MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies (original) (raw)
. Author manuscript; available in PMC: 2007 Apr 24.
Published in final edited form as: Nat Cell Biol. 2005 Jun 5;7(7):719–723. doi: 10.1038/ncb1274
Abstract
Small RNAs, including small interfering RNAs (siRNAs) and microRNAs (miRNAs) can silence target genes through several different effector mechanisms1. Whereas siRNA-directed mRNA cleavage is increasingly understood, the mechanisms by which miRNAs repress protein synthesis are obscure. Recent studies have revealed the existence of specific cytoplasmic foci, referred to herein as processing bodies (P-bodies), which contain untranslated mRNAs and can serve as sites of mRNA degradation2–7. Here we demonstrate that Argonaute proteins — the signature components of the RNA interference (RNAi) effector complex, RISC — localize to mammalian P-bodies. Moreover, reporter mRNAs that are targeted for translational repression by endogenous or exogenous miRNAs become concentrated in P-bodies in a miRNA-dependent manner. These results provide a link between miRNA function and mammalian P-bodies and suggest that translation repression by RISC delivers mRNAs to P-bodies, either as a cause or as a consequence of inhibiting protein synthesis.
RNAi was initially characterized as a post-transcriptional gene silencing mechanism in which the experimental introduction of long double-stranded RNAs (dsRNAs) induces sequence-specific destruction of homologous mRNAs (reviewed in ref. 1). RNAi pathways are initiated when dsRNAs are processed by Dicer into siRNAs of 21–26 nucleotides. siRNAs are incorporated into the effector complex RISC. In RISC, the siRNA is bound by an Argonaute protein, which uses the sequence of the siRNA to select and cleave complementary substrates (reviewed in ref. 8).
RISC can also silence gene expression by preventing protein synthesis. Genetic studies of Caenorhabditis elegans that are mutant for Dicer forged the initial link between a previously known class of small regulatory RNAs, the stRNAs, and the RNAi pathway9–13. Subsequent studies showed that stRNAs are archetypes of a large class of regulatory RNAs, known as miRNAs (reviewed in ref. 14). Although miRNA and siRNA pathways can be biochemically compartmentalized, both types of RNAs enter RISC, bind to Argonaute proteins and identify their silencing targets in conceptually similar ways. They differ, at least in animals, in that miRNAs most often pair imperfectly with their targets and are thus unable to direct Argonaute-mediated cleavage15. Instead, miRNAs repress protein synthesis in a cleavage-independent fashion8,14,15.
The mechanism by which miRNAs repress translation of their target mRNAs is unknown. Conceivably, RISC could prevent protein synthesis from miRNA targets in one of several ways. RISC could affect translation, per se, by altering rates of initiation or elongation by the ribosome. Alternatively, translation could proceed unaffected with nascent polypeptides being degraded. A third possibility is that target mRNAs could be somehow sequestered from the translational machinery.
To investigate this third model, we assessed the localization of an ectopically expressed, Myc-tagged human Argonaute protein, Myc–Ago2, which is fully functional for siRNA-mediated silencing16. Immunofluorescence indicated a pattern of Myc–Ago2 localization in discrete cytoplasmic foci, although some Ago2 may also be distributed throughout the cytoplasm (Fig. 1a). Both the size of individual foci and the number of foci varied between individual cells with an average of four to nine clear foci per cell. Endogenous Ago2 could also be detected in foci with a similar average number of three to six foci per cell (Fig. 1b). Notably, these foci were observed when the antibody was used to stain wild-type but not Ago2-mutant mouse embryo fibroblasts16, indicating that this pattern truly reflects the localization of the endogenous Ago2 protein (data not shown). Differences in numbers of apparent foci probably result from anti-Ago2 antibodies giving overall weaker signals than did monoclonals that recognize epitope-tagged proteins.
Figure 1.
