Differential Regulation of Ceramide Synthase Components LAC1 and LAG1 in Saccharomyces cerevisiae (original) (raw)
Abstract
In Saccharomyces cerevisiae, the essential ceramide synthase reaction requires the presence of one of a homologous pair of genes, LAG1 and LAC1. Mutants that lack both of these genes cannot produce ceramide and exhibit a striking synthetic growth defect. While the regulation of ceramide production is critical for the control of proliferation and for stress tolerance, little is known of the mechanisms that ensure proper control of this process. The data presented here demonstrate that the pleiotropic drug resistance (Pdr) regulatory pathway regulates the transcription of multiple genes encoding steps in sphingolipid biosynthesis, including LAC1. The zinc cluster transcriptional activators Pdr1p and Pdr3p bind to Pdr1p/Pdr3p-responsive elements (PDREs) in the promoters of Pdr pathway target genes. LAC1 contains a single PDRE in its promoter, but notably, LAG1 does not. Reporter gene, Northern blot, and Western blot assays indicated that the expression level of Lac1p is approximately three times that of Lag1p. Detailed analyses of the LAC1 promoter demonstrated that transcription of this gene is inhibited by the presence of the transcription factor Cbf1p and the anaerobic repressor Rox1p. LAG1 transcription was also elevated in _cbf1_Δ cells, indicating at least one common regulatory input. Although a hyperactive Pdr pathway altered the profile of sphingolipids produced, the loss of either LAC1 or LAG1 alone failed to produce further changes. Two other genes involved in sphingolipid biosynthesis (LCB2 and SUR2) were found to contain PDREs in their promoters and to be induced by the Pdr pathway. These data demonstrate extensive coordinate control of sphingolipid biosynthesis and multidrug resistance in yeast.
Sphingolipids are important structural components of eukaryotic membranes and are thought to provide rigidity to these structures (for a review, see reference 47). Along with this architectural role, sphingolipids and their precursor molecules are major participants in the regulation of cell proliferation and function. The first bioactive signaling lipids formed during de novo sphingolipid biosynthesis are the long-chain bases (LCBs) dihydrosphingosine and sphingosine (phytosphingosine in Saccharomyces cerevisiae) (12, 58). LCBs are substrates that are required for ceramide synthase to produce ceramide. Ceramide has been the focus of intensive research due to its control of apoptosis and its action as a growth inhibitor (6, 48). LCBs can also be phosphorylated to produce phosphorylated LCBs (LCBPs). Interestingly, sphingosine-1-phosphate, an LCBP found in mammalian cells, acts to oppose ceramide-induced apoptosis and is thought to be a stimulator of proliferation (43). The balance between ceramide and LCBP production is a crucial determinant of cell fate and has been dubbed the ceramide/LCBP rheostat (66).
Studies of sphingolipid metabolism in S. cerevisiae have provided a wealth of information on the structure and function of this pathway (for reviews, see references 11, 53, and 58). Genetic approaches with S. cerevisiae permitted the isolation of a large number of genes encoding enzymes that influence sphingolipid biosynthesis. Physiological studies established that stress conditions lead to a transient elevation of LCB and LCBP levels along with a sustained increase in ceramide concentration (13, 27, 63). While moderate increases in LCBPs are required for normal stress tolerance, the high-level accumulation of these compounds is toxic (43, 44, 63). Similarly, ceramide production in S. cerevisiae must also be limited, as mutants lacking the IPC synthase step (ceramide to inositolphoshorylceramide [IPC]) (52) may be lethal due to the excessive production of ceramide (59).
While the sphingoid base kinases of S. cerevisiae have been known for several years, only recently have the genes required for ceramide synthase activity been described. These two homologous genes, designated LAG1 and LAC1, were first identified on the basis of Lag1p influencing the aging of yeast (15, 28). Later work demonstrated that the presence of either Lag1p or Lac1p was required for the normal delivery of glycosylphosphatidylinositol-anchored proteins to the plasma membrane (2). Additionally, a _lag1_Δ _lac1_Δ mutant was found to have a major growth defect that others correlated with an inability to synthesize ceramide (2, 21, 59). A strain lacking only Lag1p or Lac1p did not show a growth defect, ceramide synthesis block, or glycosylphosphatidylinositol-anchored protein delivery delay.
Despite the identification of most, if not all, of the genes and proteins involved in the key steps of the yeast sphingolipid pathway (reviewed in references 11 and 53) (Fig. 1), very little is known about the regulation of this pathway. Recent experiments have provided evidence that casein kinase 2 is required for normal ceramide synthase function (35), while the only report on the transcriptional regulation of genes in the sphingolipid pathway in any organism demonstrates the involvement of the multiple, or pleiotropic, drug resistance (Pdr) pathway in the activation of IPT1 gene transcription (23). Ipt1p is required for the last step in yeast sphingolipid synthesis, the production of the plasma membrane component mannosyldiinositolphosphorylceramide [M(IP)2C] (14). Pdr1p and Pdr3p belong to the Zn2Cys6-cluster transcription factors and are the master regulators of multidrug resistance in yeast (reviewed in references 36, 50, and 73). These transcription factors activate the expression of the multidrug ATP-binding cassette transporters Pdr5p, Snq2p, and Yor1p, which are involved in the detoxification of a plethora of xenobiotics, including some anticancer drugs, azole antifungals, and most other classes of currently available antifungals used for human treatment (37).
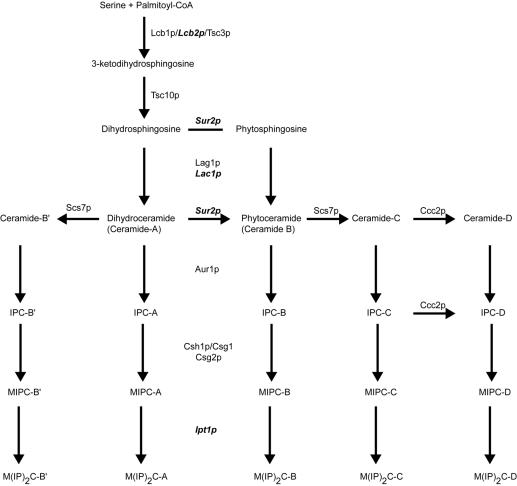
FIG. 1.
Biosynthesis of sphingolipids in S. cerevisiae. A simplified diagram listing key steps in sphingolipid synthesis is shown. Proteins known to be involved at each step are listed by genetic designation. Gene products that are responsive to Pdr pathway regulation are indicated in bold italics.
Here we provide evidence that the influence of the Pdr pathway extends to several other steps in S. cerevisiae sphingolipid biosynthesis. The LAC1, but not LAG1, gene is a transcriptional target of the Pdr pathway and is responsive to oxygen levels due to repression by the anaerobic transcriptional repressor Rox1p (reviewed in reference 29). Loss of the pleiotropically acting transcription factor Cbf1p causes the transcription of both LAC1 and LAG1 to increase. Finally, genes encoding components of the serine palmitoyltransferase (LCB2) and dihydrosphingosine hydroxylase (SUR2) were also found to be regulated by the Pdr pathway. These data provide strong support for the idea that a major in vivo role of Pdr transcriptional control is to provide coordinate control of membrane composition through the regulation of sphingolipid biosynthesis and the membrane transporter proteins that function in the resulting lipid environment.
MATERIALS AND METHODS
Strains, media, and β-galactosidase assay.
The S. cerevisiae strains used for this study are listed in Table 1. Yeast cells were grown on rich YPD (2% yeast extract, 1% peptone, 2% glucose) (Becton Dickinson or Difco) medium or on synthetic complete medium lacking appropriate auxotrophic components (60). Yeast strains were transformed by the lithium acetate method (18). β-Galactosidase assays were performed as described previously, with _o_-nitrophenyl-β-d-galactopyranoside (ONPG) (19) or chlorophenol red β-d-galactopyranoside (Roche) as a substrate as described in the Clontech manual (Clontech, Palo Alto, Calif.). Growth was monitored by measuring the optical density at 595 nm on a BioTek EL800 microplate reader.
TABLE 1.
