Fanconi Anemia Proteins Are Required To Prevent Accumulation of Replication-Associated DNA Double-Strand Breaks (original) (raw)
Abstract
Fanconi anemia (FA) is a multigene cancer susceptibility disorder characterized by cellular hypersensitivity to DNA interstrand cross-linking agents such as mitomycin C (MMC). FA proteins are suspected to function at the interface between cell cycle checkpoints, DNA repair, and DNA replication. Using replicating extracts from Xenopus eggs, we developed cell-free assays for FA proteins (xFA). Recruitment of the xFA core complex and xFANCD2 to chromatin is strictly dependent on replication initiation, even in the presence of MMC indicating specific recruitment to DNA lesions encountered by the replication machinery. The increase in xFA chromatin binding following treatment with MMC is part of a caffeine-sensitive S-phase checkpoint that is controlled by xATR. Recruitment of xFANCD2, but not xFANCA, is dependent on the xATR-xATR-interacting protein (xATRIP) complex. Immunodepletion of either xFANCA or xFANCD2 from egg extracts results in accumulation of chromosomal DNA breaks during replicative synthesis. Our results suggest coordinated chromatin recruitment of xFA proteins in response to replication-associated DNA lesions and indicate that xFA proteins function to prevent the accumulation of DNA breaks that arise during unperturbed replication.
The hereditary syndrome Fanconi anemia (FA) belongs to a group of caretaker gene diseases characterized by genomic instability and increased susceptibility to cancer. A hallmark of FA is cellular hypersensitivity to DNA interstrand cross-links (ICLs), suggesting a defect in the DNA damage response (18, 19, 48). Twelve FA complementation groups have been identified, and the majority of the corresponding genes have been cloned (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCJ, FANCL, and FANCM) (22, 23, 25, 43, 55, 59, 60, 66, 87, 93). Although the function of the FA proteins is unknown, identification of BRCA2 (breast cancer-associated gene 2) as FANCD1 and of FANCJ as the _BRCA1_-associated helicase gene Brip1/BACH1, suggests convergence of the FA/BRCA pathway with a larger network of proteins involved in DNA repair (7, 51-53, 95). This is underscored by the discovery that FANCM is related to archaeal Hef, a protein that binds and processes irregular arrangements of DNA in branched structures resembling replication forks (50, 71, 72).
According to current models, the FA pathway consists of an upstream nuclear core complex, including FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM, required for the activation of its target, FANCD2 (24, 34-36, 59, 60, 66). FANCD2 is monoubiquitinated during S phase and in response to various types of DNA damage, including DNA ICLs, DNA double-strand breaks (DSBs), and replication fork stalling (36, 89). DNA damage-induced monoubiquitination of FANCD2 is also reduced in cells from Seckel syndrome patients with a defect in the ataxia telangiectasia- and _RAD3_-related gene, ATR (1), suggesting that the FA pathway is under at least partial control of the ATR kinase. Monoubiquitination of FANCD2 is required for its association with chromatin and localization into nuclear foci containing BRCA1, RAD51, MRE11-RAD50-NBS1, replication protein A (RPA), PCNA, and BRCA2 (37, 42, 44, 65, 68, 89). FANCD2 is also phosphorylated in response to different types of DNA damage (68, 78, 90), and it is suspected that FANCD2 phosphorylation is part of two separate pathways that are controlled by one of the two checkpoint kinases, ATR or ATM (ataxia telangiectasia mutated) (68, 78, 90).
Several findings support the idea that FA proteins function during the S phase of the cell cycle. ICLs, the major genotoxic challenge for FA cells, are processed through generation of DNA DSB intermediates, which are generated specifically during S phase (5, 21, 28, 82) and repaired by the process of homologous recombination (HR) (83). The hypothesis that FA proteins are likely to function in ICL removal via HR repair during S phase is supported by evidence that FANCD1/BRCA2 is a central component of the HR repair mechanism (47, 79) and interacts with both RAD51 recombinase and FANCD2 (10, 20, 44, 98). Additional evidence supports a role for FANCA, FANCC, FANCG, and FANCD2 in HR (69, 70, 100, 101). New evidence from the DT40 model strongly implicates the FA downstream protein BRIP1/BACH1 helicase in DNA interstrand cross-link repair (7). Furthermore, the FA core complex proteins are part of BRAFT, a larger complex containing the Blm helicase, topoisomerase IIIα, and RPA (61), which supports the hypothesis of a function for FA proteins in replication-associated repair mechanisms.
To elucidate FA protein function(s) in the DNA damage response during replication, we established cell-free assays using Xenopus egg extracts that have been used to understand the role of other DNA repair proteins such as Mre11, Blm, ATR, and ATM during replication (12-17, 38, 39, 49, 54, 56, 85, 102). We have cloned the Xenopus laevis homologs of several of the FA proteins (generally termed xFA) and show that these proteins are recruited to chromatin in response to DNA lesions encountered by the replication machinery. Our findings suggest that the xFA proteins are required to prevent accumulation of DNA breaks that arise not only in response to exogenous DNA damage, but also during unperturbed replication.
MATERIALS AND METHODS
Antibodies.