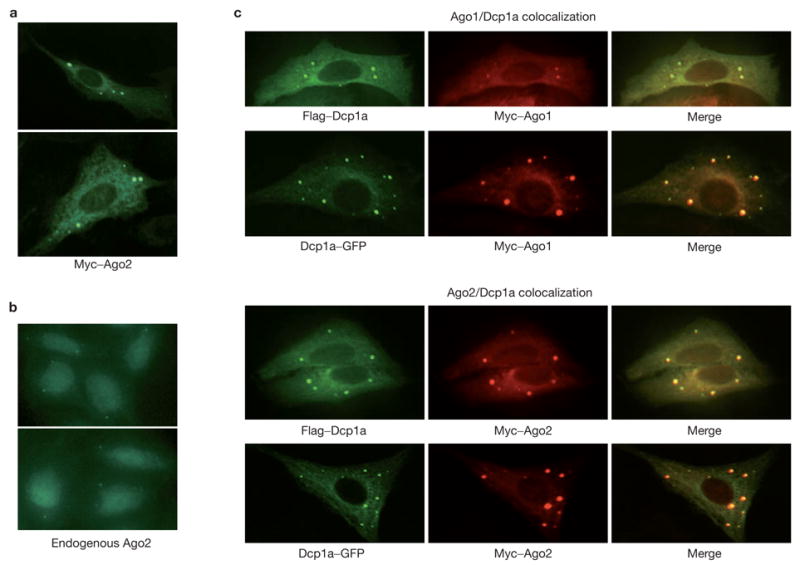
Argonaute proteins localize to mammalian P-bodies. (a) Myc-tagged Ago2 protein was expressed in U2-OS cells. Ago2 protein localized to discrete cytoplasmic foci by staining with FITC-conjugated anti-Myc. (b) Endogenous Ago2 protein was localized in U2-OS cells by staining with a rabbit anti-Ago2 antibody. (c) Argonaute proteins colocalized with either GFP- or Flag-tagged Dcp1a, a signature component of the mammalian P-bodies. Argonaute proteins were visualized using a Rhodamine-Red-conjugated anti-Myc. Dcp1a was visualized either by GFP or FITC-conjugated anti-Flag.
Ago2 appeared in foci irrespective of its ability to cleave target RNAs, as demonstrated by the localization of catalytically incompetent mutants (see Supplementary Information, Fig. S1). Similar localization patterns were also seen for all other mammalian Argonaute proteins that have been shown to bind miRNAs (Ago1, -3 and -4; see Supplementary Information, Fig. S2). In contrast, members of the second Argonaute subfamily, the Piwi family, have not been shown to interact with miRNAs and do not localize to cytoplasmic foci when ectopically expressed (for example, Hiwi; data not shown).
The pattern and size of the Argonaute-containing foci were reminiscent of the cytoplasmic P-bodies (also termed Dcp bodies or GW bodies) that have previously been observed in both yeast and animal cells2–5,7,17. To test the possibility that Argonaute-containing foci were P-bodies, we asked whether Ago proteins colocalized with known P-body components. Dcp1a is a component of the decapping enzyme and has been demonstrated to reside in P-bodies in both yeast and mammals2–5,7. A comparison of the localization pattern of Myc–Ago1 or Myc–Ago2 with that of either Flag epitope- or green fluorescent protein (GFP)-tagged Dcp1a showed a remarkable overlap (Fig. 1c). In some cases, the relative intensity of the Dcp1a and Ago proteins in an individual focus varied, but close inspection reveals that essentially all Dcp1a and Ago foci overlap.
Because Argonaute proteins were localized to P-bodies, we asked whether we could observe a biochemical interaction between Ago and other P-body components. We could easily detect the presence of either GFP- or Flag-tagged Dcp1a in immunoprecipitates of Myc-tagged Ago1 or Ago2 (Fig. 2a). Moreover, when Dcp1a–GFP was immunoprecipitated with GFP antibody, Ago1 and Ago2 were co-immunoprecipitated (Fig. 2a). Interaction was also observed between Myc–Ago1/Myc–Ago2 and a second subunit of the decapping enzyme, Dcp2 (Fig. 2b). The physical interaction between Argonautes and Dcp1a or Dcp2 proteins could be a protein–protein interaction, or could be the result of these proteins interacting with a common mRNA or P-body structure. To distinguish between these possibilities, we treated lysates, prior to immunoprecipitation, with RNaseA, which both degrades mRNAs and destroys P-body integrity6,18. Even after such treatment, the interaction between Myc–Ago2 and GFP–Dcp1a or Flag–Dcp2 was preserved (Fig. 2c, d). These results indicate that mammalian Ago subfamily proteins are not only concentrated in P-bodies, but also interact physically with the P-body components Dcp1a and Dcp2 in a manner that is independent of ribonucleoprotein (RNP) or P-body integrity.
