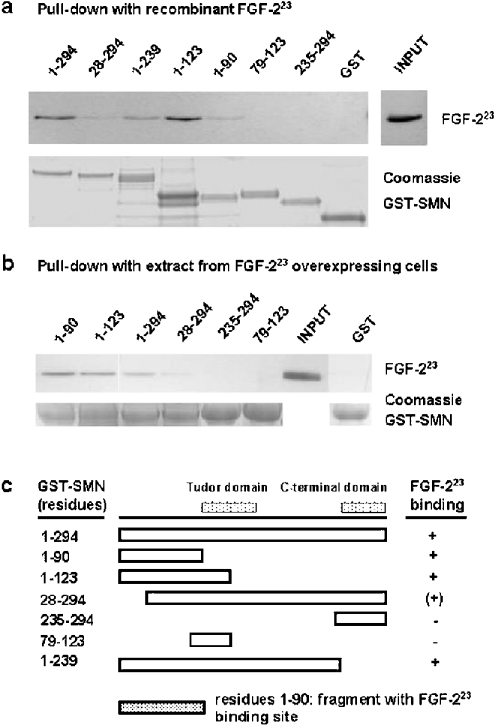
To test if the observed SMN–FGF-223 interaction is due to a direct interaction, we performed pull-down assays with recombinant proteins. The binding of FGF-223 to SMN was analysed by pull-down assays with SMN mutants. Full-length and various deleted forms of human SMN were expressed as fusions with GST and immobilized on glutathione–agarose beads. After incubation with recombinant FGF-223, beads were washed with IP buffer and PBS, and the presence of bound FGF-223 was analysed by gel electrophoresis and Western blot with a specific monoclonal antibody (Figure 3a). The pull-down revealed direct binding of FGF-223 to full-length SMN (residues 1–294) and C-terminal-truncated SMN (1–239). No binding could be observed to the C-terminal oligomerization domain of SMN, namely residues 235–294. A SMN mutant (79–123), containing most of the primary structure of the Tudor domain, was not recognized by FGF-223. While the complete Tudor domain spans residues 91–151 [37], the probed sequence represents the N-terminal two thirds of its length, and probably does not represents the correct conformation. However, the complete Tudor domain is included in other fragments, e.g. 28–294 (see below). FGF-223 bound to the 1–90 and 1–123 SMN fragments respectively, indicating an interaction with the N-terminal part of SMN. Interestingly, the N-terminal-truncated mutant 28–294 (exon 1) was bound to a lower extent compared with full-length SMN. Mutations in exon 1 are associated with SMA [8]. Taken together, these data demonstrate binding of FGF-223 to an N-terminal fragment 1–90, but with an influence of the first 28 residues on binding (Figure 3c). This is in a sequence range where RNA binding has been mapped previously (residues 28–91) [38]. The FGF-223-binding region also overlaps to a small extent with the Gemin2 (SIP1) binding domain, which is contained in exons 1/2 and 3 of SMN [11]. Additionally, the FGF-223-binding region corresponds to the p53-binding site (residues 29–51) on SMN exon 2 [39] The results of the pull-down binding assays with recombinant FGF-223 are corroborated by similar binding experiments with extracts from FGF-223-overexpressing Schwann cells (Figure 3b). Immobilized GST–SMN mutant proteins were incubated with extract, and were extensively washed with high-salt and detergent-rich buffer. The data identify the same FGF-223-binding region on SMN as the direct binding assay with bacterially expressed FGF-223 (Figure 3b).
Figure 3. FGF-223 binds directly to a N-terminal sequence of SMN.
