Structural basis of family-wide Rab GTPase recognition by Rabenosyn-5 (original) (raw)
. Author manuscript; available in PMC: 2006 Feb 2.
Published in final edited form as: Nature. 2005 Jul 21;436(7049):415–419. doi: 10.1038/nature03798
Abstract
Rab GTPases regulate all stages of membrane trafficking, including vesicle budding, cargo sorting, transport, tethering, and fusion1, 2. In the inactive (GDP-bound) conformation, accessory factors facilitate the targeting of Rab GTPases to intracellular compartments3–8. Following nucleotide exchange to the active (GTP-bound) conformation, Rab GTPases interact with functionally diverse effectors including lipid kinases, motor proteins, and tethering complexes. How effectors distinguish between homologous Rab GTPases represents an unresolved problem with respect to the specificity of vesicular trafficking. Using a structural proteomic approach, we have determined the specificity and structural basis underlying the interaction of the multivalent effector Rabenosyn-5 with the Rab family. The results demonstrate that even structurally similar effectors can achieve highly selective recognition of distinct subsets of Rab GTPases exclusively through interactions with the switch and interswitch regions. The observed specificity is determined at a family-wide level by structural diversity in the active conformation, which governs the spatial disposition of critical conserved recognition determinants, and by a small number of both positive and negative sequence determinants that allow further discrimination between Rab GTPases with similar switch conformations.
Many Rab GTPases, including Rabs 4, 5, 7, 9, 11, 14, 21 and 22, have overlapping localizations within the endosomal system2, 9–15. The sequential endocytic and recycling functions of Rab5 and Rab4 are further linked by multivalent effectors, such as Rabenosyn-5, Rabaptin-5, and Rabip4, which interact with both Rab proteins through separate domains16–18. Multiple Rab partners have also been identified for other effectors13, 19. Although effectors engage conserved and non-conserved regions of Rab GTPases20–22, what determines specificity at the Rab family level remains unknown.
To better understand how structural variability in the switch regions contributes to Rab-effector recognition, crystal structures were determined for 11 Rab GTPases bound to GppNHp and/or GDP (Supplementary Figure 1 and Supplementary Tables 1–3). Combined with earlier studies (Supplementary Table 3), at least one structure is available for the active conformation of 14 Rab GTPases, spanning 6 subfamilies23, 24 and representing a third of the Rab family, excluding isoforms. With the exception of Rab21, both switch regions adopt a unique active conformation independent of crystal packing (Supplementary Figure 2). The inactive switch conformations, however, are either disordered or dictated by crystal contacts. In Rab21, the switch II region remains poorly ordered even in the active conformation.
To compare the active structures, superpositions were generated for all pair-wise combinations (Supplementary Figure 3). Whereas the most similar elements coincide with conserved motifs required for nucleotide and Mg2+ binding, structural variability is a hallmark of the switch II region, the interswitch region, and the α3/β5 loop. Although the latter regions are typically mobile, the structural diversity in the switch II region is primarily the consequence of non-conservative substitutions25. Consistent with evolutionary pressure on functional sites, non-phylogenetic relationships occur at the local structural level. For example, the switch regions of Rab4 (subfamily II) adopt an active conformation similar to Rab5 and Rab22 (subfamily V). Conversely, the active switch conformations are dissimilar for Rab7 and Rab9 (subfamily VII) as well as Rab4 and Rab11 (subfamily II). These local relationships contribute to Rab-effector recognition and are reminiscent of the non-phylogenetic functional specificity reported for the Ras superfamily26.