Figure 2.
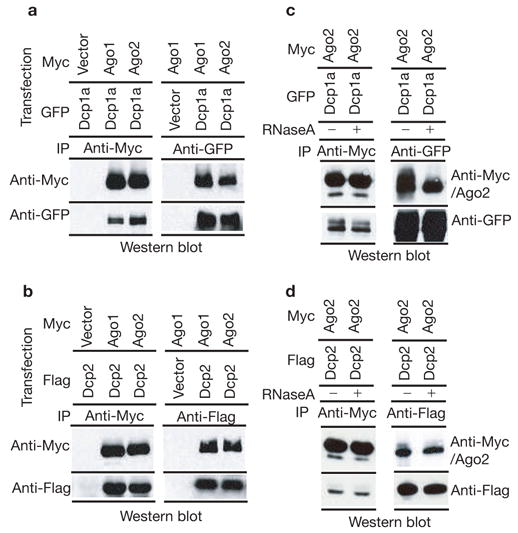
Argonaute proteins bind components of mammalian P-bodies. Human 293T cells were transfected with Myc-tagged Ago1 or Ago2 in combination with either GFP-tagged Dcp1a or Flag-tagged Dcp2 expression plasmids (as indicated). (a) Ago1 or Ago2 (anti-Myc) or Dcp1a (anti-GFP) immunoprecipitates were western blotted with anti-Myc or anti-GFP antibodies as indicated. (b) Ago1 or Ago2 (anti-Myc) or Dcp2 (anti-Flag) immunoprecipitates were western blotted with anti-Myc or anti-Flag antibodies as indicated. (c) Extracts were treated with RNaseA before immunoprecipitation of Ago2 (anti-Myc) or Dcp1a (anti-GFP). Immunocomplexes were western blotted with anti-Myc or anti-GFP antibodies as indicated. (d) Extracts were treated with RNaseA before immunoprecipitation of Ago2 (anti-Myc) or Dcp2 (anti-Flag). Immunocomplexes were western blotted with anti-Myc or anti-Flag antibodies as indicated.
The Argonaute proteins could accumulate in P-bodies because of protein–protein interactions, independent of their ability to function in RNAi. Alternatively, the Argonaute proteins could be targeted to P-bodies in a manner that depends upon intact interactions with small RNAs or even upon successful miRNA–mRNA recognition. We generated two mutant Ago2 proteins using the crystal structure of the human Ago1 PAZ domain bound to an siRNA-like duplex as a guide19. These mutants, Ago2-PAZ9 and Ago2-PAZ10, contain either nine or ten point mutations within the PAZ domain of Ago2. Both mutants show a substantially reduced ability to interact with small RNAs or to cleave target mRNAs (Fig. 3a). Notably, both mutants failed to accumulate in P-bodies (Fig. 3b). These results are consistent with the notion that Argonaute requires interaction with a small RNA for its accumulation in P-bodies. This outcome could suggest specific recognition of Ago proteins loaded with miRNA/siRNAs, either with or without target mRNAs, by a factor that mediates P-body localization. Alternatively, the mutations could have markedly altered the overall structure of the Ago2 proteins such that they were completely non-functional. Contrary to the latter possibility, both Ago2-PAZ9 and Ago2-PAZ10 mutants were expressed at normal levels and retained the ability to interact with Dcp1a and Dcp2 (Fig. 3a, c). This latter observation argues that the Ago–Dcp1a or Ago–Dcp2 interactions are not sufficient to recruit Ago proteins to the mammalian P-bodies. Thus, the simplest interpretation of the failure of Ago2-PAZ9 or Ago2-PAZ10 to localize to P-bodies is that miRNA binding to Ago2 is required for its accumulation within P-bodies.
Figure 3.