(a) For in vitro binding experiments (pull-downs) SMN-deletion mutants were expressed as fusion proteins with GST, bound to glutathione–agarose beads and incubated with recombinant FGF-223. After the binding reaction, beads were washed with IP buffer and PBS, and the bound FGF-223 was detected by SDS/PAGE and Western blot with anti-FGF-2 antibody. To demonstrate equal loading, GST–SMN mutants were stained with Coomassie Blue. (b) Pull-down experiment similar to that of (a), but, instead of recombinant FGF-2, lysates from Schwann cells overexpressing DsRed-tagged FGF-223 were used. After 1 h of binding, beads were washed extensively with IP buffer (enriched with 1 M NaCl, 0.1% Tween 20 and 0.1% Nonidet P40) and the bound protein was analysed by SDS/PAGE and Western blot with anti-FGF-2 antibody. As a control, the same amount of beads employed as in the binding reactions (visual estimation after Coomassie Blue staining) were Coomassie-Blue-stained after SDS/PAGE. The aligned bands are shown as loading controls. Numbers denote the amino acid residues of the SMN mutants. (c) Structure of SMN-deletion mutants (boxes) and results of pull-downs. C-terminal end of binding site: FGF-223 bound to SMN 1–90 as the minimal fragment of the N-terminus. The 28–294 SMN fragment was only weakly bound, indicating the importance of the sequence from residues 1–28 as a part of the binding site. The compared data demonstrate FGF-223 binding to a N-terminal sequence of SMN, which is coded by exons 1–3 of the human SMN gene. The sequence codes by exons 2 and 3 demonstrates homology with the so-called HMGB-box of HMGB proteins and HMGB-box-containing proteins, and was shown previously to interact in vitro with nucleic acid homopolymers [38]. The relative binding intensity is indicated as +, (+) and −. The positions of the Tudor domain and the C-terminal domain, which is deleted in most cases of SMA, are shown.
U2 snRNA binding to SMN is not influenced by FGF-223
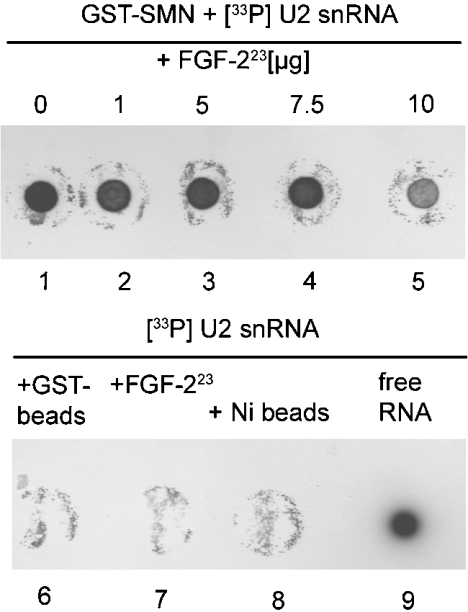
Previously, it has been shown that SMN is able to bind U1, U2, U4 and U5 snRNAs in a sequence-specific manner [40,41]. The mapped binding of FGF-223 to the RNA-binding region of SMN raises the question of whether FGF-223 is able to compete with RNA for binding to SMN. To test this possibility, we performed in vitro binding assays with immobilized full-length GST–SMN on agarose beads, added radioactively labelled U2 snRNA (used as a model snRNA for this competition experiment), unlabelled competitor [tRNA and poly(dG-dC)·poly(dG-dC) DNA respectively] and either none or increasing amounts of recombinant FGF-223 (Figure 4). After extensive washing of the beads, the bound RNA fraction was analysed on dot blots. As expected, SMN is able to bind specifically to U2 snRNA (Figure 4, lane 1). No non-specific binding to beads or FGF-223 was detectable (Figure 4, lanes 6–8). The data revealed no influence of increasing amounts of FGF-223 on U2 snRNA binding to SMN (Figure 4, lanes 2–5). Only at a very high amount of added FGF-223, could a slight decrease be detected. Importantly, immobilized FGF-223 was not able to bind U2 snRNA (Figure 4).
Figure 4. U2 snRNA binding to SMN is not influenced by FGF-223.
Immobilized full-length SMN as a fusion with GST was incubated with labelled U2 snRNA, unlabelled non-specific competitor [tRNA (results not shown) or poly (dG-dC)·poly(dG-dC)] and no or increasing amounts of recombinant FGF-223 (lanes 1–5). After incubation, beads were washed, the RNA was recovered by TRIzol® isolation and detected after dot blotting of bound fractions. Only a very high amount of FGF-223 was able to disrupt RNA–SMN binding. No non-specific U2 snRNA binding to beads could be detected (lanes 6 and 8). Immobilized FGF-223 did not bind to U2 snRNA.