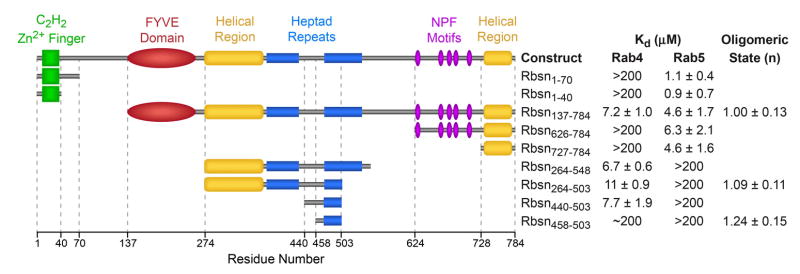
Rabenosyn-5 (Rbsn) contains distinct Rab4 and Rab5 binding sites within residues 264–500 and 627–784, respectively17. As shown in Figure 1 and Supplementary Figure 4, the Rab5 binding domain maps to a predicted helical region at the C-terminus (Rbsn728–784). A BLAST search detected significant homology with residues 458–503 in the Rab4 binding region though not with other proteins. Rbsn458–503 overlaps the second of two heptad repeats, yet this region and Rbsn728–784 are monomeric. Although Rbsn458–503 binds Rab4, the affinity is 10–100 fold weaker than that of a longer construct, Rbsn440–503, which represents the minimal Rab4 binding domain. The structure of Rbsn458–503 reveals a helical hairpin similar to an anti-parallel coiled coil (Supplementary Figure 4). In the complexes with Rab GTPases discussed below, an equivalent structure is observed for the homologous core. These observations suggest two important questions: i) how specific are the Rabenosyn-5 Rab binding domains?; and ii) how do they distinguish structurally similar Rab GTPases?
Figure 1.
Mapping of the Rabenosyn-5 Rab GTPase binding domains. Mean Kd values and standard deviations were obtained from 2–4 SPR experiments using purified Rab GTPases and Rabenosyn-5 constructs. The oligomeric state (n = 1 for monomer; n = 2 for dimer) was determined from sedimentation equilibrium experiments over the concentration range from 5 – 50 mM. Values represent the mean and standard deviation for 3 measurements.
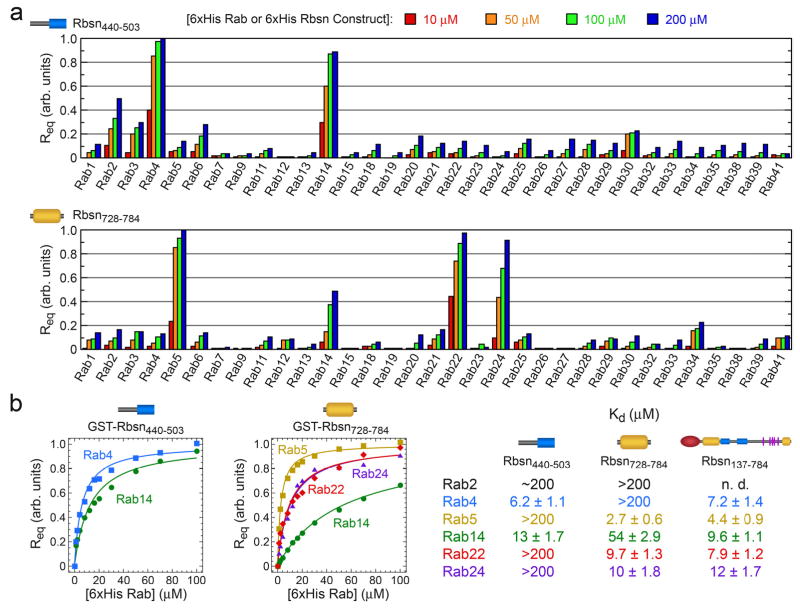
The specificities of Rbsn440–503 and Rbsn728–784 were profiled against 33 purified Rab GTPases (Figure 2 and Supplementary Figure 5). Rbsn440–503 binds with similar affinity to Rab4 and Rab14 and weakly to Rab2 whereas Rbsn728–784 exhibits the highest affinity for Rab5, 3 fold lower affinity for Rab22 and Rab24, and binds weakly to Rab14. Other interactions are too weak for detection (Kd > 200 mM). Thus, Rbsn440–503 and Rbsn728–784 selectively recognize distinct subsets of Rab GTPases, despite similar core structures. Although further studies are required to determine the in vivo significance, it is noteworthy that both Rab14 and Rab22 extensively co-localize with Rab5 and have been implicated endosomal trafficking12, 13, 27, 28. Given that the cellular functions of effectors including Rabenosyn-5 may be mediated by interactions with subsets of Rab GTPases, it will be of interest to consider the specificity for the entire Rab-family when evaluating the molecular basis by which effectors regulate trafficking events.