S. cerevisiae strains used for this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BY4742 | _MAT_α his3_-Δ_1 leu2_-Δ_0 lys2_-Δ_0 ura3_-Δ_0 | Research Genetics |
| BY4742 Δ_rox1_ | BY4742 _rox1_-Δ::kanMX4 | Research Genetics |
| BY4742 Δ_lac1_ | BY4742 _lac1_-Δ::kanMX4 | Research Genetics |
| BY4742 Δ_cbf1_ | BY4742 _cbf1_-Δ::kanMX4 | Research Genetics |
| SEY6210 | _MAT_α leu2-3,-112 ura3-52 lys2-801 trp1_-Δ_901 his3_-Δ_200 suc2_-Δ_9 Mel− | Scott Emr |
| SEY6210 ρ0 | SEY6210 [ρ0] | 47 |
| SEY6210 Δ_pdr1_ | SEY6210 pdr1_-Δ_2::hisG | 47 |
| SEY6210 Δ_pdr3_ | SEY6210 pdr3_-Δ_1::hisG | 47 |
| SEY6210 Δ_pdr1_ Δ_pdr3_ | SEY6210 pdr1_-Δ_2::hisG pdr3_-Δ_1::hisG | 47 |
| SEY6210 ρ0 Δ_pdr1_ | SEY6210 [ρ0] pdr1_-Δ_2::hisG | 47 |
| SEY6210 ρ0 Δ_pdr3_ | SEY6210 [ρ0] pdr3_-Δ_1::hisG | 47 |
| SEY6210 ρ0 Δ_pdr1_ Δ_pdr3_ | SEY6210 [ρ0] pdr1_-Δ_2::hisG pdr3_-Δ_1::hisG | 47 |
| MK0201-2 | SEY6210 _lac1_-Δ::kanMX4 | This study |
| MK0112-1 | SEY6210 mPDRE-LAC1 | This study |
| MK0220-22 | SEY6210 _lag1_-Δ::dpl200 | This study |
| MK0228-7-2 | SEY6210 _lag1_-Δ::dpl200 mPDRE-LAC1 | This study |
| MK0211-1 | SEY6210 [ρ0] _lac1_-Δ::kanMX4 | This study |
| MK0221-2 | SEY6210 [ρ0] _lag1_-Δ::dpl200 | This study |
To introduce the XhoI mutation of the LAC1 Pdr1p/Pdr3p-responsive element (PDRE) into the yeast genome, we used the pop in-pop out strategy (56). First, the BamHI/NotI fragment of pMK0101-7 was cloned into the same sites of the integrative plasmid pRS306 (62). This plasmid was linearized with XbaI and transformed into the SEY6210 strain, with selection for URA3+ transformants. The presence of the mutation was verified by restriction analysis of colony PCR products amplified with primers CGC CAT TCG CCA TTC AGG (lac1-06BL) and AGG GCT TGG CTT TAT TGT CG (lac1-06U). To remove the integrating plasmid and wild-type LAC1 promoter sequences, we counterselected the URA3 marker from the correct transformants on 5-fluoroorotic-acid-containing plates as described previously (3). Since this procedure yields a large population of clones reverting to the wild-type PDRE, the clones were again analyzed for the presence of the XhoI mutation by colony PCR with primers AGG GCT TGG CTT TAT TGT CG (lac1-06U) and TCC TGG AGG GCT GTC ATT G (lac1-06L). The resulting strain was named MK0112-1.
To introduce the _lac1_Δ allele, we amplified the corresponding region from the genomic DNA of BY4742 _lac1-_Δ::kanMX with primers TTC GTT ACC GTT TTT CTG TTC CAT AC (lac1-07U) and TTA GCG GCA CGA AAG CAA G (lac1-07L) and transformed it into the wild-type SEY6210 strain, producing MK0201-2. Transformants were verified by colony PCR with primers GTC TTT TCC GGT CAT TCC AAC AA (lac1-09U) and ACA ATT CGG TAT CGC TGC TTC TG (lac1-09L).
The LAG1 deletion cassette was generated by PCR with the primers (LAG1 sequence is in lowercase) tag aca gtg ttg aga gtg aac tcc aag ata cag aga aac tga aga aat aac gac aac ATG GTT TTC CCA GTC ACG ACG TT (lag1-05U) and ttg gcc ttc ata cag ggg gga aat cat atg atg ata cgt att ctc ctt aag ata cgt TTA TGT GGA ATT GTG AGC GGA TA (lag1-05L) and the plasmid pDDB57 (71), provided by Aaron Mitchell (Columbia University, New York, N.Y.). After the transformation of strain SEY6210 to the Ura+ phenotype, clones were verified by colony PCR with primers CAG CCC AGG GGT AGT GAA AAA CGA TGA A (lag1-02U) and CGC TTG GCC AGA AAT AAC GAA AAA TGA ACA (lag1-02L). Positive clones were treated with 5-fluoroorotic-acid-containing medium, and Ura− colonies were isolated. The correct loopout of the CaURA3 marker was verified by PCR.
Immunological methods.
Protein extracts were prepared by glass bead lysis as previously described (31). Protein concentrations were determined by use of a Bio-Rad protein assay kit as recommended by the supplier. Equal amounts of protein were resuspended in sodium dodecyl sulfate (SDS) loading buffer prior to analysis by Western blotting (69). Blots were incubated with an anti-hemagglutinin (HA) (Babco) or anti-Vph1p (Molecular Probes) antibody and were developed with an enhanced chemiluminescence detection system (Pierce).
RNA isolation, radiolabeling, and Northern blotting.
Total yeast RNA was isolated as described by Schmitt et al. (57). The RNA was fractionated through a 1% agarose-formaldehyde gel, capillary blotted onto a nylon membrane (Nytran Super Charge; Schleicher & Schuell), and hybridized by standard procedures (55). The LAC1 hybridization probe was generated by PCR amplification of a segment of the LAC1 coding sequence from genomic DNA with primers lac1-08U (AGC CAA GCC CTT CAA ACA A) and lac1-08L (AAC TTT CAT CAC TTT CTT CGT CTG). The resulting product was labeled with [α-32P]dCTP by a random priming procedure using the Radprime DNA labeling system (Invitrogen). The LAG1 hybridization probe was generated by PCR amplification of almost the complete open reading frame from genomic DNA with primers lag1-03U (TTA ATG CAA AAA CAA GAA GAC GAA ACT) and lag1-03L (GGC GCA GAT AGA TCC AAA AGA AC). This product was labeled with [α-32P]dCTP by PCR with the same primers according to a standard protocol (55).
Construction of reporter plasmids.
For the generation of a LAC1 promoter-lacZ reporter plasmid, the corresponding region was amplified from the genomic DNA with primers lac1f forward (ccg gag atc tct CAT TTT CTG TCG CTC TTA TGA TCC [with a new BglII site shown in lowercase letters]) and lac1 f rev (cgg gat ccG TCG ACA TAG CTC TTG TTT ATT GAT ACT G [with a new BamHI site shown in lowercase letters]). The BamHI/BglII fragment of the resulting PCR product was cloned into the BamHI site of the pSEYC102 reporter plasmid (17), yielding pMK0105-3. It was also cloned into pCR2.1 TOPO (Invitrogen), yielding pCR2.1 TOPOw1, for subsequent use in footprinting. A clone with the reverse insert orientation and the CAP1 promoter in frame with lacZ was designated pMK0105-2. For the generation of a _LAG1_-lacZ reporter plasmid, the promoter was amplified as a BamHI fragment (indicated in each primer in lowercase letters) with primers lag1 hse1-08U (ctg gat cCA TGT TGT CGT TAT TTC TTC AGT TTC TCT G) and lag1 hse1-08L (gat gga tcC ATT TTT GTC AAC GGT TTC CTT AGA TAG) and then cloned into the BamHI site of pSEYC102, resulting in pMK0301-L18.
For construction of the _SUR2_-lacZ reporter plasmid, the promoter was amplified with the primers SUR2 forward (ccg aat TCG AAT CAC ATC CTA C [with the introduced EcoRI site in lowercase]) and SUR2 Reverse (ccg gaT CCA TCG TAT ATT TTC T [with the introduced BamHI site in lowercase]). The EcoRI/BamHI fragment was then cloned between the EcoRI and BamHI sites of pSEYC102, yielding the pSEYC-SUR2 plasmid. For construction of the _LCB2_-lacZ reporter plasmid, the promoter was amplified with primers LCB2 forward (GGA AAA TTA AAG TTT) and LCB2 reverse (att cca gga tcC ATA ATC ATT ACT TTT) (mismatched positions are shown in lowercase). The resulting PCR product was cloned into pCR2.1 TOPO. The EcoRI/BamHI fragment was then cloned between the same sites of pSEYC102, yielding the pSEYC-LCB2 plasmid.
DNase I footprinting.
DNase I protection experiments were performed essentially as described before (30). The Pdr1p and Pdr3p DNA binding domains were overproduced in Escherichia coli as described previously (30, 32). Purified Rox1p fused to maltose binding protein was provided by Richard Zitomer (State University of New York at Albany). For Pdr1p and Pdr3p binding experiments, an XbaI/NotI fragment corresponding to the region from the internal XbaI site in the LAC1 promoter to −754 bp upstream was isolated. It was labeled at the XbaI site with [γ-32P]ATP by T4 polynucleotide kinase. For Rox1p footprint analysis, an XbaI/SpeI fragment, also labeled at the same XbaI site but extending across the LAC1 ATG, was used.
Deletion mutagenesis of LAC1 promoter.