Generation of anti-human FANCD2 rabbit polyclonal antisera was described previously (41). Anti-xFANCD2 polyclonal rabbit antibodies were raised against an equal mixture of three keyhole limpet hemocyanin-bound xFANCD2 peptides (Global Peptides) corresponding to amino acids 1 to 18, 890 to 908, and 1425 to 1443. The antisera were affinity purified against the three non-keyhole limpet hemocyanin-bound peptides immobilized on an AminoLink Plus column (Pierce) according to the manufacturer's instructions. Anti-xFANCA and anti-xFANCF polyclonal rabbit antibodies were raised against a chimeric protein containing an N-terminal glutathione S_-transferase (GST) tag (GST/pDEST 15) with a C-terminal region of xFANCA (amino acids 1205 to 1383) or full-length xFANCF (amino acids 1 to 340) and purified from bacteria as previously described (96). Affinity purification columns were made with chimeric HIS-xFANCA1205-1383 and HIS-xFANCF1-340 (HIS/pDEST 17), purified as described in the QIA_experessionist protocol (QIAGEN) and immobilized on an AminoLink Plus column. (For xFA antibody characterization, see Fig. S2 in the supplemental material.) The antibodies for neutralization of xATR and immunodepletion of xATR-interacting protein (xATRIP) have been described previously (8, 57).
Preparation of Xenopus egg extracts.
Extracts were prepared from Xenopus eggs according to the method of Murray (67). In brief, eggs were dejellied in 2% cysteine, pH 7.8; washed three times in XB buffer (10 mM KCl, 1 mM MgCl2, 100 nM CaCl2, 10 mM HEPES, 5 mM EGTA, 1.75% [wt/vol] sucrose, pH 7.8); and washed three times in CSF-XB buffer (XB buffer containing 5 mM EGTA and 2 mM MgCl2). Eggs were crushed by low-speed centrifugation (10,000 × g; 10 min), and the cytoplasmic fraction was cleared by centrifugation (16,000 × g; 20 min) after the addition of energy mix (15 mM creatine phosphate, 2 mM ATP, 2 mM MgCl2), cytochalasin B (10 μg/ml), cycloheximide (100 μg/ml), and Pefabloc (100 μg/ml). To release extracts from M to S phase, CaCl2 was added to a final concentration of 0.4 mM, and the extracts were incubated for 20 min at 23°C. Mitomycin C (MMC; 5 to 150 μM) and caffeine (4 mM) were added to S-phase extracts immediately prior to the addition of sperm chromatin and incubated for the time indicated; aphidicolin (50 ng/μl) was added at the time points indicated. For complete inhibition of replication initiation, recombinant Xenopus geminin (17) was added to S-phase extracts and incubated at 23°C for 15 min prior to the addition of sperm chromatin.
Preparation of nuclei and chromatin fractions.
At given time points, identical aliquots (50 to 100 μl) of egg extract containing 1,000 pronuclei (sperm heads)/μl were each diluted in nuclear isolation buffer (40 mM HEPES, 100 mM KCl, 20 mM MgCl2) or chromatin isolation buffer (40 mM HEPES, 100 mM KCl, 20 mM MgCl2, 0.2% Triton X-100) and purified through a 30% (wt/vol) sucrose cushion. Samples were centrifuged for 20 min at 6,000 × g; the nuclear and chromatin pellets, respectively, were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
DNA replication assay.
Replication of sperm chromatin in S-phase egg extracts was monitored as described previously (15). Briefly, sperm chromatin was added to S-phase extracts at 1,000 pronuclei/μl. Reaction aliquots were pulse-labeled with [α-32P]dGTP (Amersham) at time windows of 0 to 30 min, 30 to 60 min, 60 to 90 min, and 90 to 120 min at 23°C. Reactions were stopped with 1% SDS-40 mM EDTA (pH 7.8) and digested with proteinase K (1 mg/ml) at 37°C for 1 h. DNA was extracted with phenol-chloroform and electrophoresed on a 1% agarose gel.
Immunodepletion.
To immunodeplete from S-phase extracts, 200 μl of preswelled and washed (50% slurry in phosphate-buffered saline) Sepharose 4B beads (Amersham) were rotated overnight at 4°C with 500 μl of phosphate-buffered saline and 100 μl of xFANCD2, xFANCA affinity-purified antisera, anti-xATRIP, or the corresponding preimmune sera. The beads were pelleted from solution by centrifugation at 2,500 rpm for 10 min at 4°C and washed three times in XB buffer. A total of 100 μl of extract was added to the beads. The extract-bead mixture was rotated for two rounds at 4°C for 40 min.
TUNEL assay.
A total of 50 μl of xFANCA, xFANCD2, or preimmune serum-depleted S-phase egg extracts were incubated with 10,000 pronuclei/μl for 120 min at 23°C. The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed as previously described (15).
Immunoblotting.
Protein samples were separated on 3 to 8% Tris-acetate gels or 4 to 12% NuPAGE Bis-Tris gels (Invitrogen) and transferred to Immobilon P membranes (Millipore).
After a blocking in 5% milk for 1 h, membranes were incubated with the following primary antibodies: anti-hFANCD2, anti-xFANCA, anti-xFANCF, anti-xATM (81), anti-xTOP3α, anti-hORC2, anti-hRPA70, and anti-hPCNA (Santa Cruz). Horseradish peroxidase-conjugated rabbit or mouse secondary antibody (Jackson Laboratories) was used. Protein bands were visualized using an ECL Plus system (Amersham). Antibodies against xTOPIIIα, ORC2, and RPA70 were a kind gift from W. Dunphy.
Online supplemental material.