Figure 2.
Quantitative family-wide analysis of Rab GTPase-effector specificity. a, Initial screen for the interaction of 6xHis (or GST) fusions of Rab GTPases with GST (or 6xHis) fusions of Rbsn440–503 and Rbsn728–784. For each potential interaction, the equilibrium SPR signal (Req) was measured at four concentrations of the 6xHis Rab GTPase or 6xHis Rbsn construct. b, Concentration dependence of the equilibrium SPR signal (Req) for the binding of 6xHis Rab GTPases to GST fusions of Rbsn440–503 and Rbsn728–784. Mean Kd values and standard deviations for 2–4 independent experiments are tabulated on the right.
Crystallization screens for the high affinity combinations yielded crystals for the Rab4-Rbsn440–503 and Rab22-Rbsn728–784 complexes. The structures reveal similar modes of interaction, with the helical hairpins engaging equivalent residues in the switch and interswitch regions (Figure 3 and Supplementary Figure 6). Surface areas of 2136 Å2 and 1312 Å2 are buried at the respective interfaces. The significantly larger contact area in the Rab4-Rbsn440–503 complex evidently compensates for the weak binding of Rab4 to the helical hairpin core.
Figure 3.
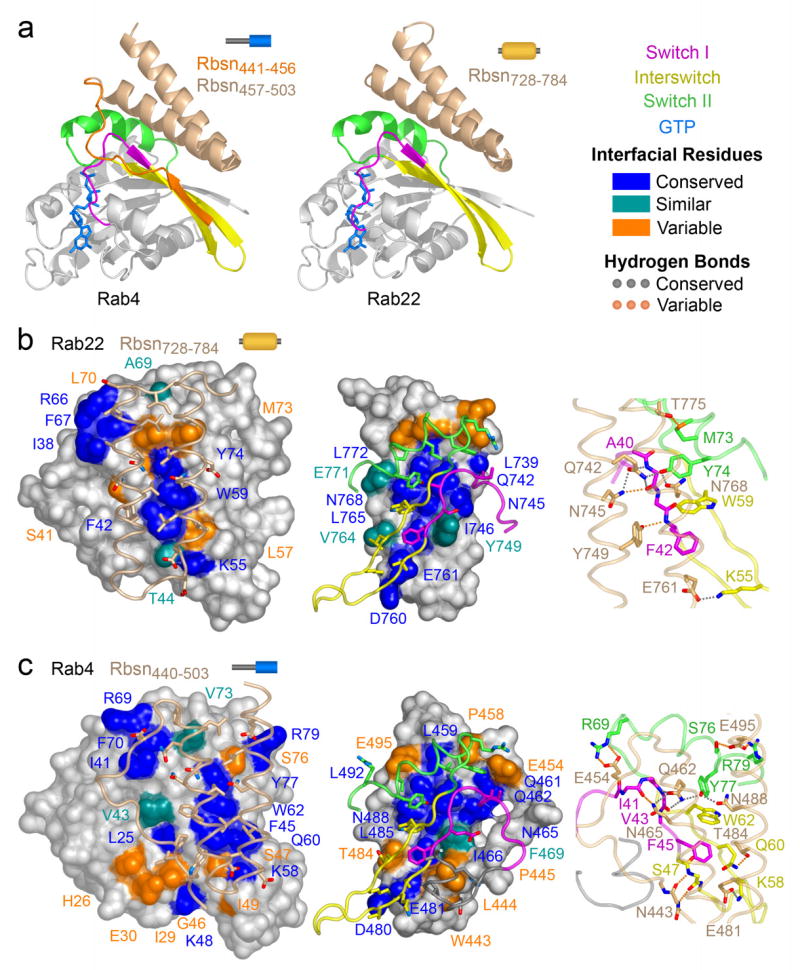
Structural basis of Rab recognition by Rabeonsoyn-5. a, Ribbon rendering of GTP-bound Rab4 Q67L and Rab22 Q64L in complex with the minimal Rab binding domains of Rabenosyn-5. b, Conservation and variability in the Rab22-Rbsn728–784 interface. Spheres covered by a semitransparent surface represent Rab22 (left panel) or Rbsn728–784 (right panel). Hydrogen bonding interactions are depicted in the right panel. c, Conservation and variability in the Rab4-Rbsn440–503 interface. Spheres covered by a semitransparent surface represent Rab4 (left panel) or Rbsn440–503 (right panel). Hydrogen bonding interactions are depicted in the right panel.