Deletion mutations in the _LAC1_-lacZ reporter pMK0105-3 plasmid were generated by digestion with the following restriction enzymes and by blunt end religation after Klenow treatment when necessary: for pMK0105-3-Δ1-2, SmaI/XbaI; for pMK0105-3-Δ2-9, SmaI/SacII; for pMK0105-3-Δ3-3, PmlI; for pMK0105-3-Δ4-6, SmaI/PmlI; for pMK0105-3-Δ5-1, XbaI/BspEI; for pMK0105-3-Δ6-1, XmaI/BspEI; for pMK0105-3-Δ7-22, SacII/XbaI; for pMK0105-3-Δ8-18, PmlI/SacII. For mutation of the LAC1 PDRE to an XhoI restriction site, the promoter was amplified with two primer sets: lac1f forward with the primer GTC TAG CAT GAT CAG GTA CAA TGC and primer CCA ACA AAG TTG CTA GAT CCt CGa gAC CAT TTT CGT CAT T (mutant residues are in lowercase) with lac1f rev. The resulting PCR products were diluted, mixed together, and amplified again with the flanking primers to give the complete, mutated promoter. The mutant promoter was then cloned into pCR2.1 TOPO to give pMK0101-7. The BamHI/BglII fragment of pMK0101-7 was cloned into the BamHI site of pSEYC102, yielding pMK0103-3. A complete deletion of the LAC1 PDRE was generated in a similar way, except that a different mutagenic primer, CCA ACA AAG TTG CTA GA-ACC ATT TTC GTC ATT (deletion site indicated by a dash), was used to produce pMK0104-4. The same strategy was used to alter the Rox1p binding site to an AflII restriction site, except that the primers used were Lac1-12U (GAT TTT TAA GTT ATT TTT CTc TTa agT CCC TCC TTG GCT TGT TGC) and Lac1-12L (GCA ACA AGC CAA GGA GGG Act tAA gAG AAA AAT AAC TTA AAA ATC) (the position of the new AflII site is indicated in lowercase).
Epitope tagging.
The LAC1 gene was amplified from genomic DNA with the primers TTT AcT AgT GAA ACA ATT CGG TAT CGC TGC TTC TGA, introducing a SpeI site (in lowercase) and TTT GTC GGT TGA TCG CTA TTG TTT TTA TTA, containing the endogenous SalI site. After SalI-SpeI digestion, the fragment was cloned under the control of the wild-type LAC1 promoter into the pCR2.1 TOPOw1 clone, resulting in pMK0201-13. Next, the entire gene was cut out from pMK0201-13 by SpeI and NotI and cloned into SpeI- and NotI-cut pRS313 to generate pMK0204-7. For the introduction of the N-terminal 3× HA epitope, the pMK0204-7 plasmid was amplified with primers lac1-14U (gac gtc ata cgg ata gcc cgc ata gtc agg aac atc gta tgg gta CAT AGC TCT TGT TTA TTG ATA CTG TGT CTA TCT AAA TG) and lac1-14L (ccg gac tat gca gga tcc tat cca tat gac gtt cca gat tac gct TCG ACA ATA AAG CCA AGC CCT TC) (epitope tag sequence in lowercase) by the Exsite procedure (Stratagene) to generate pMK0204-7-3Ha. The LAG1 gene was amplified with primers Lag1-06U (CTT CCT TCA CCA AAT CGC AAA CAT CTA A) and Lag1-06L (CAA GGG TCC CAA CTA TCG CAA TCA T) and cloned into pCR2.1 TOPO to generate pMK0211-2. This clone was then digested with XhoI and SpeI and cloned into the same sites of pRS314 to generate pMK0213-1. For the introduction of the N-terminal 3× HA epitope, the pMK0213-1 plasmid was amplified with primers lag1-07U (gac gtc ata cgg ata gcc cgc ata gtc agg aac atc gta tgg gta CAT GTT GTC GTT ATT TCT TCA GTT TCT CTG) and lag1-07L (ccg gac tat gca gga tcc tat cca tat gac gtt cca gat tac gct ACA TCA GCT ACG GAC AAA TCT ATC G) by the Exsite procedure (Stratagene) to give pMK0213-1-3Ha.
Sphingolipid analysis.
Ten optical density units (at 600 nm) of exponentially growing cells was harvested, and the cells were resuspended in 1 ml of synthetic complete medium minus Leu and labeled by the addition of 100 μCi of [3H]serine. The cells were incubated for 40 min at 30°C, diluted with prewarmed fresh medium, and incubated for another 80 min. Labeling was terminated by the addition of NaF and NaN3. The cells were harvested and broken with glass beads, and lipids were extracted with chloroform-methanol (1:1 [vol/vol]). Phospholipids were deacylated by treatment with a mild base and sphingolipids were analyzed by thin-layer chromatography using solvent system A (chloroform-methanol-NH4OH [90:70:1 {vol/vol}]) for the analysis of IPC species or B (chloroform-methanol-acetic acid-water [16:6:4:1.6 {vol/vol}]) for the analysis of ceramide species. Radioactivity was detected and quantified by two-dimensional radioscanning on a Berthold Tracemaster 40 instrument. The positions of the various labeled sphingolipid metabolites were determined by comparison to known standards as described previously (16).
RESULTS
Control of expression of a component of ceramide synthase by Pdr1p/Pdr3p.
The work of several laboratories has established that the expression of genes conferring the multiple or pleiotropic drug-resistant phenotype on cells is primarily responsive to the actions of two different but related Zn2Cys6-cluster-containing transcription factors, Pdr1p and Pdr3p (1, 7). Gain-of-function mutant forms of these transcriptional regulatory proteins lead to the overproduction of target genes, such as the ATP-binding cassette transporter protein Pdr5p, which in turn dramatically increase resistance to a broad range of unrelated drugs (49). Pdr1p and Pdr3p bind to a DNA element (PDRE) that is present in the promoter regions of target genes and acts to stimulate the transcription of these loci (reviewed in references 36, 50, and 73).
Along with membrane transporters such as Pdr5p, the IPT1 gene encoding the enzyme inositol phosphotransferase has been found by recent experiments to be transcriptionally regulated by Pdr1p and Pdr3p (23). IPT1 contains a single PDRE in its promoter and is also induced in response to the loss of the mitochondrial genome in a Pdr3p-dependent fashion. Since Ipt1p is required for sphingolipid biosynthesis, we examined the promoters of genes encoding other steps in the pathway to determine if any of these might also be regulated by Pdr1p and Pdr3p. A search of 1,000 bp upstream of all known genes corresponding to steps in sphingolipid biosynthesis for a consensus PDRE (TCCGCGGA) found three promoters, in addition to IPT1, with at least seven of eight matches: they were LCB2, SUR2, and LAC1. LCB2 encodes a subunit of the serine palmitoyltransferase enzyme catalyzing the first committed step in sphingolipid biosynthesis (51), while SUR2 directs the production of a hydroxylase enzyme that modifies the LCB that is ultimately present in ceramide (22). LAC1, along with its homologue LAG1, encodes a protein that is required for ceramide synthase activity (21, 59). Interestingly, LAG1 does not contain a PDRE in its promoter, suggesting that the expression of LAC1 and LAG1 is differentially controlled. To compare the transcriptional regulation of these two homologous genes, we assessed the mRNA levels of these two loci.
LAC1, but not LAG1, transcription is responsive to changes in Pdr1p/Pdr3p activity.
Total RNAs were prepared from an isogenic series of strains containing various alleles of PDR1 and/or PDR3 in addition to a strain lacking mitochondrial DNA (ρ0). Equal amounts of RNA were electrophoresed through formaldehyde gels, transferred to nitrocellulose membranes, and incubated with radioactive probes specific for LAC1 or LAG1. 18S RNA was visualized by staining to ensure equal loading (Fig. 2).
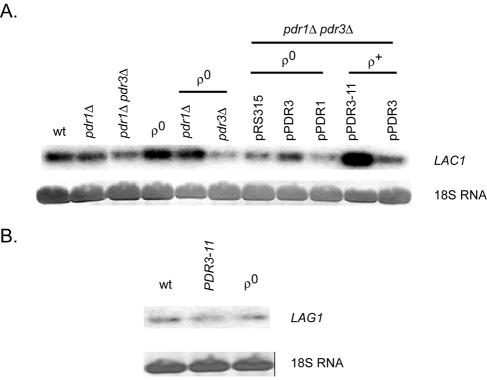
FIG. 2.
Northern blot analysis of LAC1 and LAG1 transcription. Total RNAs were prepared from the indicated strains, electrophoresed through formaldehyde-agarose gels, and transferred to a nylon membrane. 18S RNA was visualized by staining to ensure equal loading, and LAC1 or LAG1 transcripts were detected by probing the blot with 32P-labeled DNA fragments from each coding region. (A) LAC1 mRNA levels are shown with relevant genetic backgrounds indicated. Plasmids carrying wild-type PDR1 (pPDR1), PDR3 (pPDR3), or hyperactive PDR3 (pPDR3-11) are listed along with the empty vector control (pRS315). (B) LAG1 mRNA levels are shown.
The steady-state levels of LAC1 mRNA were found to increase fivefold in the presence of a hyperactive allele of PDR3 (_PDR3_-11) and to be elevated twofold in ρ0 cells. The increase of LAC1 mRNA in ρ0 cells required the presence of wild-type PDR3, consistent with the idea that LAC1 is a downstream target of Pdr3p-dependent retrograde regulation, as seen previously for IPT1 (23) and PDR5 (25). The loss of PDR1 and PDR3, either individually or together, had no major influence on LAC1 expression in ρ+ cells.
In contrast to the response of LAC1 to Pdr pathway activity changes, LAG1 mRNA levels were not significantly influenced by either the loss of the mitochondrial genome or the presence of a _PDR3_-11 allele. Additionally, comparisons of LAC1 and LAG1 hybridization signals suggested that LAC1 was expressed at a higher steady-state level than LAG1. To confirm that these observed changes in mRNA levels were reflected in protein production and to more directly compare the expression of Lac1p and Lag1p, we prepared epitope-tagged alleles of each gene and analyzed their expression by Western blotting.