Figure S1 in the supplemental material shows regional homologies between human and Xenopus laevis homologs of FANCA, FANCD2, FANCF, and FANCL. Figure S2 in the supplemental material shows characterization of antibodies recognizing the Xenopus FANCA, FANCD2, and FANCF proteins. Figure S3A and C in the supplemental material show that critical steps of the FA pathway—interaction between FA core complex members and induction of FANCD2 following DNA damage—are conserved in Xenopus. Figure S3B and Table S1 in the supplemental material show that treatment of a Xenopus cell line, XTC-2, with MMC causes chromosomal aberrations comparable to those observed with human cells.
RESULTS
Conservation of the FA pathway in Xenopus laevis.
We identified Xenopus homologs of several human FA genes (see Fig. S1 in the supplemental material). A downstream effector of the FA pathway, xFANCD2, has an overall homology of 70% compared with that of human FANCD2; both known modification target sites, i.e., K561 for monoubiquitination (K563 in Xenopus) and S222 for phosphorylation (S224 in Xenopus) are conserved (Fig. 1) (36, 90). Furthermore, interactions between FA core complex members and the DNA damage-induced appearance of FANCD2-L (36) occur in Xenopus cells and egg extracts (see Fig. S3 in the supplemental material). We infer from these data that the critical components and the activation of the FA pathway are conserved in Xenopus.
FIG. 1.
Homology between Xenopus and human FANCD2. Full-length Xenopus FANCD2 (xFANCD2) was aligned with full-length human FANCD2 (hFANCD2) with MacVector 7.1.1 software. Amino acid identity is indicated by dark shading; similar amino acids are indicated by light shading. Two known critical residues, S222 and K561 (indicated by arrows), are conserved between human and Xenopus FANCD2.
xFA proteins associate with chromatin in a replication initiation-dependent manner.
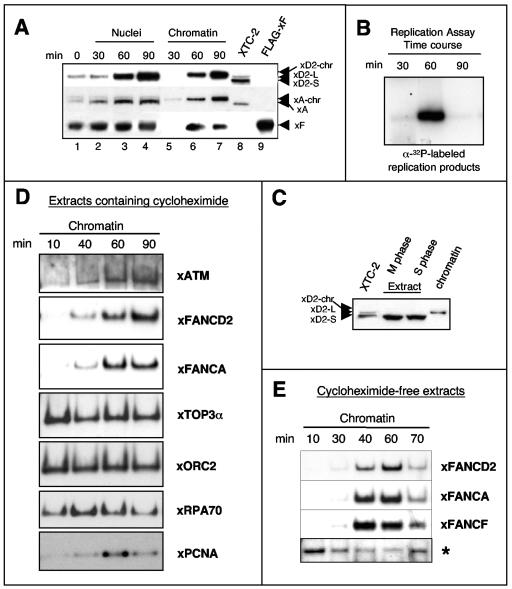
To analyze localization and chromatin binding behavior of xFA proteins, we used extracts prepared from Xenopus eggs. These extracts are arrested at the end of meiosis (M-phase extract) and contain only negligible amounts of DNA. Upon chemical activation, the extracts are released into S phase in tight synchrony. When sperm chromatin is added to S-phase extracts, the DNA decondenses and a nuclear membrane forms (20 min), followed by one round of semiconservative chromosomal replication. Following addition of sperm chromatin, we reisolated nuclei and chromatin fractions at time points before, during, and after replication and assayed for the presence of xFANCA, xFANCF, and xFANCD2. These proteins accumulated in the newly formed nuclei and associated with chromatin during replication (Fig. 2A), which typically occurred between 30 to 60 min (Fig. 2B) (67). A slow-mobility form of xFANCD2 was predominant in whole nuclei (Fig. 2A, lanes 1 to 4), as well as in the nuclear chromatin fractions (Fig. 2A, lanes 5 to 7), at the indicated time points. Since the extracts were precisely synchronized throughout S phase and replication of added sperm chromatin started shortly after nuclear membrane formation, it is likely that xFANCD2-S was converted to its modified form (FANCD2-L) immediately upon nuclear import and then recruited to replicating chromatin. Only the small isoform of xFANCD2 was detected in M-phase and S-phase extracts (Fig. 2C), where only negligible amounts of DNA were present. This observation implies that modification of xFANCD2 during S phase is dependent on the presence of chromatin, consistent with previous evidence that chromatin binding is associated with monoubiquitination of FANCD2 (65, 98). Chromatin-bound FA proteins are labeled with the suffix “-chr” in the text to provide a frame of reference because egg extracts and nuclear chromatin reisolated from egg extracts typically contain only a single isoform of xFANCD2. Egg extracts have a form comparable to FANCD2-S in cellular extracts, while chromatin replicated in egg extracts contained FANCD2-chr, comparable to FANCD2-L on chromatin isolated from cells. The mobility changes for FANCD2-chr and FANCA-chr compared with chromatin-bound counterparts in cell extracts (Fig. 2A, compare lanes 7 and 8) are likely due to large amounts of chromatin and protein in the egg extract samples.