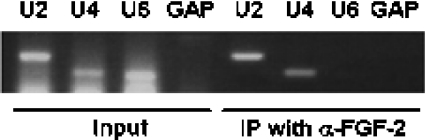
FGF-223 is associated with snRNA from snRNPs
SMN is an assembly factor for spliceosomal proteins in the cytoplasm and able to assemble Sm proteins in a ring-shape structure on the snRNAs (U snRNAs). To analyse whether FGF-223 is associated with the RNA from snRNPs, we performed co-immunoprecipitations of the FGF-223 complex, and tested for the presence of U2, U4 and U6 snRNAs. After immobilization of the immunoprecipitated complexes on Protein-A–agarose, RNA was purified by a combination of two methods (TRIzol® and column chromatography), and was subsequently analysed by reverse transcription PCR. RNA from untreated lysates was purified in parallel with the samples and was used as input control. RNA was reverse-transcribed by primers specific for the probed snRNAs. Additionally, the presence of GAPDH genomic DNA respectively was controlled by specific primers to test for DNA-contamination in the immunoprecipitated material. No genomic DNA contamination could be detected (Figure 5). The immunoprecipitations revealed the presence of U2 and U4 snRNAs in the FGF-2 complexes (Figure 5), indicating binding of FGF-2 to snRNPs. Interestingly, no U6 RNA, although present in the input, has been found to be associated with FGF-2. In contrast with the other U snRNAs, U6 snRNA is kept in the nucleus and does not enter the cytoplasm for snRNP assembly. The partial base-pairing of U4/U6 snRNAs is a late step in splicing assembly and is located in the nucleus [42]. Additionally, U6 RNA is not associated with Sm proteins, but with LSm proteins.
Figure 5. FGF-223 is associated with RNA from snRNPs.
FGF-223 complexes were immunoprecipitated from lysates of Schwann cells. RNA was purified by a combination of two methods, reverse-transcribed with specific primers for snRNAs (U2, U4 and U6 snRNAs respectively) and subsequently analysed by PCR. As a control, RNA from input material was used and reverse-transcribed to show the presence of all U snRNAs in the input, as well as to have a loading-control. Additionally, primers for the housekeeping gene GAPDH were employed as combined controls for DNA contamination and non-specifically bound RNA molecules respectively. U2 and U4, but not U6, snRNAs are associated with the FGF-223 complex, indicating a role for this FGF-2 isoform in the association with snRNPs from the cytosol. Unlike U2 and U4 snRNAs, U6 snRNAs do not leave the nucleus for snRNP assembly. Controls with GAPDH does not show non-specifically bound mRNA (IP, immunoprecipitation) or DNA contamination (input).
DISCUSSION
A number of mitogenic growth factors, growth factor receptors and growth-regulatory proteins have been reported to be localized to intracellular structures, e.g. FGF-1, FGF-2 and ciliary neurotrophic factor (see [43] for an overview), suggesting that intranuclear functions are a general phenomenon of some growth factors. In the present study, the direct binding of a growth factor to a component of the splicing machinery, namely to the SMN protein, is shown for the first time. Additionally, as a functional consequence of FGF-223 binding to SMN, this is the first described growth factor to be associated with snRNPs.
A specific interaction of SMN with FGF-223, but not with low-molecular-mass FGF-2 (FGF- 218) was initially observed by co-immunoprecipitation studies in rat Schwann cells [24]. In the present study, we extended our analyses by demonstrating that the interaction is direct and is not dependent on a molecular bridge like an additional RNA molecule or protein. RNase A digestion of immunoprecipitated material shows no indication for such a role of RNA in the SMN–FGF-2 complex. Although RNA in protein–RNA complexes could be resistant to RNases, a direct interaction has been verified by pull-down experiments in the present study. In addition, the binding site of SMN that is necessary for FGF-223 interaction was mapped. Finally, FGF-223 is associated with U2 and U4, but not with U6, snRNAs.