Comparison of Lac1p and Lag1p protein expression.
Previous experiments have demonstrated that N-terminally tagged forms of both Lac1p and Lag1p are functional (2). Using an inverse PCR strategy (ExSite), we placed a 3× HA epitope tag at the extreme amino terminus of both Lac1p and Lag1p. Each 3× HA-tagged allele was carried on a low-copy-number plasmid and was introduced into the isogenic series of strains used for the analysis of mRNA levels described above. Appropriate transformants were grown to mid-log phase, and whole-cell extracts were prepared. Equal amounts of each extract were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by blotting with antibodies against the HA epitope or the vacuolar ATPase subunit Vph1p as a loading control (Fig. 3).
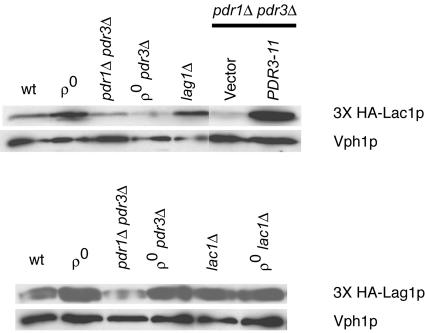
FIG. 3.
Western blot analysis of HA-tagged Lac1p and Lag1p. Whole-cell protein extracts were prepared from the indicated strains, and equal amounts of each extract were electrophoresed by SDS-PAGE. Each strain contained a low-copy-number plasmid expressing a 3× HA-tagged form of either Lac1p (top) or Lag1p (bottom). Where indicated, a low-copy-number plasmid carrying hyperactive PDR3 (PDR3-11) or the empty vector (pRS315) was also present. After electrophoresis, the proteins were transferred to nitrocellulose and the membrane was probed with an anti-HA antibody. A monoclonal antibody directed against the vacuolar membrane protein Vph1p was used to ensure equal loading and transfer.
The levels of 3× HA-tagged Lac1p protein were induced in ρ0 cells compared to ρ+ cells, as described above for Northern blotting. This induction also required the presence of a functional copy of PDR3. Interestingly, the loss of the LAG1 gene gave rise to a small but reproducible increase in 3× HA-Lac1p. We were unable to detect increased β-galactosidase expression upon the introduction of the LAC1-lacZ fusion gene into the _lag1_Δ background (data not shown), and further experiments are required to determine the basis of this potential regulation. The presence of the _PDR3_-11 allele strongly induced 3× HA-tagged Lac1p, similar to the effect of this hyperactive mutant on the LAC1 mRNA level.
Western blotting for 3× HA-Lag1p indicated that the expression of this protein was not influenced by any changes in Pdr pathway activity. The slight decrease of 3× HA-Lag1p seen in _pdr1_Δ _pdr3_Δ cells was not reproducible. Unlike the increase in protein levels when 3× HA-Lac1p was blotted from a _lag1_Δ strain, no change in 3× HA-Lag1p expression was seen when it was analyzed from a _lac1_Δ background. A comparison of the relative intensities of these equivalently tagged proteins supported the notion that Lac1p was approximately 300% more abundant than Lag1p when normalized to Vph1p expression levels. The combined results of Northern and Western blotting analyses indicate that LAC1 expression reaches a higher steady-state level than LAG1 expression and that only LAC1 is responsive to the Pdr regulatory pathway.
Pdr pathway activity controls LAC1 transcription through a single PDRE.
To determine if the observed changes in LAC1 expression occurred via alterations in LAC1 promoter activity, we constructed a fusion gene between E. coli lacZ and the LAC1 promoter. A fragment corresponding to position −754 to the LAC1 ATG was translationally fused to lacZ and carried on a low-copy-number URA3 vector. This reporter plasmid was then introduced into an isogenic series of strains with various levels of Pdr pathway activity. Low-copy-number plasmids expressing gain-of-function forms of PDR3 were used to assess the response of LAC1-lacZ to hyperactive forms of this transcriptional regulator. The ability of the fusion gene to be induced in ρ0 cells was also tested.
The LAC1-lacZ fusion plasmid faithfully reflected the expression patterns seen for both native LAC1 mRNA and its encoded protein (Fig. 4). The loss of the mitochondrial genome increased _LAC1_-dependent β-galactosidase activity twofold, while the presence of hyperactive forms of Pdr3p caused a larger increase, of approximately fivefold, compared to the enzyme levels produced in wild-type cells. The removal of either PDR3 only or both PDR1 and PDR3 had a negligible effect on the expression of LAC1-lacZ in ρ+ cells, although the introduction of a _pdr3_Δ allele into ρ0 cells blocked the normal induction seen in this genetic background. These data provide strong support that the promoter activity of LAC1 is elevated in response to changes in Pdr pathway function.
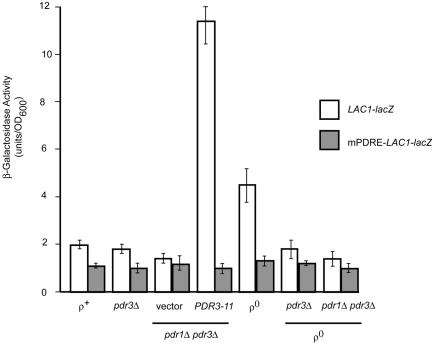
FIG. 4.
A single PDRE mediates Pdr pathway regulation of LAC1. Low-copy-number plasmids carrying either a wild-type LAC1 promoter fusion to lacZ (LAC1-lacZ) or a mutant promoter lacking the PDRE (mPDRE-LAC1-lacZ) were introduced into the strains listed at the bottom of the figure (relevant genetic markers are shown). An empty vector plasmid (pRS315) or the same vector carrying the hyperactive _PDR3_-11 allele were present where indicated. Transformants were grown to mid-log phase, and β-galactosidase activities were determined as described in Materials and Methods.
To determine if the target site for Pdr pathway influence on LAC1 was provided by the PDRE located at position −433, we constructed a mutant form of this binding site that no longer functioned as a Pdr1p or Pdr3p binding site (see below) in the context of an otherwise normal LAC1 promoter. This mutant promoter (designated mPDRE-LAC1) was then fused to lacZ and introduced into the same series of strains that was used to analyze the wild-type LAC1-lacZ fusion gene. Notably, the mPDRE-LAC1-lacZ fusion gene was no longer able to be induced either by the presence of the _PDR3_-11 allele or in ρ0 cells. The loss of the LAC1 PDRE eliminated the influence of the Pdr pathway on _LAC1_-dependent β-galactosidase activity and confirmed that the effects of Pdr3p were most likely mediated through this promoter element.
Both Pdr1p and Pdr3p bind to the single PDRE in the LAC1 promoter.
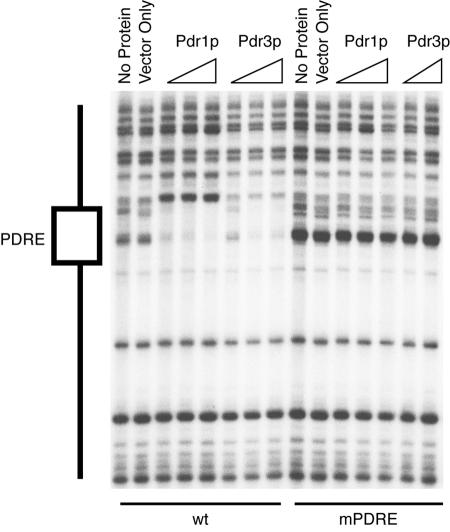
The replacement of the wild-type LAC1 PDRE with the mPDRE blocked the influences of Pdr1p and Pdr3p on LAC1-lacZ expression. To confirm that both Pdr1p and Pdr3p were able to bind to the wild-type PDRE but not the mPDRE, we performed DNase I protection experiments with both versions of the LAC1 promoter. The DNA binding domains of both Pdr1p and Pdr3p were produced in bacteria, as described previously (32), and were incubated with a 32P-labeled fragment from LAC1 containing the wild-type or mutant form of the PDRE. Protein-DNA complexes were then treated with DNase I and electrophoresed in polyacrylamide-urea gels. The cleavage products were visualized by autoradiography (Fig. 5).
FIG. 5.
Pdr1p and Pdr3p bind to LAC1 PDRE in vitro. DNA fragments containing the PDRE region from either the wild-type LAC1 promoter (wt) or the site-directed mutant form (mPDRE) were radiolabeled with 32P as described in Materials and Methods. Each probe was incubated with no added proteins, with protein extracts from bacterial cells carrying an empty expression vector (vector only), or with the same vector expressing the DNA binding domain of Pdr1p or Pdr3p. Protein-DNA complexes were then treated with DNase I followed by electrophoresis through denaturing acrylamide-urea gels as described previously (32). The location of the PDRE (indicated to the left) was determined by comparison to a Maxam-Gilbert chemical cleavage reaction and restriction digestion (not shown).