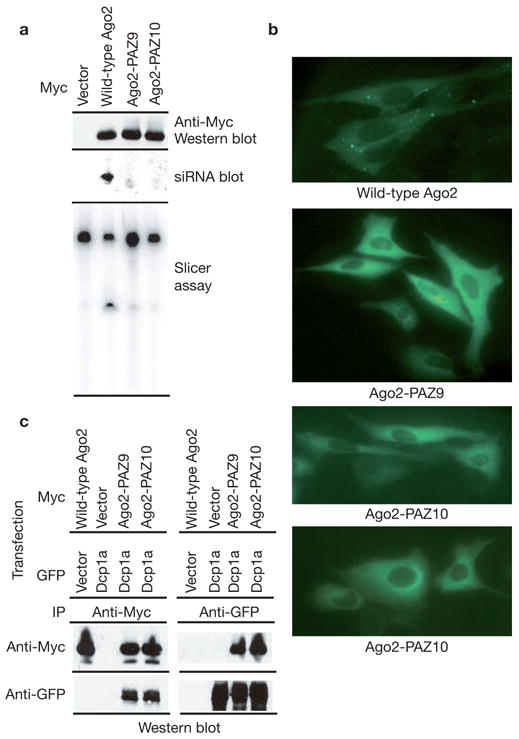
Accumulation of Argonaute proteins in P-bodies requires an intact siRNA-binding domain. (a) Wild-type and mutant Ago2 proteins, Ago2-PAZ9 and Ago2-PAZ10 (as indicated), were expressed as Myc–epitope fusions in cells transfected with a luciferase siRNA. Western blotting with an anti-Myc antibody was used to measure protein expression (upper panel). Small RNA binding was measured by northern blotting of RNA extracted from immunocomplexes (middle panel). Nuclease activity was also measured against a complementary substrate (bottom panel). (b) Subcellular localization of wild-type and non-small RNA binding mutant Ago2 proteins was determined by immunostaining with anti-Myc antibodies. (c) Myc-tagged wild-type or mutant Ago2 proteins were co-expressed with GFP-tagged Dcp1a. Ago2 (anti-Myc) or Dcp1a (anti-GFP) immunoprecipitates were analysed by western blotting with anti-Myc or anti-GFP antibodies, as indicated.
The presence of Argonaute proteins within P-bodies suggests that miRNA-mediated repression of protein synthesis might result in the targeting of miRNA–mRNA complexes to P-bodies. To test this hypothesis, we asked whether an mRNA targeted for translational repression by a miRNA would accumulate within P-bodies. We generated luciferase mRNA expression constructs with and without a portion of the C. elegans lin-41 3′ UTR, which is a target of the let-7 miRNA. Both the let-7 target and its control counterpart also contained 24 binding sites for the MS2 coat protein in their 3′ UTRs (Fig. 4a, b). These MS2-binding sites allowed us to follow the localization of these mRNAs by co-expression of an MS2–YFP–NLS fusion protein. In cells that lack a target mRNA, the fusion protein remains localized to the nucleus, reducing background cytoplasmic fluorescence. However, in the presence of an mRNA containing appropriate binding sites, a fraction of the fusion protein is carried into the cytoplasm, where its localization reports the location of the target mRNA20.
Figure 4.