FIG. 2.
xFA proteins accumulate in nuclei and associate with chromatin in a replication-dependent manner. (A) Nuclear accumulation and chromatin binding of FA proteins during replication. Sperm chromatin was added to S-phase egg extracts and reisolated at indicated time points during replication. Nuclear or chromatin fractions were analyzed by SDS-PAGE and immunoblotting. Xenopus XTC-2 cell lysates (see the supplementary material) were used as a control for xFANCD2-S (short form), xFANCD2-L (long form), and xFANCA; a lysate containing Flag-tagged xFANCF (overexpressed in 293 EBNA cells) was used as a positive control. The suffix “-chr” indicates the chromatin-bound protein isoforms from egg extracts as described in the text. (B) Replication assay. Throughout the experimental procedure described in the legend to panel A, semiconservative replication was monitored by pulsing replicating extract aliquots with [α-32P]GTP at time windows 0 to 30, 30 to 60, and 60 to 90 min. (C) Isoforms of xFANCD2 detectable in M and S phase and in chromatin fractions from replicating nuclei. An isoform of xFANCD2 with relatively slower mobility associates with chromatin during replication. Electrophoretic mobility was compared between xFANCD2 isolated from XTC-2 cell lysates, DNA-free mitotic egg extracts (M phase), and DNA-free S-phase egg extracts (S phase) and from nuclear sperm chromatin replicating in S-phase egg extracts (chromatin). (D) Comparison of chromatin binding patterns between xFA proteins and other DNA replication-associated proteins. Sperm chromatin was added to cycloheximide-containing S-phase egg extracts, and chromatin fractions were reisolated at the times shown and analyzed for the indicated proteins by immunoblotting. (E) xFA proteins dissociate from chromatin in nonarrested extracts following replication. Sperm chromatin was added to S-phase egg extracts in the absence of cycloheximide. Chromatin fractions were reisolated at the indicated time points and analyzed for xFANCA, xFANCD2, and xFANCF by immunoblotting. A nonspecific band indicated by an asterisk was used as a loading control.
Unlike other DNA replication proteins such as xRPA70 or xPCNA, the xFA proteins remained associated with chromatin once the bulk of replication was completed (Fig. 2D, lane 90). On the other hand, the chromatin binding pattern of the checkpoint signaling kinase xATM, which monitors origin firing during normal replication (85), closely resembled the temporal binding pattern of both xFANCA and xFANCD2. To further investigate the observed chromatin association pattern of xFA proteins, we used egg extracts prepared in the absence of cycloheximide. In classically prepared Xenopus egg extracts, cycloheximide blocks accumulation of cyclin B, which is required for the transition from S to M phase. Thus, in the presence of cycloheximide, extracts were halted in a G2-like state after DNA replication (Fig. 2D), whereas absence of cycloheximide allows the extracts to exit S phase (63, 67). Interestingly, in cycloheximide-free extracts, xFA proteins dissociated from chromatin once replication was over (Fig. 2E). Thus, the release of xFA proteins from chromatin was not triggered by the completion of replicative DNA synthesis alone but occurred only when extracts were allowed to exit S phase (compare Fig. 2D and E).
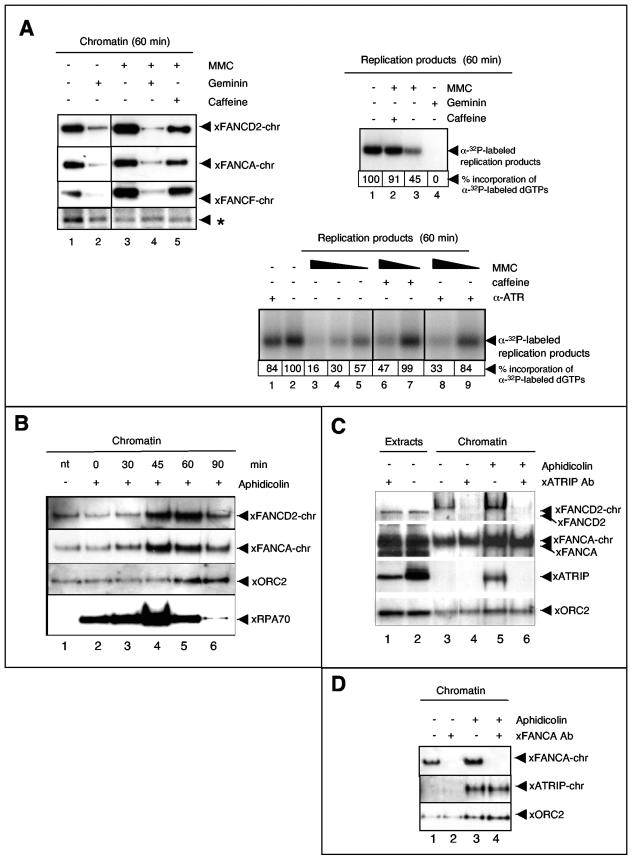
Accumulating evidence suggests that FA proteins function in the repair of specific DNA lesions that are encountered during transit through S phase (40, 70, 82, 89, 92). To determine if xFA proteins bound to chromatin in a replication-dependent manner, we blocked replication initiation by adding geminin to S-phase egg extracts. Geminin prevents assembly of prereplication complexes and thus inhibits replication initiation but does not affect chromosome decondensation or nuclear membrane formation (88). As shown in Fig. 3A, top left, accumulation of xFA core complex proteins and xFANCD2 on chromatin (compare lanes 1 and 2) was drastically inhibited in the presence of geminin, demonstrating that recruitment of FA proteins to chromatin occurs in a strictly replication initiation-dependent manner.
FIG.3.