The binding epitope in Rbsn728–784 is divided into two hydrophobic pockets by a polar ridge bisecting the long axis. One pocket buries invariant residues (Phe 42 and Trp 75) from the switch I and interswitch regions of Rab22. The second pocket, which is capped by residues 733PIEEEL738 at the N-terminus of α1, docks variable residues in switch II (Leu 70 and Met 73) and partially buries a cluster of conserved residues at the switch interface (Ile 38, Arg 66, and Phe 67). The side chains of residues lining the polar ridge mediate hydrogen bonding interactions with the backbone of Ala 41 in switch I and side chain hydroxyl of Tyr 74 in switch II. Additional polar interactions at the periphery involve conserved (Lys 55) and variable (Met 73) residues in the interswitch and switch regions.
In Rbsn440–503, the N terminus of α1 is foreshortened by Pro 458, which packs against Val 73 in the switch II region of Rab4 and, in conjunction with an ionic interaction between Glu 454 and Arg 69, directs the N terminal region into a flexible loop that connects with the β1 strand encoded by residues 441EGWLP445. The latter engage the β2 strand of Rab4 via two main chain hydrogen bonds, allowing Trp 443 and Pro 445 to pack against the invariant Phe 45 in switch I while Leu 444 occupies a hydrophobic pocket on the α1 helix of Rab4. This pocket exists as a consequence of a glycine residue following Phe 45. In most Rab GTPases, including Rab5 and Rab22, this glycine is replaced by a bulky side chain. Finally, several substitutions contribute to differences in the interface with the helical hairpin.
To identify compositional determinants of the observed specificity, we analyzed the effects of mutations within or proximal to the binding interfaces (Figure 4). In Rab5, broadly conserved residues were substituted with alanine whereas residues selectively conserved in Rab5, Rab22 and Rab24 were replaced with the consensus residue for other Rab GTPases. Although most substitutions have little effect or otherwise enhance the affinity for Rbsn728–784, 10 fold decreases are observed for the A57D and M89S substitutions. The corresponding residues in Rab22 (Ser 41 in switch I and Met 73 in switch II) are located within or proximal to the interface with Rbsn728–784. Met 73 is buried in a hydrophobic pocket flanked by variable residues in the helical hairpin. Interestingly, the M89A substitution is without effect, suggesting that the M89S defect reflects sequestration of a polar residue in a non-polar environment. Conversely, the effects of the Ala 57 substitutions correlate with side chain volume rather than polarity, consistent with a packing defect.
Figure 4.