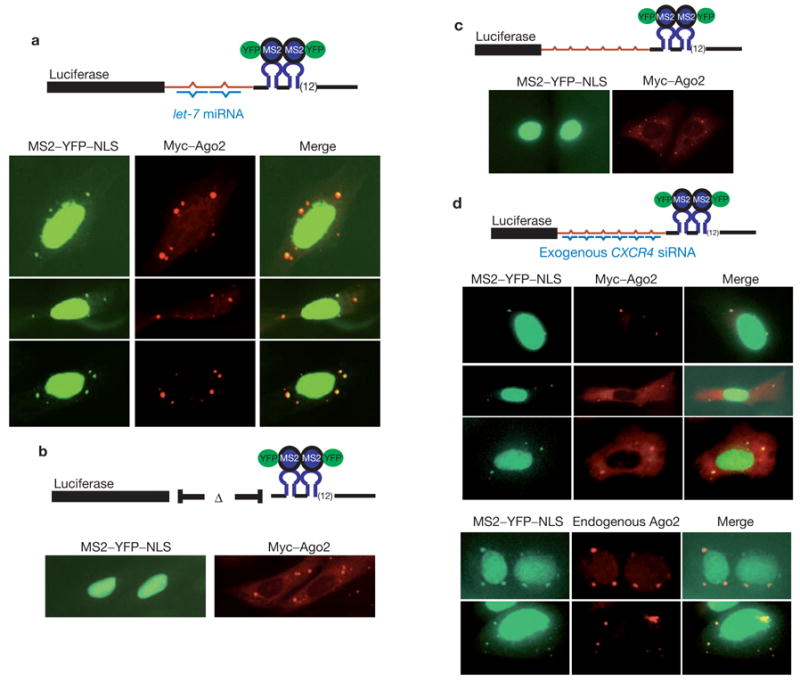
miRNA-dependent localization of target mRNAs to mammalian P-bodies. (a) Plasmids expressing the let-7 target, Myc–Ago2 protein and MS2–YFP–NLS were cotransfected into U2-OS cells. Reporter mRNA was visualized indirectly using the fusion protein. Ago2 protein was visualized using Rhodamine-Red-conjugated anti-Myc. (b) Analyses were identical to in a except that the target mRNA did not contain the lin-41 3′ UTR fragment. (c) The CXCR4 target was co-expressed with MS2–YFP–NLS and Myc–Ago2. Detection was as in a. (d) The CXCR4 reporter was co-expressed with MS2–YFP–NLS and with (upper) or without (lower) Myc–Ago2 in cells that were also transfected with the CXCR4 siRNA. Endogenous Ago2 was visualized with a rabbit anti-Ago2 antibody. Diagrammatic representation targets are shown next to each panel.
Expression of the let-7 target in U2-OS cells, which endogenously express abundant endogenous let-7 miRNA, provided two observations. First, the construct containing the lin-41 3′ UTR fragment generates ~twofold less luciferase than the control transcript that does not contain the sites. Both the site-dependence and the magnitude of the change in the reporter are consistent with previously observed regulation of similar reporters by let-7 (ref. 21). A second, and critical, observation was that when the let-7 target was expressed in U2-OS cells, we observed discrete cytoplasmic foci of the MS2–YFP–NLS fusion protein that colocalized with Myc–Ago2 (Fig. 4a). Colocalization was not unique to Ago2 but was also observed for other Argonaute subfamily members (see Supplementary Information, Fig. S3). This observation indicated that the let-7 target mRNA concentrated in cytoplasmic P-bodies. Notably, expression of a target mRNA that lacked the let-7 binding sites showed no cytoplasmic foci of the MS2–YFP–NLS protein, indicating that this mRNA was not concentrated in P-bodies (Fig. 4b).
The aforementioned data indicate that an mRNA can be localized to P-bodies in a manner that is dependent on the presence of a miRNA-binding site. However, because let-7 is endogenously expressed, we could not ensure that localization was miRNA directed. Thus, we constructed a target that contains both the MS2-binding region and multiple imperfect complements of a CXCR4 siRNA that has been shown to act as a miRNA mimetic, repressing target expression in the absence of slicer cleavage22 (Fig. 4c, d). Co-expression of the CXCR4 target with the fluorescent MS2 protein in the absence of the CXCR4 siRNA showed no clear foci (Fig. 4c). However, when the siRNA was co-delivered, we observed that the CXCR4 target was concentrated in cytoplasmic foci that colocalized with Myc–Ago2, Myc–Ago3 and endogenous Ago2 proteins (Fig. 4d and see Supplementary Information, Fig. S3). Concomitant with its localization in P-bodies, the expression of the reporter was reduced by two- to threefold (data not shown). Considered together, our results indicate that miRNA-regulated mRNAs can accumulate in mammalian P-bodies in a manner that depends both on the presence of the small RNA and upon the presence of the appropriate recognition sites in the target mRNA.
Several observations in this work now indicate a connection between mammalian P-bodies and miRNA-regulated repression of mRNAs. First, Argonaute proteins localize to P-bodies. Second, localization of Ago2 to P-bodies requires the ability to bind miRNAs. Third, two different reporter mRNAs — one a target of the endogenous let-7 miRNA and one a target of an artificial miRNA mimic — accumulate in P-bodies in a manner that is dependent on the miRNA, or the miRNA-binding sites. Fourth, Ago1 and Ago2 co-immunoprecipitate with the P-body components, Dcp1a and Dcp2. Moreover, because Dcp–Ago interactions are resistant to RNaseA and occur with Ago2 mutants that do not localize to P-bodies, this interaction is likely to occur both within and outside of P-bodies and could have an early role in the targeting of miRNA targets to P-bodies. Considered together, a straightforward hypothesis based on these observations is that mRNAs undergoing miRNA-mediated translation repression accumulate within P-bodies in conjunction with an Argonaute–miRNA complex.