Chromatin recruitment of xFA proteins in response to replication fork stalling and checkpoint activation. (A, top left) S-phase extracts were treated as indicated and supplemented with sperm chromatin. Following replication (60 min), chromatin was reisolated and the same blot was assayed in series for the presence of the xFA proteins indicated. A nonspecific band, indicated by an asterisk, was used as a loading control. (Top right) MMC-treated extracts activate a caffeine-sensitive S-phase checkpoint. A replication assay was performed to monitor incorporation of [α-32P]GTP during one round of replication in extracts treated as indicated. Total amounts of replication products at 60 min following addition of sperm chromatin to nontreated or treated extracts were analyzed by agarose gel electrophoresis and exposure to X-ray film. Band density percentages were determined using a Bio-Rad Molecular PhosphorImager Fx (Kodak screens, Quantity-1 software). (Bottom) Rescue of dose-dependent, MMC-induced replication reduction by neutralizing xATR. A total of 1,000 sperm pronuclei/μl were added to replicating extracts containing different concentrations of MMC (lanes 3 to 9). Extracts were otherwise untreated or contained caffeine (lanes 6 and 7) or a neutralizing anti-xATR antibody (lanes 8 and 9). The MMC concentration was 150 μM (lanes 3, 6, and 8), 50 μM (lanes 4, 7, and 9), or 5 μM (lane 5). (B) Stalling of replication forks induces recruitment of xFA proteins to chromatin. Sperm chromatin was added to S-phase egg extracts. Extracts were either untreated (−) or aphidicolin was added (+) at the indicated time points following addition of sperm DNA: 0 min, prenuclear membrane formation; 30 min, beginning of replication; 60 min, midreplication; 90 min, late or postreplication; and 120 min, postreplication. All chromatin fractions were reisolated at 135 min following the addition of sperm DNA, analyzed by SDS-PAGE, and immunoblotted as indicated. (C) Chromatin binding of xFANCD2 is xATRIP dependent. Egg extracts were treated with beads coupled to preimmune serum (lanes 2, 3, and 5) or beads coupled to anti-xATRIP serum (lanes 1, 4, and 6). Sperm chromatin was added to preimmune- or xATRIP-depleted extracts and allowed to replicate. Extracts were either untreated or supplemented with aphidicolin (added 50 min after the addition of sperm DNA). Chromatin fractions were reisolated at 70 min and assayed by immunoblotting for the presence of xFANCD2, xFANCA, xATRIP, and xORC-2. (D) Chromatin binding of xATRIP is xFANCA independent. Egg extracts were treated with beads coupled to preimmune serum (lanes 1 and 3) or beads coupled to anti-xFANCA serum (lanes 2 and 4). Sperm chromatin was added to preimmune- or xFANCA-depleted extracts and allowed to replicate. Extracts were either untreated or supplemented with aphidicolin (added 50 min after the addition of sperm DNA). Chromatin fractions were reisolated at 70 min and assayed for the presence of xFANCA, xATRIP, and xORC-2 by immunoblotting.
Recruitment of FA proteins to chromatin during disrupted replication.
The hallmark of Fanconi cells is their hypersensitivity to DNA interstrand cross-linking agents such as MMC. As shown in Fig. 3A, top left, chromatin binding of xFANCA, xFANCF, and xFANCD2 increased in the presence of MMC (compare lanes 1 and 3), consistent with fractionation and immunofluorescence data for FA proteins in asynchronously dividing human cells (62, 80). Addition of caffeine, an inhibitor of the major checkpoint kinases, ATM and ATR (84), reversed the MMC-induced recruitment of xFA proteins to chromatin, suggesting that xFA-chromatin binding following MMC treatment was part of a checkpoint controlled by one of the caffeine-sensitive kinases ATM or ATR (Fig. 3A, top left, compare lanes 1 and 5). Importantly, even in the presence of MMC, extracts failed to support chromatin recruitment of xFA proteins when replication initiation was blocked with geminin (Fig. 3A, top left, compare lanes 3 and 4). Replication in these extracts was monitored in matched aliquots by measuring incorporation of radiolabeled nucleotides into the nascent DNA strand. The replication assay shown in Fig. 3A, top right, demonstrated that replication occurred (lane 1), was reduced by exposure to MMC (lane 3), and was rescued by addition of caffeine (lane 2). As expected, no incorporation was detected in the presence of geminin (lane 4). Taken together, these data suggest that chromatin binding of xFA proteins is specifically triggered by DNA lesions that are encountered during DNA replication.
The finding that caffeine restored wild-type-like incorporation of nucleotides during replication in MMC-treated extracts suggests that replication inhibition induced by MMC is due to activation of an S-phase checkpoint, a response that allows cells to repair damage before they proceed to the next cell division, thus preventing the transmission of mutations (4, 9, 73, 75). To further explore the effect of MMC on replication, we determined whether the reduction of replication products occurred in an MMC dose-dependent manner. As shown in Fig. 3A, bottom, DNA replication was significantly blocked at high concentrations of MMC (150 μM) (lane 3) and could not be rescued by caffeine (lane 6). In contrast, at lower MMC concentrations, replication was less inhibited (50 μM, lane 4; 5 μM, lane 5) and could be rescued in the presence of caffeine (lane 7).
To determine if the MMC-induced replication block was under control of the major replication checkpoint kinase, ATR, we added a neutralizing anti-xATR antibody (Fig. 3A, bottom, lanes 8 and 9) to egg extracts to specifically block the xATR kinase function, thereby preventing phosphorylation of the Chk1 protein that is required for activation of the S-phase checkpoint (57). Comparable to the effect observed in the presence of caffeine, inhibition of xATR did not rescue the very strong replication block in high MMC concentrations (lane 7); however, at lower MMC concentrations, reduction of replication products was restored back to wild-type levels when xATR was blocked. Thus, at lower doses the MMC-induced reduction in incorporation of nucleotides during replication is due to activation of an S-phase checkpoint that depends on xATR and results in an increase in xFA-chromatin binding.