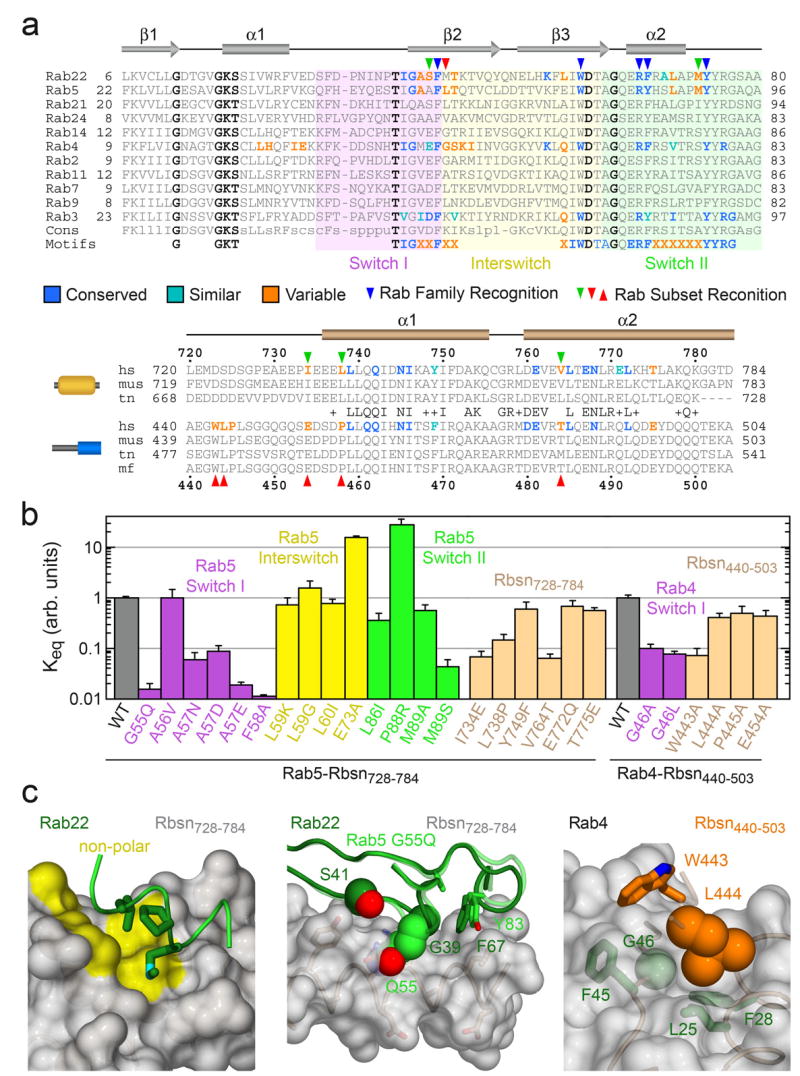
Structure-based mutational analysis of Rab and Rabeonsoyn-5 interaction specificity. a, Local alignment of representative Rab GTPases and Rabenosyn-5 homologs. Residues within the binding interfaces for Rab22-Rbsn728–784, Rab4-Rbsn440–503, Rab5-Rabaptin-522, and Rab3-Rabphilin21 are highlighted according to conservation. b, Effect of site specific substitutions on the interaction between Rab5 and Rbsn728–784 or Rab4 and Rbsn440–503. Mean values and standard deviations were calculated from 2 experiments. c, Views of the Rab22-Rbsn728–784 and Rab4-Rbsn440–503 interfaces relevant to the mutations discussed in the text. Also shown (middle panel) is the structure of the Rab5 G55Q mutant following superposition with Rab22.
The ability of Rbsn728–784 to recognize Rab5 and Rab22 yet discriminate against Rab21 derives in part from a distinctive substitution in Rab21 whereby a glutamine replaces the otherwise invariant switch I glycine. The corresponding G55Q substitution in Rab5 decreases affinity by 2 orders of magnitude. As the Cα of Gly 55 packs against residues in switch II, a glutamine substitution should alter the structure and likely the stability of the active conformation. Indeed, the structure of the G55Q mutant reveals a main chain perturbation in which Gln 55 bulges outward. Superposition with Rab22 suggests that van der Waals overlap contributes to the observed binding defect. Notably, the reverse Q53G substitution in Rab21 does not confer the ability to interact with Rbsn728–784, nor is it sufficient to allow Rab21 to adopt an active conformation similar to Rab5 or Rab22 (Supplementary Figure 2).