SMN binds to the arginine-rich N-terminus of FGF-223 which consists of four or five RGR motifs with two or three of them grouped into one cluster (Figure 1b). The primary structure between these motifs is filled with hydrophobic amino acid residues. The known three-dimensional NMR structure of FGF-218 [44] does not include the N-terminus of the higher-molecular-mass isoforms. Two sequences responsible for nuclear localization of FGF-2 have been found previously: the N-terminal sequences of FGF-221 and FGF-223 with the RGR motifs, and a sequence in the C-terminus, which is common to all isoforms, including FGF-218 [24]. Since no binding of SMN to the FGF-221 isoform, but to FGF-223, could be observed, the number or configuration of RGR motifs seems to be critical for efficient SMN protein binding. In the far-Western blot, recombinant FGF-2 isoforms were employed without the proper post-translational modifications, including arginine methylation. Therefore methylation of FGF-223 is, at least in vitro, not an absolute requirement for the interaction with SMN. However, in cells, it has been shown that methylation affects the nuclear accumulation of the growth factor [45]. FGF-223 can be methylated asymmetrically in vitro by PRMT (protein arginine methyltransferase) 1 [25], but it is not known yet if PRMT5, a symmetrically methylating arginine methyltransferase, is able to modify FGF-223. PRMT5 in the methylosome complex in the cytoplasm stimulates SMN-mediated snRNP assembly by methylating Sm proteins [46,47]. However, it has been demonstrated that SMN is also able to bind to methylated or unmethylated GAR1 respectively to a comparable extent [23]. SMN binds to the arginine/glycine domains of the snoRNP proteins GAR1 and fibrillarin, but additional domains or a particular conformation may be important for high-affinity binding [40]. Therefore arginine methylation may not be an absolute requirement of SMN for selecting binding partners.
In the present study, we also showed binding of recombinant FGF-223, as well as FGF-223 from Schwann cell lysates, to human full-length SMN protein. Rat and human SMN proteins are almost identical, with only a few different residues (results not shown). Analyses of FGF-223 binding to SMN-deletion mutants limit the interaction to a sequence (residues 1–90) in the N-terminus of SMN. This binding region overlaps with the sequence (residues 13–44) responsible for Gemin2/SIP1 binding. Other studies indicate Gemin2 binding to amino acid residues 52–91 of human SMN [48]. Exon 2 of SMN contains a domain (residues 29–51) which is able to bind in vitro to nucleic acids [38] and to p53 [39]. This element binds RNA homopolymeric poly(G) sequences, and is conserved across species. Its nucleic-acid-binding activity can be modulated by sequences of exons 3 and 4 (residues 52–91 and 92–158) respectively, with the latter including the Tudor domain [38,49]. A protein mutant mimicking a frameshift mutation of a SMA patient in the Tudor domain by a 5 bp microdeletion demonstrated reduced RNA-binding capacity [49]. Interestingly, the FGF-223 and nucleic-acid-binding domain displays homology with the known nucleic-acid-binding domain of high-mobility-group proteins of the HMGB-type (HMGB-box, formerly HMG-1/2-box) [38,50]. SMN is able to bind directly in a sequence-specific manner to the SL1 loop sequence of U1 snRNA. This interaction is independent of Sm core assembly [40]. However, deletion studies with SMN to map this interaction have not yet been performed. SMN is able to bind U2, U4 and U5 snRNAs with an affinity in the low nanomolar concentration [41]. In the present study, we demonstrate FGF-223 interaction with the RNA-binding domain of SMN and interaction of SMN with U2 snRNA. However, this interaction could not be competed by FGF-223. SMN self-associates by domains encoded by exons 3 and 7 [48]. In oligomeric conformation, U2 snRNA and FGF-223 could bind to different positions of the structure, e.g. differentially bind to central or marginal structural elements, so that no direct competition can take place, despite binding to the same sequence. Importantly, FGF-223 does not generally bind to acidic polymers, like nucleic acids, since direct binding of FGF-223 to U2 RNA could not be observed.
It is conceivable that FGF-223 could compete with Gemin2 and/or p53 respectively for binding to SMN. In this context, it is noteworthy that deletion of the Gemin2-binding domain of SMN diminished also the association of Sm proteins with SMN, suggesting a scenario where FGF-223 binding could influence Sm binding to SMN. For example, FGF-223 could destabilize the Gemin2–SMN interaction, which in turn would lead to unstable Gemin2 and subsequent reduction of its cellular levels [51,52]. In vivo, reduced Gemin2 levels result in disturbance of snRNP assembly and enhanced motoneuron degeneration [53]. With regard to the development of SMA, the SMN–FGF-223 interaction could influence the stability of SMN–Gemin2 complexes and influence the phenotypic impact of SMN mutations in the development of the disease.