To determine the influence of fork stalling during DNA replication on recruitment of FA proteins to chromatin, we added aphidicolin to replicating extracts. Aphidicolin treatment blocks replicative polymerases, while helicases continue to unwind the DNA helix, thereby generating long single-stranded DNA (ssDNA) stretches (74, 85, 97). As shown in Fig. 3B, addition of aphidicolin to replicating extracts resulted in increased chromatin association of xFA proteins, as well as the ssDNA binding protein xRPA. Interestingly, whereas chromatin binding of xRPA increased significantly when aphidicolin was added before or during ongoing replication, recruitment of xFA proteins to chromatin increased only when aphidicolin was added during ongoing replication at 45 min (midreplication) and 60 min (late replication) (compare lane 1 with lanes 4 and 5). In contrast, when aphidicolin was added to extracts before the onset of replication (0 min or 30 min; compare lane 1 with lanes 2 and 3) or after replication was finished (90 min, compare lane 1 with lane 6), chromatin binding of xFA proteins did not increase. It is also important to note that under our experimental conditions, aphidicolin does not result in detectable DNA DSBs (54). These results suggest that the xFA proteins are recruited when the moving replication fork encounters sites of DNA damage.
The xATRIP/xATR complex controls chromatin binding of xFANCD2 independently of xFANCA.
ATR plays a critical role in coordinating the response to DNA damage. Its activation is usually linked to ongoing DNA replication (39, 57, 86, 91). The current model suggests that the ATR complex consisting of the ATR kinase and its binding partner, ATRIP, control S-phase progression in response to DNA damage and replication fork stalling. ATR and ATRIP are mutually dependent partners in the cellular S-phase checkpoint and DNA damage response (2, 3, 6, 11, 30, 45, 46, 94, 103). Generation of RPA-coated ssDNA is the critical signal that triggers recruitment of the tightly associated ATRIP-ATR complex. Our results demonstrated that MMC-induced increase in xFA-chromatin binding was reversed by the ATR-ATM inhibitor caffeine (Fig. 3A, top left, and data not shown). Thus, we asked whether depletion of the xATRIP subunit of the xATR kinase affected recruitment of the xFA proteins to chromatin. As shown in Fig. 3C, recruitment of xFANCD2 to chromatin was negligible in replicating extracts depleted of xATRIP, regardless of whether replicating extracts were unchallenged or treated with aphidicolin. In contrast, the xATRIP-depleted extracts still fully supported recruitment of xFANCA to chromatin in the presence or absence of aphidicolin. Similar results were obtained when extracts contained the neutralizing xATR antibody that blocked xATR phosphatidylinositol 3-kinase function (data not shown). To further investigate the influence of ATRIP on the FA pathway, we quantitatively depleted xFANCA from replicating extracts and tested for chromatin binding of xATRIP. Since levels of chromatin-associated xATRIP during unperturbed replication were barely detectable by Western blotting (Fig. 3C and D), we added aphidicolin to induce chromatin recruitment of xATRIP. As shown in Fig. 3D, immunodepletion of xFANCA did not affect the aphidicolin-induced chromatin recruitment of xATRIP. Thus, the xATRIP-xATR complex regulates recruitment of xFANCD2 to chromatin independently of the regulation that xFANCA and other core complex proteins exert in unperturbed replication, as well as in response to replication fork stalling.
Chromatin association of xFANCD2 depends on xFANCA.
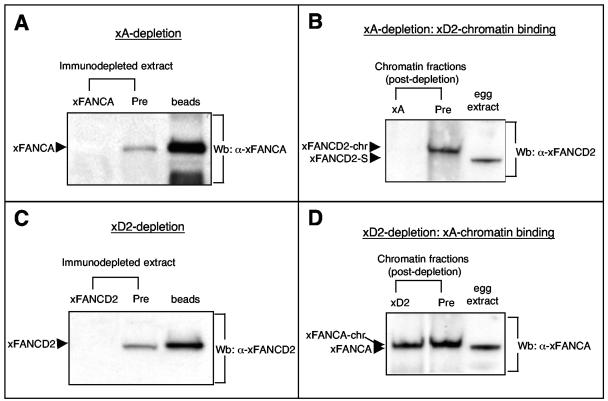
A functional FA core complex is believed to act upstream of FANCD2 by mediating its monoubiquitination (36, 89). The core complex protein xFANCA was quantitatively depleted from egg extracts to determine its influence on recruitment of xFANCD2 to chromatin (Fig. 4A). Sperm chromatin was added to the xFANCA-depleted extracts, followed by reisolation of replicated chromatin after 90 min. As shown in Fig. 4B, replication-associated binding of xFANCD2 to chromatin was abrogated in the absence of xFANCA. In contrast, immunodepletion of xFANCD2 from egg extracts (Fig. 4C) did not affect chromatin binding of xFANCA (Fig. 4D). Our results indicate that the xFANCA core complex is required for recruitment or stabilization of xFANCD2 at chromatin in response to DNA damage encountered during the replication process.
FIG. 4.