Both bacterially expressed Pdr1p and Pdr3p were able to bind to the wild-type but not the mutant form of the LAC1 PDRE. These observations provide further support for the simple model that the Pdr pathway regulation of LAC1 is mediated by the binding of either Pdr1p or Pdr3p to the single LAC1 PDRE. While the Pdr pathway input to LAC1 regulation is provided by the PDRE, we noticed that the loss of this element had a modest influence on LAC1 expression in the absence of Pdr pathway activation. This suggests that other elements contribute to LAC1 expression, and we performed deletion analyses to map the locations of these other regulatory sites.
Deletion mapping of LAC1 promoter identifies two negative regulatory sites.
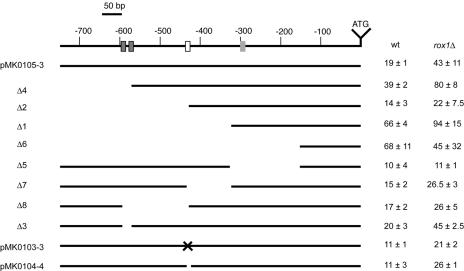
A series of both 5′ and internal deletion mutation derivatives of the LAC1 promoter was constructed in the context of the LAC1-lacZ fusion gene. These deletion mutants were transformed into wild-type or _rox1_Δ cells, and transformants were grown to mid-log phase and assayed for their levels of _LAC1_-dependent β-galactosidase expression (Fig. 6). We tested the influence of Rox1p on LAC1 expression, as a sequence inspection of the LAC1 promoter and DNA microarray analyses suggested the possibility that this anaerobic repressor protein regulates the expression of this gene (39, 67).
FIG. 6.
Deletion mapping of the LAC1 promoter. A series of 5′ and internal promoter deletion derivatives of the LAC1-lacZ plasmid were generated. The solid lines indicate the extent of the promoter that remains in each construct. The name of each mutant construct is shown on the left side of the figure. A line drawing of the wild-type LAC1 promoter region is shown at the top with numbers referring to distances from the ATG codon. The dark gray boxes show the location of the putative Cbf1p binding sites, the white box corresponds to the PDRE, and the light gray box denotes the Rox1p binding site. The PDRE was altered by a site-directed mutation in pMK010303 and was precisely deleted from pMK0104-4. Each plasmid was transformed into isogenic wild-type (BY4742) or _rox1_Δ (BY4742 _rox1_Δ) cells, and β-galactosidase activities were assayed as described in the text.
Expression of the wild-type LAC1-lacZ gene fusion was found to be approximately twofold higher in _rox1_Δ cells than in isogenic wild-type cells. Furthermore, truncation of the LAC1 promoter to 572 bp also led to an additional twofold increase in _LAC1_-dependent β-galactosidase activity in both wild-type and _rox1_Δ cells. This deletion variant removed two potential binding sites for the transcription factor Cbf1p, a DNA binding protein involved in the regulation of methionine biosynthesis and kinetochore function (reviewed in reference 68). These observations suggest that Cbf1p acts as a negative regulator of LAC1 expression, and this suggestion was supported by an analysis of LAC1-lacZ in a _cbf1_Δ strain (see below).
A truncation to 433 bp upstream of the LAC1 ATG eliminated the PDRE and reduced the response to the loss of Rox1p. Surprisingly, smaller LAC1-lacZ derivatives, containing either 324 or 153 bp of 5′ noncoding sequence, produced much higher _LAC1_-dependent lacZ activities. Only the promoter containing 324 bp of LAC1 5′ DNA exhibited any response to the loss of Rox1p, consistent with the location of the Rox1p binding site near position −300. Since an internal deletion that removed the sequence between −433 and −324 failed to produce elevated LAC1 expression, we believe that the increased β-galactosidase activity directed by the −324 5′ deletion construct is most likely due to the juxtaposition of vector sequences near the LAC1 transcription start site.
Other internal deletion derivatives were examined to further refine the locations of regulatory elements in the LAC1 promoter. Two PmlI restriction sites are present, at positions −587 and −571, in LAC1. Both of these PmlI cleavage sites are embedded within consensus (RCACRTG; R = purine) binding sites for Cbf1p (70). Deletion of the DNA between the PmlI sites restored a single consensus Cbf1p site and retained normal LAC1 expression control, suggesting that only one binding site is required for Cbf1p regulation at this target promoter. Deletion of the DNA between positions −587 and −433, removing both Cbf1p sites and the PDRE, had a composite effect on LAC1 expression. While deletion of the Cbf1p sites would be expected to increase expression, the loss of the positively acting PDRE overrode this influence, and the resulting internal deletion mutant was expressed at approximately 50% the level of wild-type LAC1-lacZ.
An internal deletion mutant promoter, lacking residues −324 to −153, no longer exhibited a response to the loss of Rox1p. Importantly, this internal deletion derivative was still normally inducible in ρ0 cells (data not shown), confirming that the promoter was still active. This result provides further support that the Rox1p target site lies within this interval of the LAC1 promoter. We tested this idea by DNase I protection assays.
Rox1p binds to LAC1 promoter.
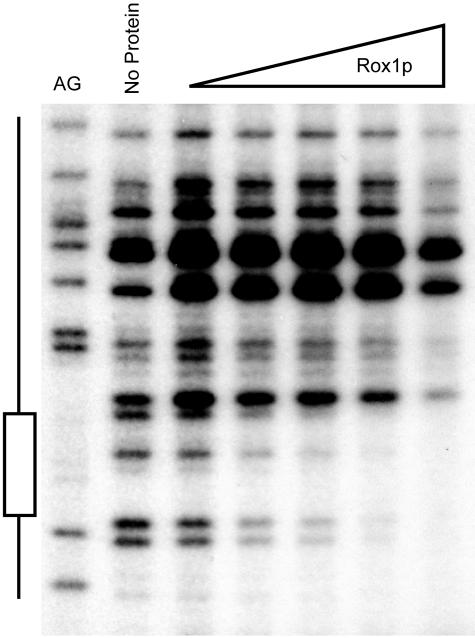
Previous DNA microarray experiments have suggested that LAC1 expression is controlled by the presence of oxygen through the action of the anaerobic repressor protein Rox1p (40). The deletion mapping analysis reported here suggests that Rox1p regulation is mediated through the LAC1 promoter and localized between positions −324 and −153. To determine if Rox1p could bind to this region of LAC1, we performed a DNase I protection analysis using bacterially expressed Rox1p. A 32P-labeled LAC1 promoter fragment was prepared and incubated with recombinant Rox1p. Protein-DNA complexes were then digested briefly with DNase I and analyzed by denaturing urea-PAGE (Fig. 7).
FIG. 7.
Recombinant Rox1p binds to an element in the LAC1 promoter. DNase I protection analysis was used as described in the text. A purine-specific chemical cleavage reaction (AG) was used to locate the Rox1p binding site (shown to the left). DNase I digestion was carried out in the absence of added protein (no protein) or with increasing amounts of Rox1p (indicated at the top).
Rox1p was found to protect a region including positions −296 to −285 from DNase I cleavage. This segment of the LAC1 promoter contains a predicted Rox1p binding site (TCT ATT GTT TCCC), although other potential Rox1p recognition sites have been noted elsewhere in LAC1 (67). To confirm that this binding site for Rox1p is required for the anaerobic regulation of LAC1, we mutated this element and tested the resulting mutant promoter for the ability to respond to Rox1p control. The expression of a LAG1-lacZ fusion was also assessed in _rox1_Δ cells to determine if this homologous gene was regulated similarly to LAC1.
Differential control of LAC1 and LAG1 by Rox1p.
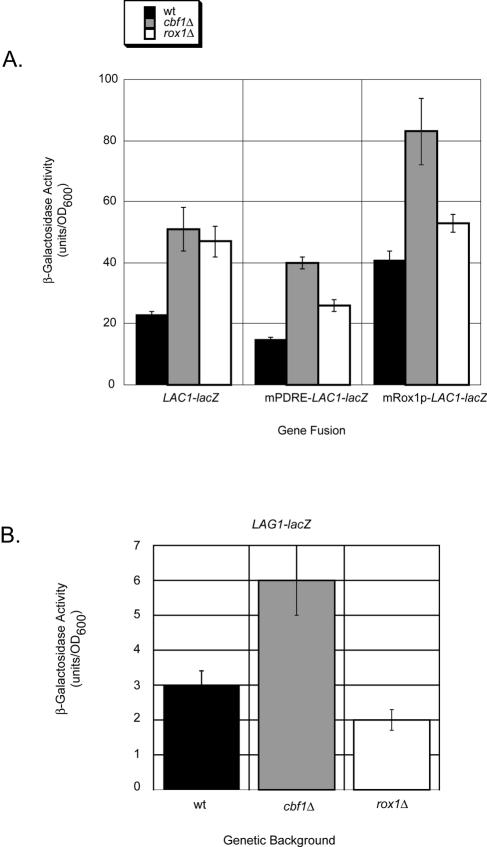
To compare the responses of the two genes encoding homologous components of ceramide synthase activity, we prepared LAG1-lacZ and _LAC1_-lacZ fusion genes. To determine if the Rox1p binding site identified by DNase I protection was responsible for the regulation of LAC1, we prepared a site-directed mutant in which 3 bp of the Rox1p element were changed to form an AflII restriction site. This mutation was constructed in the context of the LAC1-lacZ fusion gene, and plasmids carrying these three different lacZ fusion constructs (_LAG1_-lacZ, _LAC1_-lacZ, and mRox1p-LAC1-lacZ) were introduced into wild-type and _rox1_Δ cells. Transformants were grown to mid-log phase and β-galactosidase activities were measured (Fig. 8).