In the nucleus, FGF-223 localizes to nuclear gems [24] and is chromatin-associated [24,54]. However, the molecular basis of this association is still unknown. A putative interaction of FGF-223 with HMGB proteins, or with proteins with a HMGB-box, respectively, could provide the molecular basis for the observed association of high-molecular-mass FGF-2 with chromatin. Additionally, we could detect FGF-223 partly co-localized with SMN in the cytoplasm of Schwann cells (results not shown), indicating SMN–FGF-223 complex formation during snRNP assembly. To test a possible association of FGF-223 with snRNPs biochemically, we performed co-immunoprecipitations, and tested for the presence of distinct snRNAs. U2 and U4 snRNAs could be coimmunoprecipitated, strongly suggesting an association of FGF-223 with snRNPs via SMN in the cytoplasm. Concomitant with the localization of the interaction is the finding that U6 snRNA is not associated with FGF-223: U6 snRNA does not leave the nucleus for snRNP assembly [42]. The data also demonstrate that the FGF-2–U2 and U4 snRNA association is not the result of a non-specific binding during the course of the experiment. Since no FGF-223 binding to U2 snRNA could be detected in the present study, the observed snRNA–U2/U4 association can be considered as the result of an indirect interaction.
FGF-223 is a growth factor, which exaggerates its functions not only by a classical signalling pathway of an extracellular factor, but also as an intracellular/intranuclear protein. A specific nuclear role of high-molecular-mass FGF-2 in tumour progression has been elucidated recently [55]. Several other growth factors and growth factor receptors have been reported to be localized in the nucleus {e.g. FGF-1, CNTF (ciliary neurotrophic factor) [56]}. FGF-2 can enter the cell after binding to tyrosine-kinase receptors on the cell membrane and migrate to the nucleus [49,57]. In this way, interaction with snRNPs or a subfraction thereof could take place as a possibility of a cell integrating extracellular signals of a growth factor with splicing processes. The intracellular function of FGF-223 may include direct modulation of splicing assembly, alternative splicing or splicing efficiency by binding to SMN and snRNPs.
Acknowledgments
We are grateful to Hella Brinkmann for expert technical help, I. Heike for preparation of Figures and Dr M. van Griensven for providing space in the radioactive laboratory. This work was supported by the following grants to P. C.: research grants from the Deutsche Gesellschaft für Muskelkranke (DGM; German Foundation for Muscular Diseases) and the Fritz Thyssen Foundation, Köln, Germany.
References
- 1.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Jablonka S., Rossoll W., Schrank B., Sendtner M. The role of SMN in spinal muscular atrophy. J. Neurol. 2000;247(Suppl. 1):I37–I42. doi: 10.1007/s004150050555. [DOI] [PubMed] [Google Scholar]
- 3.Sendtner M. Molecular mechanisms in spinal muscular atrophy: models and perspectives. Curr. Opin. Neurol. 2001;14:629–634. doi: 10.1097/00019052-200110000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Talbot K. Spinal muscular atrophy. J. Inherit. Metab. Dis. 1999;22:545–554. doi: 10.1023/a:1005516625866. [DOI] [PubMed] [Google Scholar]
- 5.Terns M. P., Terns R. M. Macromolecular complexes: SMN – the master assembler. Curr. Biol. 2001;11:R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 6.Meister G., Eggert C., Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 7.Buhler D., Raker V., Luhrmann R., Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 8.Pellizzoni L., Kataoka N., Charroux B., Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzoni L., Baccon J., Charroux B., Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- 10.Meister G., Buhler D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 12.Meister G., Buhler D., Laggerbauer B., Zobawa M., Lottspeich F., Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 13.Charroux B., Pellizzoni L., Perkinson R. A., Shevchenko A., Mann M., Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charroux B., Pellizzoni L., Perkinson R. A., Yong J., Shevchenko A., Mann M., Dreyfuss G. Gemin4: a novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubitz A. K., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 16.Baccon J., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- 17.Pellizzoni L., Baccon J., Rappsilber J., Mann M., Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 18.Hebert M. D., Szymczyk P. W., Shpargel K. B., Matera A. G. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friesen W. J., Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J. Biol. Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 20.Friesen W. J., Massenet S., Paushkin S., Wyce A., Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 21.Brahms H., Meheus L., de Brabandere V., Fischer U., Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones K. W., Gorzynski K., Hales C. M., Fischer U., Badbanchi F., Terns R. M., Terns M. P. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- 23.Whitehead S. E., Jones K. W., Zhang X., Cheng X., Terns R. M., Terns M. P. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 2002;277:48087–48093. doi: 10.1074/jbc.M204551200. [DOI] [PubMed] [Google Scholar]
- 24.Claus P., Döring F., Gringel S., Müller-Ostermeyer F., Fuhlrott J., Kraft T., Grothe C. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J. Biol. Chem. 2003;278:479–485. doi: 10.1074/jbc.M206056200. [DOI] [PubMed] [Google Scholar]
- 25.Klein S., Carroll J. A., Chen Y., Henry M. F., Henry P. A., Ortonowski I. E., Pintucci G., Beavis R. C., Burgess W. H., Rifkin D. B. Biochemical analysis of the arginine methylation of high molecular weight fibroblast growth factor-2. J. Biol. Chem. 2000;275:3150–3157. doi: 10.1074/jbc.275.5.3150. [DOI] [PubMed] [Google Scholar]
- 26.Burgess W. H., Bizik J., Mehlman T., Quarto N., Rifkin D. B. Direct evidence for methylation of arginine residues in high molecular weight forms of basic fibroblast growth factor. Cell. Regul. 1991;2:87–93. doi: 10.1091/mbc.2.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Florkiewicz R. Z., Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc. Natl. Acad. Sci. U.S.A. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florkiewicz R. Z., Baird A., Gonzalez A. M. Multiple forms of bFGF: differential nuclear and cell surface localization. Growth Factors. 1991;4:265–275. doi: 10.3109/08977199109043912. [DOI] [PubMed] [Google Scholar]
- 29.Pasumarthi K. B., Kardami E., Cattini P. A. High and low molecular weight fibroblast growth factor-2 increase proliferation of neonatal rat cardiac myocytes but have differential effects on binucleation and nuclear morphology. Evidence for both paracrine and intracrine actions of fibroblast growth factor-2. Circ. Res. 1996;78:126–136. doi: 10.1161/01.res.78.1.126. [DOI] [PubMed] [Google Scholar]
- 30.Amalric F., Bouche G., Bonnet H., Brethenou P., Roman A. M., Truchet I., Quarto N. Fibroblast growth factor-2 (FGF-2) in the nucleus: translocation process and targets. Biochem. Pharmacol. 1994;47:111–115. doi: 10.1016/0006-2952(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 31.Huber K., Meisinger C., Grothe C. Expression of fibroblast growth factor-2 in hypoglossal motoneurons is stimulated by peripheral nerve injury. J. Comp. Neurol. 1997;382:189–198. [PubMed] [Google Scholar]
- 32.Grothe C., Unsicker K. Basic fibroblast growth factor in the hypoglossal system: specific retrograde transport, trophic, and lesion-related responses. J. Neurosci. Res. 1992;32:317–328. doi: 10.1002/jnr.490320304. [DOI] [PubMed] [Google Scholar]
- 33.Tennekoon G. I., Yoshino J., Peden K. W., Bigbee J., Rutkowski J. L., Kishimoto Y., DeVries G. H., McKhann G. M. Transfection of neonatal rat Schwann cells with SV-40 large T antigen gene under control of the metallothionein promoter. J. Cell Biol. 1987;105:2315–2325. doi: 10.1083/jcb.105.5.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghidelli S., Claus P., Thies G., Wisniewski J. R. High mobility group proteins cHMG1a, cHMG1b, and cHMGI are distinctly distributed in chromosomes and differentially expressed during ecdysone dependent cell differentiation. Chromosoma. 1997;105:369–379. doi: 10.1007/BF02529752. [DOI] [PubMed] [Google Scholar]
- 35.Müller-Ostermeyer F., Claus P., Grothe C. Distinctive effects of rat fibroblast growth factor-2 isoforms on PC12 and Schwann cells. Growth Factors. 2001;19:175–191. doi: 10.3109/08977190109001085. [DOI] [PubMed] [Google Scholar]