Order of magnitude defects are also observed for the I734E, L738P, and V764T mutations that replace residues in Rbsn728–784 with the corresponding residues in Rbsn440–503. Ile 734 and Leu 738 form one side of the non-polar pocket for residues Phe 67, Met 73, and Leu 70 in the switch II region of Rab22. Both polarity and packing are adversely impacted by the I734E substitution whereas the L738P substitution disrupts the main chain conformation and side chain packing. As Val 764 packs against Phe 42, Leu 57, and Trp 59 in the switch I and interswitch regions of Rab22, desolvation is a likely factor underlying the V764T defect.
Mutations in the N-terminal region of Rbsn440–503 have minor effects with the exception of W443A, which results in a 10 fold decrease in affinity. Trp 443 packs against non-polar residues in Rab4 including the invariant Phe 45 in switch I. To facilitate this interaction and the main chain hydrogen bonds between the β1 strand of Rbsn440–503 and the β2 strand of Rab4, the side chain of Leu 444 must pack against the Cα of Gly 46. Order of magnitude defects for the G46A and G46L mutations, combined with the lack of a significant defect for the L444A substitution, implicate Leu 444 as a negative determinant that selects against the majority of Rab GTPases.
Consistent with the preceding observations, a triple mutation (E454I, P458L, T484V) that converts predicted specificity determinants in Rbsn418–503 to the corresponding residues in Rbsn728–784 exhibits reversed specificity with a preference for Rabs 5, 22, and 24 (Supplementary Figure 7). Eliminating residues 418–447, which are required for binding to Rabs 4 and 14, has no effect on the affinity of the triple mutant for Rab5 (data not shown). A chimera in which the N terminus of Rbsn418–503 is fused to the helical hairpin of Rbsn728–784 exhibits a comparable reversal of specificity resulting in a preference for Rabs 4 and 14, which is further enhanced by an amino acid substitution (V764T) in the helical hairpin.
Comparison with the Rab5-Rabaptin22, Rab3-Rabphilin21 and other GTPase-effector complexes29 reveals similarities in the overall modes of interaction, which are particularly striking for the effectors containing helical hairpins or coiled coils as the core GTPase binding element30. However, even structurally homologous domains can achieve selective recognition of distinct Rab GTPase subsets, and further discriminate between Rab GTPases with similar active conformations, on the basis of interactions with the switch and interswitch regions. The exquisite specificity of Rabenosyn-5 Rab binding domains is determined by structural as well as compositional diversity. This combination of factors enhances the affinity for Rab-effector subsets with complementary interfaces at the expense of combinations with incompatible structures and/or compositions. The family-wide nature of the recognition process is underscored by the conservation of positive determinants in interacting subsets and negative determinants in non-interacting Rab GTPases. In this respect, the encoding of Rab-effector recognition determinants parallels that observed for the determinants of functional specificity in small GTPases 26.
Methods
Constructs
Constructs were amplified and subcloned into pGEX vectors (Amersham Biosciences) for expression as GST fusions or into modified pET vectors (Supplementary Table 1). The modified pET vectors incorporate an N-terminal 6xHis tag with (MGHHHHHHGSLVPRGS) or without (MGHHHHHHGS) a thrombin cleavage site. Mutations were generated with the Quick Change kit (Stratagene). All constructs were verified by sequencing the entire coding region.