FIG. 8.
LAC1 and LAG1 expression control by Cbf1p and Rox1p. Low-copy-number plasmids containing the lacZ fusion genes listed were introduced into the indicated isogenic strains. Transformants were grown to mid-log phase and β-galactosidase activities were determined. (A) Expression of _LAC1_-lacZ fusion plasmids plotted on the same scale for comparison. (B) Expression of LAG1-lacZ plotted on an expanded scale to emphasize the observed change in gene expression.
LAC1-lacZ expression was induced to comparable levels upon the loss of either CBF1 or ROX1. The elimination of Pdr pathway input from the fusion gene (mPDRE-LAC1-lacZ) reduced the observed increase in β-galactosidase expression, likely owing to the loss of this positive regulatory input. Removal of the Rox1p binding site (mRox1p-LAC1-lacZ) strongly enhanced _LAC1_-dependent enzyme activity in wild-type and _cbf1_Δ cells, but not in _rox1_Δ cells, since Rox1p is already absent from this strain. LAG1-lacZ expression was induced only in cells lacking Cbf1p and was unaffected in a _rox1_Δ background, consistent with the absence of any Rox1p binding sites from the LAG1 promoter (data not shown).
Several important observations were made from this analysis. First, the use of _LAG1_-lacZ and LAC1-lacZ fusion genes faithfully replicated the differences in expression of these two genes that were seen by Western and Northern blot analyses. LAC1-lacZ was expressed at higher levels than LAG1-lacZ. Second, LAC1, but not LAG1, exhibited elevated expression in the absence of Rox1p. Third, the introduction of an AflII mutation into the Rox1p binding site of LAC1 elevated the expression of the resulting gene fusion, even in wild-type cells, and prevented the significant increase in expression seen in _rox1_Δ cells. Coupled with DNA microarray experiments (8-10) and our present LAG1-lacZ analysis (data not shown) showing that LAG1 expression was not responsive to Pdr pathway signals, these data argue that expression control of the homologous LAC1 and LAG1 genes involves multiple distinct mechanisms. Finally, Cbf1p appears to serve as a common regulator of both LAC1 and LAG1.
Additional Pdr pathway inputs in sphingolipid biosynthesis.
The evidence presented above indicates that the LAC1 gene, encoding a ceramide synthase component, but not its homologue LAG1, is regulated by the Pdr pathway as previously found for the IPT1 gene (23). A sequence inspection for the presence of potential PDREs in promoters of other genes linked to sphingolipid biosynthesis produced two additional candidate loci for Pdr control, specifically SUR2 and LCB2. SUR2 encodes a hydroxylase that is required for the production of phytoceramide (22), while LCB2 encodes one of the essential components of the serine palmitoyltransferase enzyme, which is involved in the first committed step in sphingolipid biosynthesis (4, 51). To determine if these genes are regulated by Pdr pathway activity, we constructed lacZ gene fusions with each of their promoters. The resulting _SUR2_-lacZ and LCB2-lacZ fusion genes were introduced on low-copy-number plasmids into isogenic wild-type and _rox1_Δ cells. Additionally, a vector plasmid (pRS315) containing or lacking the hyperactive _PDR3_-11 allele was introduced with each lacZ fusion gene. Doubly transformed cells were grown and analyzed for β-galactosidase activity as described above (Fig. 9).
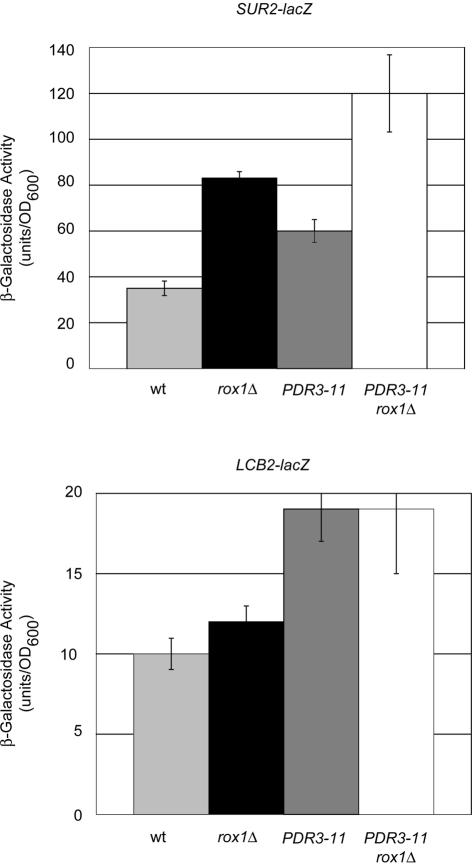
FIG. 9.
LCB2 and SUR2 response to elevated Pdr pathway activity and _rox1_Δ. _LCB2_-lacZ and SUR2-lacZ fusions carried on low-copy-number plasmids were introduced into isogenic wild-type or _rox1_Δ strains along with an empty vector (pRS315) or the same plasmid carrying the hyperactive _PDR3_-11 allele. Transformants were grown and assayed for β-galactosidase activity as described in the text.
Both _SUR2_-lacZ and LCB2-lacZ activity increased approximately twofold in the presence of _PDR3_-11. The elimination of Rox1p was found to lead to an additional twofold elevation in SUR2-lacZ expression but to have no influence on LCB2 expression. These data support the view that SUR2 transcriptional control is similar to that seen for LAC1, being influenced by both the Pdr and Rox1p regulatory systems, while LCB2 shares only the Pdr pathway regulatory component.
Changes in sphingolipid levels in response to alteration of LAC1 and LAG1 expression.
A central goal of this study was to further the link between Pdr pathway regulation and the control of sphingolipid biosynthesis. The data described above provide strong evidence that the levels of one of the ceramide synthase components (Lac1p) respond to signals that upregulate the Pdr pathway. To determine if this Pdr-dependent regulation could be seen to affect sphingolipid biosynthesis through LAC1 activation, we measured the levels of ceramide and IPC produced in a series of strains containing different LAC1 and LAG1 alleles, either in the presence or in the absence of a hyperactive form of Pdr3p. Four strains were used that varied in their LAC1 and LAG1 gene dosage: the wild type, a _lac1_Δ mutant, a _lag1_Δ mutant, or a strain containing a mutant LAC1 allele that was wild type except that its promoter had been engineered to lack the PDRE (see Materials and Methods). Each strain was transformed with a low-copy-number plasmid carrying either PDR3 or the hyperactive _PDR3_-11 mutant. Transformants were grown in selective medium and then labeled with [3H]serine to allow for the detection of IPC and ceramides. Lipid extracts were prepared and resolved by thin-layer chromatography, and labeled species were detected by autoradiography. The results of this analysis are shown in Fig. 10.
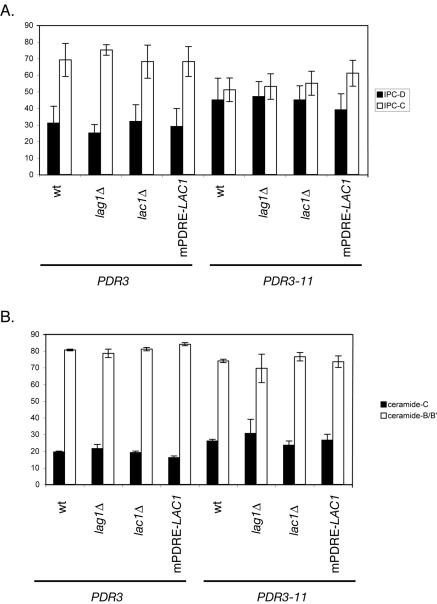
FIG. 10.
Pdr-dependent changes in sphingolipid and ceramide composition. Cells of the indicated genotype expressing either PDR3 or the hyperactive _PDR3_-11 allele were labeled with [3H]serine for 2 h at 30°C, lipids were extracted and deacylated, and sphingolipids and ceramides were analyzed by thin-layer chromatography. Mol% values for the indicated lipid species are indicated on the ordinate. (A) Relative abundance of IPC-C and IPC-D species; (B) composition of free ceramide species.
The introduction of the _PDR3_-11 allele into all four genetic backgrounds caused the levels of IPC-C and IPC-D to equalize, while in the presence of wild-type PDR3 nearly twofold more IPC-C than IPC-D was present. The major cause of this change was due to an increase in IPC-D levels, although this change occurred irrespective of changes in LAC1 and LAG1. A direct examination of free ceramide (the product of the ceramide synthase reaction) indicated a trend toward an increase in ceramide-C production in the presence of _PDR3_-11, but this effect was small. These modest effects on sphingolipid synthesis are consistent with those reported by others who used the GAL promoter to drive the high-level production of Lag1p (21) and likely reflect the elaborate regulation of sphingolipid production. Possible reasons underlying the ability of the cell to tolerate these changes in the levels of sphingolipid biosynthetic enzymes while preventing major changes in sphingolipid levels are discussed below.