Replication-associated chromatin binding of xFANCD2 requires the presence of xFANCA. (A) S-phase egg extracts were incubated either with bead-coupled anti-xFANCA antibody (xA) or with bead-coupled preimmune serum (Pre) and assayed for presence of xFANCA by immunoblotting. Right lane, xFANCA binds to anti-xFANCA beads during the depletion process. (B) Sperm chromatin was added to xA- or predepleted extracts, reisolated following replication, and assayed for the presence of xFANCD2-chr by immunoblotting. (C and D) Replication-associated chromatin binding of xFANCA does not require the presence of xFANCD2. (C) S-phase egg extracts were incubated either with bead-coupled anti-xFANCD2 antibody (xD2) or with bead-coupled preimmune serum (Pre) and assayed for presence of xFANCD2 by immunoblotting. Right lane, xFANCD2 binds to anti-xFANCD2-beads during the depletion process. (D) Sperm chromatin was added to xD2- or predepleted extracts, reisolated following replication, and assayed for the presence of xFANCA-chr by immunoblotting.
xFANCA and xFANCD2 are required to prevent accumulation of DNA DSBs during replication.
Accumulating evidence points to a role for the FA pathway in repair of DNA DSBs (26, 27, 69, 82, 100, 101). Our results implied that xFA proteins are specifically recruited to chromatin during unperturbed replication and when exogenous DNA damage is encountered during replication. Thus, we first asked whether the absence of xFANCA (as a representative core complex member) or xFANCD2 (as an effector of the FA core complex) affects the overall kinetics of DNA replication.
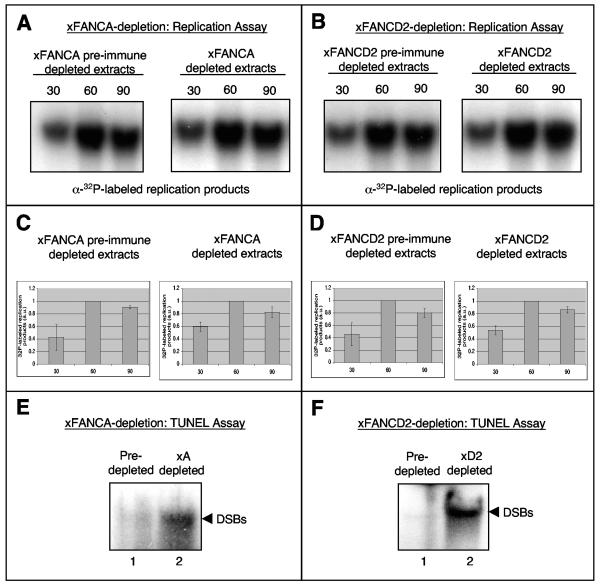
Following immunodepletion of xFANCA or xFANCD2 from egg extracts, we monitored timing and levels of nucleotide incorporation during chromosomal replication. No gross difference in replication kinetics was observed between mock- and xFANCA-depleted extracts (Fig. 5A and C) or between mock- and xFANCD2-depleted extracts (Fig. 5B and D). Thus, neither xFANCA nor xFANCD2 was required for initiation or elongation in the replication process itself. We then used a TUNEL assay to investigate whether the xFA proteins might be necessary to prevent the accumulation of DNA DSBs that are known to arise during the course of normal replication (15, 54). Chromosomal DNA was added to preimmune serum-, xFANCA-, or xFANCD2-depleted extracts, followed by one round of replication and isolation of replication products. Putative DNA DSBs in the postreplicative chromatin were labeled using terminal transferase and radioactive deoxynucleotides. We detected strong signals for incorporation of [32P]dGTP in replication products from extracts depleted of either xFANCA (Fig. 5E) or xFANCD2 (Fig. 5F) compared to mock-depleted extracts. Thus, absence of xFANCA or xFANCD2 from egg extracts results in accumulation of DNA breaks during unperturbed DNA replication.
FIG. 5.
xFA proteins are required to prevent accumulation of DNA breaks during unperturbed replication. (A and B) xFANCA and xFANCD2 are not essential for DNA replication. Genomic DNA replication was monitored by 30-min pulses of [α-32P]GTP. (A) Nucleotide incorporation into replicating chromatin in extracts incubated with protein A beads bound to preimmune serum (xA preimmune serum-depleted extracts) or extract incubated with protein A beads bound to anti-xFANCA antibodies (xA-depleted extracts). (B) Incorporation into replicating chromatin in extracts incubated with protein A beads bound to preimmune serum (xD2 preimmune serum-depleted extracts) or extract incubated with protein A beads bound to anti-xFANCD2 antibodies (xD2-depleted extracts). (C and D) Charted averages (including error bars) of the incorporation of radiolabeled nucleotides by band density from three identical replication assays (indicated) including the assays from panels A and B. (E and F) Depletion of xFANCA or xFANCD2 causes DNA breaks during normal replication. (E) Egg extracts were treated with beads coupled to xFANCA-preimmune serum (lane 1) or beads coupled to anti-xFANCA serum (lane 2). (F) Likewise, extracts were treated with either beads coupled to xFANCD2 preimmune serum (lane 1) or beads coupled to anti-xFANCD2 serum (lane 2). Sperm chromatin was added to treated extracts; after 120 min, TUNEL assays were performed. Samples were subjected to agarose gel electrophoresis and phosphorimaging.
DISCUSSION
We established cell-free assays for functional evaluation of the FA network proteins, which play critical but largely unknown roles in mechanisms that preserve genomic stability.