- 36.Guichet A., Copeland J. W., Erdelyi M., Hlousek D., Zavorszky P., Ho J., Brown S., Percival-Smith A., Krause H. M., Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature (London) 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 37.Selenko P., Sprangers R., Stier G., Buhler D., Fischer U., Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 38.Lorson C. L., Androphy E. J. The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum. Mol. Genet. 1998;7:1269–1275. doi: 10.1093/hmg/7.8.1269. [DOI] [PubMed] [Google Scholar]
- 39.Young P. J., Day P. M., Zhou J., Androphy E. J., Morris G. E., Lorson C. L. A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J. Biol. Chem. 2002;277:2852–2859. doi: 10.1074/jbc.M108769200. [DOI] [PubMed] [Google Scholar]
- 40.Yong J., Pellizzoni L., Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yong J., Golembe T. J., Battle D. J., Pellizzoni L., Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brow D. A., Vidaver R. M. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA. 1995;1:122–131. [PMC free article] [PubMed] [Google Scholar]
- 43.Jans D. A., Hassan G. Nuclear targeting by growth factors, cytokines, and their receptors: a role in signaling? BioEssays. 1998;20:400–411. doi: 10.1002/(SICI)1521-1878(199805)20:5<400::AID-BIES7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 44.Moy F. J., Seddon A. P., Bohlen P., Powers R. High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:13552–13561. doi: 10.1021/bi961260p. [DOI] [PubMed] [Google Scholar]
- 45.Pintucci G., Quarto N., Rifkin D. B. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol. Biol. Cell. 1996;7:1249–1258. doi: 10.1091/mbc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meister G., Eggert C., Buhler D., Brahms H., Kambach C., Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 48.Young P. J., Man N. T., Lorson C. L., Le T. T., Androphy E. J., Burghes A. H., Morris G. E. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum. Mol. Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- 49.Bertrandy S., Burlet P., Clermont O., Huber C., Fondrat C., Thierry-Mieg D., Munnich A., Lefebvre S. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum. Mol. Genet. 1999;8:775–782. doi: 10.1093/hmg/8.5.775. [DOI] [PubMed] [Google Scholar]
- 50.Wisniewski J. R., Schulze E. Insect proteins homologous to mammalian high mobility group protein 1: characterization and DNA-binding properties. J. Biol. Chem. 1992;267:17170–17177. [PubMed] [Google Scholar]
- 51.Wang J., Dreyfuss G. A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem. 2001;276:9599–9605. doi: 10.1074/jbc.M009162200. [DOI] [PubMed] [Google Scholar]
- 52.Wang J., Dreyfuss G. Characterization of functional domains of the SMN protein in vivo. J. Biol. Chem. 2001;276:45387–45393. doi: 10.1074/jbc.M105059200. [DOI] [PubMed] [Google Scholar]
- 53.Jablonka S., Holtmann B., Meister G., Bandilla M., Rossoll W., Fischer U., Sendtner M. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10126–10131. doi: 10.1073/pnas.152318699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun G., Doble B. W., Sun J. M., Fandrich R. R., Florkiewicz R., Kirshenbaum L., Davie J. R., Cattini P. A., Kardami E. CUG-initiated FGF-2 induces chromatin compaction in cultured cardiac myocytes and in vitro. J. Cell Physiol. 2001;186:457–467. doi: 10.1002/1097-4652(2000)9999:999<000::AID-JCP1044>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 55.Thomas-Mudge R. J., Okada-Ban M., Vandenbroucke F., Vincent-Salomon A., Girault J. M., Thiery J. P., Jouanneau J. Nuclear FGF-2 facilitates cell survival in vitro and during establishment of metastases. Oncogene. 2004;23:4771–4779. doi: 10.1038/sj.onc.1207638. [DOI] [PubMed] [Google Scholar]
- 56.Olsnes S., Klingenberg O., Wiedlocha A. Transport of exogenous growth factors and cytokines to the cytosol and to the nucleus. Physiol. Rev. 2003;83:163–182. doi: 10.1152/physrev.00021.2002. [DOI] [PubMed] [Google Scholar]
- 57.Bossard C., Laurell H., Van den Berghe L., Meunier S., Zanibellato C., Prats H. Translokin is an intracellular mediator of FGF-2 trafficking. Nat. Cell Biol. 2003;5:433–439. doi: 10.1038/ncb979. [DOI] [PubMed] [Google Scholar]