DISCUSSION
These data provide several new insights into the interface of the Pdr pathway with sphingolipid biosynthesis in S. cerevisiae. Previous work has established that the IPT1 gene, encoding the final step in sphingolipid biosynthesis (14), is regulated by Pdr pathway signals (23). In this work, we provide evidence that three other genes involved in sphingolipid production are also under the transcriptional influence of Pdr pathway activity. Since a large number of membrane transporter proteins have already been demonstrated to be controlled by Pdr1p/Pdr3p activity status (24, 30, 42, 72), these findings support the idea that the Pdr pathway may act to coordinate the synthesis of both these integral membrane transporters and components of their lipid milieu.
Our data provide the first characterization of the transcriptional control of ceramide synthase in any organism. The regulation of the ceramide synthase component encoded by LAC1 was found to be fairly complex. These experiments provide evidence for LAC1 transcriptional control being responsive to three distinct gene regulatory pathways, namely Pdr, Rox1p, and Cbf1p. These three different regulatory systems allow LAC1 expression to be adjusted to the activity of multidrug resistance genes, oxygen, and potentially methionine biosynthesis. The precise role of Cbf1p in gene expression is somewhat murky, but clear data indicate a role for this protein in the expression of genes involved in methionine biosynthesis, and _cbf1_Δ cells are auxotrophic for this amino acid (38).
In comparison to the pattern of LAC1 gene regulation, LAG1 shares only the influence of Cbf1p. Additionally, in reporter gene, Western blot, and Northern blot analyses, LAC1 expression was approximately threefold higher than LAG1 expression. Based on these data, we expected Lac1p to be the predominant contributor to ceramide synthase activity in S. cerevisiae. However, direct biochemical measurements of ceramide synthase activity in single mutant _lag1_Δ or _lac1_Δ cells indicate that Lag1p provides the majority of this enzymatic function (20). The discrepancy between the expression and biochemical data may be explained simply by Lag1p exhibiting a higher specific activity than Lac1p. However, as pointed out previously (20, 21), the results of the biochemical assay must be interpreted with care since the actual substrate specificities of Lag1p and Lac1p are not known. Whatever the nature of the differences between Lag1p and Lac1p, the expression of only one of these proteins is sufficient for a normal growth rate (2).
It is interesting that of all the steps in de novo sphingolipid biosynthesis, the ceramide synthase reaction is the only one in which redundancy is found. This is likely related to the complex regulation that acts to precisely modulate production of the bioactive signaling lipid ceramide (reviewed in reference 26). Not only does ceramide have a potent regulatory effect, but the LCB precursors to ceramide (dihydrosphingosine and phytosphingosine) also have several downstream effects, including the inhibition of amino acid transport and G1 arrest (5, 64). An excessive production of ceramide has been postulated to underlie the lethality of an _aur1_Δ strain that lacks the enzyme required for the conversion of ceramide to IPC (59). While transient elevations in ceramide are required for stress responses (13), sustained eightfold increases are correlated with a loss of viability (52, 59).
The fact that ceramide and its precursors have these potent signaling effects likely explains two related difficulties in the study of LAC1 and LAG1. We tested a large number of different strains that possessed various levels of expression of either Lag1p or Lac1p yet found relatively subtle changes in sphingolipid biosynthesis (Fig. 10 and unpublished data). Others have driven Lag1p or Lac1p expression from the strong GAL promoter yet also did not detect significant increases in either ceramide synthase activity or large changes in the levels of sphingolipid intermediates (21). Our experiments used authentic regulatory signals that we have found to modulate the expression of several genes in the pathway yet were similarly unable to elicit major changes in pathway intermediates. Additionally, we tested a large battery of drugs (>20 different compounds) that are known substrates for Pdr-mediated resistance in strains with variable LAC1 and LAG1 expression but failed to detect significant phenotypic changes (data not shown).
These difficulties most likely stem from the elaborate buffering of ceramide synthesis by multiple mechanisms in S. cerevisiae in particular and in eukaryotes in general (see references 11 and 53 for recent reviews). The substrates for the ceramide synthase reaction may be withdrawn from the pathway by either enzymatic breakdown (54) or transport out of the cell by the recently described Rsb1p membrane protein (34). Interestingly, microarray experiments indicate that the expression of RSB1 is also regulated by the Pdr pathway (8). Ceramide can also be directly degraded by the action of two different ceramidase enzymes (45, 46). Finally, it has been suggested that the ABC transporter Yor1p (33), another Pdr target gene (24), can transport sphingolipid intermediates (45). These multiple regulatory systems effectively limit the changes in ceramide and other sphingolipid pathway intermediates that can be elicited. This is physiologically crucial since the levels of LCBs and ceramide have major effects on proliferation, and ultimately, viability (recently reviewed in reference 41).
Taken together, these data suggest the possibility that a major role of the Pdr pathway in vivo is to control the location and synthesis of sphingolipids. The list of Pdr-regulated genes involved in sphingolipid homeostasis includes four that are involved in biosynthesis (LAC1, SUR2, LCB2, and IPT1) and two that are involved in the transport of these lipids (RSB1 and YOR1). Intriguingly, the RTA1 gene (65) also exhibits Pdr pathway regulation (8) and is an RSB1 homologue. Further work will be required to confirm this hypothesis, but the body of evidence linking Pdr pathway function and sphingolipid biosynthesis makes this a compelling model. Similar findings have been reported for mammalian cells (reviewed in reference 61), suggesting that this coordinate control of multidrug resistance and sphingolipid production is conserved throughout evolution.
Acknowledgments
We thank Bob Dickson and Lina Obeid for important discussions. We thank Richard Zitomer and Aaron Mitchell for providing reagents and for assisting in early stages of this work.
This work was supported in part by NIH grant GM49825, grant 631-065925 from the Swiss National Science Foundation, and the Polish State Committee for Scientific Research (KBN) funds for the Wroclaw Medical University and grant 6 P05A 01221.
REFERENCES
- 1.Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262**:**16871-16879. [PubMed] [Google Scholar]
- 2.Barz, W. P., and P. Walter. 1999. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell 10**:**1043-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke, J. D., F. Lacroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine 5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197**:**345-346. [DOI] [PubMed] [Google Scholar]
- 4.Buede, R., C. Rinker-Schaffer, W. J. Pinto, R. L. Lester, and R. C. Dickson. 1991. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 173**:**4325-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, N., C. Mao, J. Heitman, Y. A. Hannun, and L. M. Obeid. 2001. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J. Biol. Chem. 276**:**35614-35621. [DOI] [PubMed] [Google Scholar]
- 6.Cuvillier, O. 2002. Sphingosine in apoptosis signaling. Biochim. Biophys. Acta 1585**:**153-162. [DOI] [PubMed] [Google Scholar]
- 7.Delaveau, T., C. Jacq, and J. Perea. 1992. Sequence of a 12.7 kb segment of yeast chromosome II identifies a PDR-like gene and several new open reading frames. Yeast 8**:**761-768. [DOI] [PubMed] [Google Scholar]
- 8.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470**:**156-160. [DOI] [PubMed] [Google Scholar]
- 9.Devaux, F., E. Carvajal, S. Moye-Rowley, and C. Jacq. 2002. Genome-wide studies on the nuclear _PDR3_-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 515**:**25-28. [DOI] [PubMed] [Google Scholar]
- 10.Devaux, F., P. Marc, C. Bouchoux, T. Delaveau, I. Hikkel, M. C. Potier, and C. Jacq. 2001. An artificial transcription activator mimics the genome-wide properties of the yeast Pdr1 transcription factor. EMBO Rep. 2**:**493-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson, R. C., and R. L. Lester. 2002. Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1583**:**13-25. [DOI] [PubMed] [Google Scholar]
- 12.Dickson, R. C., and R. L. Lester. 1999. Yeast sphingolipids. Biochim. Biophys. Acta 272**:**347-357. [DOI] [PubMed] [Google Scholar]
- 13.Dickson, R. C., E. E. Nagiec, M. Skrzypek, P. Tillman, G. B. Wells, and R. L. Lester. 1997. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272**:**30196-30200. [DOI] [PubMed] [Google Scholar]
- 14.Dickson, R. C., E. E. Nagiec, G. B. Wells, M. M. Nagiec, and R. L. Lester. 1997. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J. Biol. Chem. 272**:**29620-29625. [DOI] [PubMed] [Google Scholar]
- 15.D'mello, N. P., A. M. Childress, D. S. Franklin, S. P. Kale, C. Pinswasdi, and S. M. Jazwinski. 1994. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J. Biol. Chem. 269**:**15451-15459. [PubMed] [Google Scholar]
- 16.Eisenkolb, M., C. Zenzmaier, E. Leitner, and R. Schneiter. 2002. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol. Biol. Cell 13**:**4414-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emr, S. D., A. Vassarotti, J. Garret, B. C. Geller, M. Takeda, and M. G. Douglas. 1986. The amino terminus of the yeast F1-ATPase beta-subunit precursor functions as a mitochondrial import signal. J. Cell Biol. 102**:**523-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350**:**87-96. [DOI] [PubMed] [Google Scholar]