Expression and Purification
BL21(DE3) Codon Plus-RIL cells (Stratagene) transformed with expression plasmids were cultured in 2xYT containing 100 mg/L ampicillin. For co-expression of modified pET15b-Rbsn728–784 and modified pET28a-Rab22 Q64L, the cultures were supplemented with 50 mg/L kanamycin. Cells were grown at 22°C to an OD600 of 0.4, induced with 0.05 mM IPTG for 16 h, and disrupted by sonication in 50 mM Tris, pH 7.5 or pH 8.5, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 0.1 mM PMSF, and 0.2 mg/ml lysozyme. After supplementing with 0.5% Triton X-100, the lysates were centrifuged at 35,000xg for 40 min. For 6xHis fusions, supernatants were loaded onto Ni2+-NTA agarose (Qiagen), washed with 50 mM Tris, pH 7.5 or pH 8.5, 500 mM NaCl, 2 mM MgCl2, 15 mM imidazole, 0.1% 2-mercaptoethanol, and eluted with a gradient of 15–300 mM imidazole. For GST fusions, the supernatants were loaded onto glutathione-Sepharose (Amersham Biosciences), washed with 50 mM Tris, pH 7.5 or pH 8.5, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, and eluted with 10 mM reduced glutathione. Fusion tags were removed by digestion with thrombin (Haematologic Technologies) or PreScission protease (Amersham Biosciences) and the cleaved proteins isolated over glutathione-Sepharose or Ni2+-NTA agarose. Constructs were further purified by ion exchange over Source Q or Source S followed by gel filtration over Superdex-75 (Amersham Biosciences). Complexes were prepared by co-purification following co-expression (Rab22 Q64L/Rbsn728–784) or by mixing in a 1:1 molar ratio followed by gel filtration over Superdex-75 (Rab4 Q67L/Rbsn440–503).
Nucleotide Exchange
Rab GTPases at 2–10 mg/mL were incubated for 6 h with a 25 fold excess of GppNHp in 25 mM Tris, pH 7.5 or pH 8.5, 150 mM NaCl, 5 mM EDTA, 0.1% 2-mercaptoethanol, and alkaline phosphatase (100 IU/μmol GTPase). After supplementing with 10 mM MgCl2, excess nucleotide was removed by gel filtration over Superdex-75 (>2 mg GTPase) or a Pierce D-Salt column (<2 mg GTPase).
Surface Plasmon Resonance
SPR experiments were performed on a BIAcore X instrument. Anti-GST was coupled to CM5 sensor chips using reagents and protocols supplied by the manufacturer. All proteins were dialyzed into 10 mM Tris, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.005% Tween 20 and centrifuged at 13000xg. Sample and reference flow cells were loaded with 800 nM GST fusion or GST, respectively. A flow rate of 0.02 mL/min was used for all injections. Binding and dissociation were monitored following injection of 6xHis fusions at varying concentration. Following curve alignment and subtraction of the reference sensogram, the equilibrium signal (Req) at each concentration was extracted by fitting to a Langmuir binding model. Dissociation constants (Kd) were obtained from a fit to Req = Rmax [6xHis fusion]/(Kd + [6xHis fusion]).
Sedimentation Equilibrium
6xHis Rabenosyn-5 constructs were dialyzed against 50 mM Tris, pH 7.5, 100 mM NaCl, and centrifuged to equilibrium in a Beckman Optima XLI. The absorbance at 230 or 280 nm was measured as a function of the radial distance (r) from the axis of rotation. Data were analyzed by fitting with the function A(r) = Co + Ci exp(−n sm (ro2 − r2) / 2), where Co and Ci are constants, n represents the oligomeric state, ro is last data point, and sm is calculated with SEDINTERP using the monomer molecular mass.
Crystallization and Structure Determination
Individual proteins and complexes were crystallized in hanging drops with or without microseeding. Crystals were transferred to a cryostabilizer solution, flash frozen, and maintained at 100°K in a nitrogen cryostream (Oxford Cryosystems). Diffraction data were collected at NSLS beamline X25 (complexes and Rab2) and on Rigaku RUH3R generators equipped with Osmic mirrors and Mar 30 cm or R-axis IV detectors. Data were processed with Denzo and scaled with Scalepack. Structures were solved by molecular replacement using AMoRe, Molrep, or Phaser. Crystallographic models were refined by simulated annealing in CNS, automated atom updating with Arp/wArp, minimization with Refmac5, and model building in O. Except where otherwise noted, all programs were used as implemented in CCP4. Additional information related to the structure determination and refinement is compiled in Supplementary Table 2 and Supplementary Figure 1. Molecular graphics were rendered with PyMol.