Our results are consistent with several previous studies that suggest a connection between P-bodies and the RNAi machinery. Homologues of Xrn1p, a 5′ to 3′ exonuclease that is concentrated in P-bodies, have been shown in plants and in Drosophila melanogaster S2 cells to degrade the 3′ products that arise from small RNA-directed cleavage of mRNAs23,24. Strikingly, Xrn1p is required for efficient RNAi in C. elegans25. Additionally, mutation of Xrn4 in Arabidopsis thaliana induced an RNAi response against certain transgenes, presumably because of the accumulation of aberrant RNAs26. Our results may also provide an explanation for slicer-independent reduction in the abundance of proposed miRNA targets in mammals27. Sequestration of translationally repressed mRNAs to P-bodies and the direct interaction between Argonautes and the Dcp1/Dcp2 complex might promote decapping of targets, thereby enhancing their degradation, which might also explain the miRNA-dependent increase in mRNA decay seen for AU rich element (ARE)-containing mRNAs28.
An issue that remains to be resolved is the precise nature of the functional connection between miRNA-mediated regulation and P-bodies. In one model, miRNAs — presumably in conjunction with Argonaute proteins — could repress the translation of their target genes by affecting some aspect of translation, per se. As a consequence, the targeted mRNAs could be recognized by an unknown machinery and taken to the P-body for sequestration and perhaps ultimate disposal. In this regard, it should be noted that, at least in yeast, defects in translation elongation prevent mRNAs from entering P-bodies, and defects in translation initiation promote mRNAs entering P-bodies5–7. Thus, the presence of miRNA-targeted mRNAs in P-bodies argues that if RISC inhibits translation directly, it is likely to do so at the level of translation initiation. An alternative model is that localization to the P-body is a direct consequence of the interaction between the Argonaute protein/miRNA complex and its target, perhaps by Argonaute promoting the assembly of a translationally repressed mRNP targeted to a P-body. In this model, it is the localization to the P-body that is, itself, responsible for preventing protein synthesis, because the translation machinery is excluded from this structure in both yeast and mammals6,29. A hybrid model suggests that RISC-mediated translation repression involves an initial inhibition of translation per se, which then leads to targeting of the mRNA to a P-body, where sequestration from the translation machinery reinforces silencing.
METHODS
DNA constructs
Myc-tagged Ago expression plasmids were as described in ref. 16. GFP-tagged Dcp1a, Flag-tagged Dcp1a and Dcp2 plasmids were as described in ref. 4. Ago2-PAZ9 and Ago2-PAZ10 mutants contained multiple point mutations in the PAZ domain of Ago2 (R277A, K278A, Y279A, F294A, Y311A, F312G, T337A, Y338A, L339A, −/+ H271A). Mutations were introduced by site-directed mutagenesis using the QuickChange Kit from Stratagene (La Jolla, CA). The mRNA reporters were constructed by modifying the pcDNA3-firefly luciferase construct as described in Fig. 4. The let-7 miRNA recognition sites were derived from C. elegans lin-41 3′ UTR. The CXCR4 siRNA recognition sites were derived from a CXCR4 siRNA-response luciferase reporter as described in ref. 22.
Cell culture and transfection
Human U2-OS, HeLa and 293T cells were cultured in DMEM (10% FBS) in a 37 °C incubator with 5% CO2. Cell transfections were performed using Mirus (Madison, WI) TransIT-LT1 reagent for DNA plasmids and Invitrogen (Carlsbad, CA) Oligofectamine reagent for siRNAs. Procedures for immunoprecipitation, immunoblotting, RNA extraction, small RNA blotting and slicer assay were described previously16. To assess the RNA dependence of protein–protein interactions, lysates were treated with RNAseA at 0.5 μg μl−1 for 20 min at room temperature before immunoprecipitation procedures as described in ref. 18.