- 19.Guarente, L. 1983. Yeast promoter and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101**:**181-191. [DOI] [PubMed] [Google Scholar]
- 20.Guillas, I., J. C. Jiang, C. Vionnet, C. Roubaty, D. Uldry, R. Chuard, J. Wang, S. M. Jazwinski, and A. Conzelmann. 2003. Human homologues of LAG1 reconstitute acyl-CoA-dependent ceramide synthesis in yeast. J. Biol. Chem. 278**:**37083-37091. [DOI] [PubMed] [Google Scholar]
- 21.Guillas, I., P. A. Kirchman, R. Chuard, M. Pfefferli, J. C. Jiang, S. M. Jazwinski, and A. Conzelmann. 2001. C26-CoA-dependent synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20**:**2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haak, D., K. Gable, T. Beeler, and T. Dunn. 1997. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272**:**29704-29710. [DOI] [PubMed] [Google Scholar]
- 23.Hallstrom, T. C., L. Lambert, S. Schorling, E. Balzi, A. Goffeau, and W. S. Moye-Rowley. 2001. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276**:**23674-23680. [DOI] [PubMed] [Google Scholar]
- 24.Hallstrom, T. C., and W. S. Moye-Rowley. 1998. Divergent transcriptional control of multidrug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 273**:**2098-2104. [DOI] [PubMed] [Google Scholar]
- 25.Hallstrom, T. C., and W. S. Moye-Rowley. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275**:**37347-37356. [DOI] [PubMed] [Google Scholar]
- 26.Hannun, Y. A., and L. M. Obeid. 2002. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277**:**25847-25850. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins, G. M., A. Richards, T. Wahl, C. Mao, L. Obeid, and Y. Hannun. 1997. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272**:**32566-32572. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, J. C., P. A. Kirchman, M. Zagulski, J. Hunt, and S. M. Jazwinski. 1998. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 8**:**1259-1272. [DOI] [PubMed] [Google Scholar]
- 29.Kastaniotis, A. J., and R. S. Zitomer. 2000. Rox1 mediated repression. Oxygen dependent repression in yeast. Adv. Exp. Med. Biol. 475**:**185-195. [PubMed] [Google Scholar]
- 30.Katzmann, D. J., P. E. Burnett, J. Golin, Y. Mahe, and W. S. Moye-Rowley. 1994. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14**:**4653-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katzmann, D. J., E. A. Epping, and W. S. Moye-Rowley. 1999. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 19**:**2998-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katzmann, D. J., T. C. Hallstrom, Y. Mahe, and W. S. Moye-Rowley. 1996. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 271**:**23049-23054. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann, D. J., T. C. Hallstrom, M. Voet, W. Wysock, J. Golin, G. Volckaert, and W. S. Moye-Rowley. 1995. Expression of an ATP-binding cassette transporter encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol. Cell. Biol. 15**:**6875-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihara, A., and Y. Igarashi. 2002. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoind long-chain base release. J. Biol. Chem. 277**:**30048-30054. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, S. D., and M. M. Nagiec. 2003. Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot. Cell 2**:**284-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolaczkowska, A., and A. Goffeau. 1999. Regulation of pleiotropic drug resistance in yeast. Drug Resist. Update 2**:**403-414. [DOI] [PubMed] [Google Scholar]
- 37.Kolaczkowski, M., A. Kolaczowska, J. Luczynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4**:**143-158. [DOI] [PubMed] [Google Scholar]
- 38.Kuras, L., H. Cherest, Y. Surdin-Kerjan, and D. Thomas. 1996. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15**:**2519-2529. [PMC free article] [PubMed] [Google Scholar]
- 39.Kwast, K. E., L. C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184**:**250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowry, C. V., and R. S. Zitomer. 1984. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc. Natl. Acad. Sci. USA 81**:**6129-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maceyka, M., S. G. Payne, S. Milstien, and S. Spiegel. 2002. Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim. Biophys. Acta 1585**:**193-201. [DOI] [PubMed] [Google Scholar]
- 42.Mahe, Y., A. Parle-McDermott, A. Nourani, A. Delahodde, A. Lamprecht, and K. Kuchler. 1996. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol. Microbiol. 20**:**109-117. [DOI] [PubMed] [Google Scholar]
- 43.Mandala, S. M., R. Thornton, I. Galve-Roperh, S. Poulton, C. Peterson, A. Olivera, J. Bergstrom, M. B. Kurtz, and S. Spiegel. 2000. Molecular cloning and characterization of a lipid phosphohydrolase that degrades sphingosine-1-phosphate and induces cell death. Proc. Natl. Acad. Sci. USA 97**:**7859-7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao, C., J. D. Saba, and L. M. Obeid. 1999. The dihydrosphingosine-1-phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 342**:**667-675. [PMC free article] [PubMed] [Google Scholar]
- 45.Mao, C., R. Xu, A. Bielawska, and L. M. Obeid. 2000. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae: an enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 275**:**6876-6884. [DOI] [PubMed] [Google Scholar]
- 46.Mao, C., R. Xu, A. Bielawska, Z. M. Szulc, and L. M. Obeid. 2000. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 275**:**31369-31378. [DOI] [PubMed] [Google Scholar]
- 47.Masserini, M., and D. Ravasi. 2001. Role of sphingolipids in the biogenesis of membrane domains. Biochim. Biophys. Acta 1532**:**149-161. [DOI] [PubMed] [Google Scholar]
- 48.Mathias, S., L. A. Pena, and R. N. Kolesnick. 1998. Signal transduction of stress via ceramide. Biochem. J. 335**:**465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyers, S., W. Schauer, E. Balzi, M. Wagner, A. Goffeau, and J. Golin. 1992. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr. Genet. 21**:**431-436. [DOI] [PubMed] [Google Scholar]
- 50.Moye-Rowley, W. S. 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acids Res. Mol. Biol. 73**:**251-279. [DOI] [PubMed] [Google Scholar]
- 51.Nagiec, M. M., J. A. Baltisberger, G. B. Wells, R. L. Lester, and R. C. Dickson. 1994. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. USA 91**:**7899-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagiec, M. M., E. E. Nagiec, J. A. Baltisberger, G. B. Wells, R. L. Lester, and R. C. Dickson. 1997. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272**:**9809-9817. [DOI] [PubMed] [Google Scholar]
- 53.Obeid, L. M., Y. Okamoto, and C. Mao. 2002. Yeast sphingolipids: metabolism and biology. Biochim. Biophys. Acta 1585**:**163-171. [DOI] [PubMed] [Google Scholar]
- 54.Saba, J. D., F. Nara, A. Bielawska, S. Garrett, and Y. A. Hannun. 1997. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J. Biol. Chem. 272**:**26087-26090. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Scherer, S., and R. W. Davis. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76**:**4951-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A simple and rapid method for preparation of RNA from S. cerevisiae. Nucleic Acids Res. 18**:**3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneiter, R. 1999. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. Bioessays 21**:**1004-1010. [DOI] [PubMed] [Google Scholar]
- 59.Schorling, S., B. Vallée, W. P. Barz, H. Riezman, and D. Oesterhelt. 2001. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell 12**:**3417-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman, F., G. Fink, and J. Hicks. 1979. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 61.Sietsma, H., R. J. Veldman, and J. W. Kok. 2001. The involvement of sphingolipids in multidrug resistance. J. Membr. Biol. 181**:**153-162. [DOI] [PubMed] [Google Scholar]
- 62.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122**:**19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skrzypek, M. S., M. M. Nagiec, R. L. Lester, and R. C. Dickson. 1999. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J. Bacteriol. 181**:**1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skrzypek, M. S., M. M. Nagiec, R. L. Lester, and R. C. Dickson. 1998. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J. Biol. Chem. 273**:**2829-2834. [DOI] [PubMed] [Google Scholar]
- 65.Soustre, I., Y. Letourneux, and F. Karst. 1996. Characterization of the Saccharomyces cerevisiae RTA1 gene involved in 7-aminocholesterol resistance. Curr. Genet. 30**:**121-125. [DOI] [PubMed] [Google Scholar]
- 66.Spiegel, S., and S. Milstien. 2000. Sphingosine-1-phosphate: signaling inside and out. FEBS Lett. 476**:**55-57. [DOI] [PubMed] [Google Scholar]
- 67.Ter Linde, J. J., and H. Y. Steensma. 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19**:**825-840. [DOI] [PubMed] [Google Scholar]
- 68.Thomas, D., and Y. Surdin-Kerjan. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61**:**503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76**:**4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wieland, G., P. Hemmerich, M. Koch, T. Stoyan, J. Hegemann, and S. Diekmann. 2001. Determination of the binding constants of the centromere protein Cbf1 to all 16 centromere DNAs of Saccharomyces cerevisiae. Nucleic Acids Res. 29**:**1054-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16**:**65-70. [DOI] [PubMed] [Google Scholar]
- 72.Wolfger, H., Y. Mahé, A. Parle-McDermott, A. Delahodde, and K. Kuchler. 1997. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 418**:**269-274. [DOI] [PubMed] [Google Scholar]
- 73.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2001. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 152**:**375-389. [DOI] [PubMed] [Google Scholar]