Supplementary Material
Supp fig 1
Supp fig 2
Supp fig 3
Supp fig 4
Supp fig 5
Supp fig 6
Supp fig 7
Supp table 1
Supp table 2
Supp table 3
Acknowledgments
We are grateful to Marino Zerial for a full length clone of Rabenosyn-5, Ellie Kittler and Maria Zapp for assistance with SPR experiments, and Anna Delprato for Rab5 mutants. SPR data were collected in UMASS Center for AIDS Research Molecular Biology Core (UMASS CFAR 5P30 AI42845). This work was supported by NIH grant GM56324.
Footnotes
Competing interests statement. The authors declare that they have no competing financial interests.
References
- 1.Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 2.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 3.Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425:856–859. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- 4.Rak A, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 5.Calero M, et al. Dual prenylation is required for Rab protein localization and function. Mol Biol Cell. 2003;14:1852–1867. doi: 10.1091/mbc.E02-11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rak A, et al. Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell. 2004;117:749–760. doi: 10.1016/j.cell.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 9.Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombardi D, et al. Rab9 functions in transport between late endosomes and the trans Golgi network. Embo J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesa R, Salomon C, Roggero M, Stahl PD, Mayorga LS. Rab22a affects the morphology and function of the endocytic pathway. J Cell Sci. 2001;114:4041–4049. doi: 10.1242/jcs.114.22.4041. [DOI] [PubMed] [Google Scholar]
- 13.Kauppi M, et al. The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci. 2002;115:899–911. doi: 10.1242/jcs.115.5.899. [DOI] [PubMed] [Google Scholar]
- 14.Junutula JR, et al. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson JC, et al. A role for the small GTPase Rab21 in the early endocytic pathway. J Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- 16.Vitale G, et al. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. Embo J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Renzis S, Sonnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- 18.Fouraux MA, et al. Rabip4' is an effector of rab5 and rab4 and regulates transport through early endosomes. Mol Biol Cell. 2004;15:611–624. doi: 10.1091/mbc.E03-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuda M. Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J Biol Chem. 2003;278:15373–15380. doi: 10.1074/jbc.M212341200. [DOI] [PubMed] [Google Scholar]
- 20.Pfeffer SR. Structural Clues to Rab GTPase Functional Diversity. J Biol Chem. 2005;280:15485–15488. doi: 10.1074/jbc.R500003200. [DOI] [PubMed] [Google Scholar]
- 21.Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhu G, et al. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol. 2004;11:975–983. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
- 23.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 24.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 25.Merithew E, et al. Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition. J Biol Chem. 2001;276:13982–13988. doi: 10.1074/jbc.M009771200. [DOI] [PubMed] [Google Scholar]
- 26.Heo WD, Meyer T. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 2003;113:315–328. doi: 10.1016/s0092-8674(03)00315-5. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Guo J, Miki T, Tachibana M, Gahl WA. Molecular cloning of two novel rab genes from human melanocytes. Gene. 1996;174:129–134. doi: 10.1016/0378-1119(96)00509-4. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Gabin AG, Cammer M, Almazan G, Charron M, Larocca JN. Role of rRAB22b, an oligodendrocyte protein, in regulation of transport of vesicles from trans Golgi to endocytic compartments. J Neurosci Res. 2001;66:1149–1160. doi: 10.1002/jnr.1253. [DOI] [PubMed] [Google Scholar]
- 29.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 30.Panic B, Perisic O, Veprintsev DB, Williams RL, Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol Cell. 2003;12:863–874. doi: 10.1016/s1097-2765(03)00356-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp fig 1
Supp fig 2
Supp fig 3
Supp fig 4
Supp fig 5
Supp fig 6
Supp fig 7
Supp table 1
Supp table 2
Supp table 3