Immunofluorescence
Cultured cells were fixed using 3% paraformaldehyde (in PBS) for 12 min at room temperature. The cells were then permeabilized in PBS containing 0.2% Triton-X100 for 6 min at 4 °C. FITC- or Rhodamine-Red-conjugated secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA). Antibody incubations were done at room temperature in PBS containing 0.5% BSA as blocking agent. The cells were examined using an Axioskop (Carl Zeiss, Thornwood, NY) fluorescent microscope. In transfected cells, not all cells showed foci; for example, in experiments following localization of miRNA targets, roughly 20% of cells showed that miRNA targets are in foci. This is probably because of the need to achieve appropriate expression levels of a target RNA and two exogenous proteins.
BIND identifiers
Four BIND identifiers (www.bind.ca) are associated with this manuscript: 295721, 295722, 295723 and 295724.
Supplementary Material
Supp fig 1-3
Figure S1 Localization of catalytically inactive Ago2 mutants. Catalytically inactive Ago2 mutants, H634A, D597A and D669A were expressed in U2-OS cells as myc-tagged proteins and localized in cells using FITC-conjugated anti-myc
Figure S2 Localization of catalytically inactive Argonaute subfamily proteins. Myc-tagged Ago1, Ago3 and Ago4 proteins were immunolocalized in human U2-OS cells using FITC-conjugated anti-myc antibodies.
Figure S3 Co-localization of Ago3 with miRNA targets. (a) The luciferase-lin41 UTR fusion reporter was co-expressed in U2-OS cells with an MS2-YFP-NLS fusion protein and myc-Ago3. Reporter mRNA was visualized indirectly with YFP. Ago3 was localized using a Rhodamine Red conjugated anti-myc antibody. (b) The luciferase-CXCR4 reporter was expressed in cells that were also transfected with the CXCR4 siRNA and a plasmid directing the expression of myc-Ago3. Analysis was as in a.
Acknowledgments
We thank members of the Hannon laboratory for helpful discussions, S. Hearn from the CSHL microscopy shared resource for assistance, and S. Janicki (CSHL), J. Lykke-Andersen (University of Colorado) and T. Achsel (University of Wurzburg) for reagents. J.L. is supported by a Special Fellow award from the Leukemia and Lymphoma Society. This work was supported by grants from the NIH (to G.J.H. and R.P.). R.P. is an investigator at the Howard Hughes Medical Institute.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
Footnotes
Note added in proof: After this study had been accepted for publication, Sen and Blau30 reported similar observations of the localization of human Ago2 to cytoplasmic P-bodies.
Note: Supplementary Information is available on the Nature Cell Biology website.
References
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 3.van Dijk E, et al. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 9.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 17.Eystathioy T, et al. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tharun S, et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 19.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 22.Doench JG, Petersen CP, Sharp P. A siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newbury S, Woollard A. The 5′-3′ exoribonuclease xrn-1 is essential for ventral epithelial enclosure during C. elegans embryogenesis. RNA. 2004;10:59–65. doi: 10.1261/rna.2195504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 27.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 28.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 29.Andrei MA, et al. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nature Cell Biol. 2205;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp fig 1-3
Figure S1 Localization of catalytically inactive Ago2 mutants. Catalytically inactive Ago2 mutants, H634A, D597A and D669A were expressed in U2-OS cells as myc-tagged proteins and localized in cells using FITC-conjugated anti-myc
Figure S2 Localization of catalytically inactive Argonaute subfamily proteins. Myc-tagged Ago1, Ago3 and Ago4 proteins were immunolocalized in human U2-OS cells using FITC-conjugated anti-myc antibodies.
Figure S3 Co-localization of Ago3 with miRNA targets. (a) The luciferase-lin41 UTR fusion reporter was co-expressed in U2-OS cells with an MS2-YFP-NLS fusion protein and myc-Ago3. Reporter mRNA was visualized indirectly with YFP. Ago3 was localized using a Rhodamine Red conjugated anti-myc antibody. (b) The luciferase-CXCR4 reporter was expressed in cells that were also transfected with the CXCR4 siRNA and a plasmid directing the expression of myc-Ago3. Analysis was as in a.