Analysis of the Xenopus homologs of FANCD2, FANCA, FANCF, and FANCL revealed regions of strong homology compared to their human counterparts, suggesting that they might contain some as-yet-unrecognized domains. Importantly, the known FA pathway characteristics are conserved in Xenopus including (i) conservation of the functional modification target sites of FANCD2, K561 (monoubiquitination), and S222 (phosphorylation); (ii) interactions between core complex members xFANCA and xFANCF (24, 34, 59, 61); and (iii) induction of an xFANCD2-L isoform following DNA damage. Thus, Xenopus egg extracts provide a cell-free system for functional analysis of the FA proteins during DNA replication and repair.
The downstream target of the FA pathway, FANCD2, is monoubiquitinated during S phase (FANCD2-L form), and this FANCD2-L isoform associates with S-phase chromatin (65, 89). Taking advantage of the fact that proteins can be analyzed in the highly synchronized Xenopus egg extracts in M and S phases even in the absence of DNA, we found that entry of the egg extract into S phase alone does not induce the xFANCD2-L form. In contrast, xFANCD2 is very quickly modified upon its import into nuclei that form once sperm chromatin is added to S-phase extracts, suggesting that formation of FANCD2-L is a DNA-dependent process. Following import of xFANCD2, xFANCA, and xFANCF into sperm nuclei, the proteins are recruited to chromatin during unperturbed replication. Chromatin binding of all three xFA proteins increases further in the presence of MMC, consistent with previous work showing that FANCD2-L and FA core complex proteins are chromatin bound during S phase and in response to DNA damage (36, 62, 64, 80, 89). Interestingly, when untreated egg extracts are prohibited from exiting S phase following DNA replication (i.e., they halt in a G2-like state), the xFA proteins stay bound to chromatin, unlike other replication-associated proteins such as xRPA70 or xPCNA. This observation supports a model in which the FA proteins participate in specific DNA repair events that occur during late replication and after the bulk of replication is completed (28, 70, 92, 100). The fact that xFA proteins dissociate from chromatin following replication when extracts are capable of exiting S phase suggests that the release from chromatin requires signaling associated with the G2-M transition of the cell cycle.
Previous reports show S-phase- and DNA damage-dependent chromatin association of several FA proteins (36, 62, 64, 80, 89). However, whether chromatin assembly of the FA proteins is directly associated with the replication process has not been evaluated. We found that in extracts containing the replication initiation inhibitor geminin, xFA proteins did not associate with chromatin even in the presence of MMC. Thus, chromatin association of the xFA proteins is strictly replication dependent. A remaining question is why DNA replication is required for recruitment of FA proteins. Our finding implies that the xFA proteins are selectively recruited to DNA lesions that either arise during normal replication or are recognized in a replication context following exogenous DNA damage.
In agreement with this idea, treatment of extracts with an inhibitor of replicative DNA polymerases, aphidicolin, also resulted in increased recruitment of xFA proteins to chromatin. Aphidicolin treatment leads to the generation of ssDNA regions due to uncoupling of the replicative helicase from the DNA polymerase. These ssDNA regions are generated regardless of whether aphidicolin is added to the egg extract before or during ongoing replication (as monitored by comparing chromatin-bound levels of the ssDNA binding protein xRPA). Interestingly, increased binding of xFA proteins to chromatin is only triggered when aphidicolin is added during the ongoing replication process, suggesting that the generation of ssDNA alone might not be sufficient for recruitment of the xFA proteins. In this regard, Cimprich and coworkers recently showed that functional uncoupling of MCM helicase and DNA polymerase occurs in response to several forms of DNA damage and that the subsequent accumulation of ssDNA is required but not sufficient to trigger the ATR-controlled checkpoint response (8). Further studies will be required to determine if the stalled replication fork itself or subsequently generated DNA intermediates and possibly additional factors trigger recruitment of the xFA proteins.
Analysis of MMC-treated extracts revealed that the MMC-induced increase in xFA-chromatin binding coincided with a reduction in replicative products. We demonstrated that this is due to activation of an intra-S-phase checkpoint that is controlled by the caffeine-sensitive checkpoint kinase ATR. In extracts containing MMC at concentrations between 5 and 50 μM, caffeine restored replication products back to wild-type levels and blocked the MMC-induced increase of xFA-chromatin association. Similarly, neutralization of the xATR kinase domain with a specific antibody inhibited checkpoint activation and subsequent reduction of replication products. In contrast, at higher MMC concentrations (150 μM), the replication block could not be overcome by caffeine or xATR neutralization. It is possible that at such high MMC concentrations the DNA lesions typically induced by this agent (intra- and interstrand cross-links) reach a level at which each replicon contains a cross-link. This could cause quantitative inhibition of replication by physical impairment of the DNA polymerase-helicase to get through the cross-links, despite the caffeine-xATR-induced block of checkpoint activation. We conclude from these findings that the additional recruitment of xFA proteins to chromatin in response to exogenous DNA damage occurs as part of an intra-S-phase checkpoint controlled by ATR. In agreement with this, Andreassen et al. recently showed that the MMC-induced increase in FANCD2 monoubiquitination, a step required for its targeting to chromatin, is dependent on ATR (1).
Moreover, we could demonstrate that chromatin recruitment of xFANCD2 is abrogated in extracts that either contained the ATR-neutralizing antibody or were depleted of xATRIP, a functional subunit of the xATR kinase. Strikingly, depletion of xATRIP had no effect on chromatin binding of xFANCA, raising the question of whether xATRIP was positioned between xFANCA (and perhaps the entire core complex) and the downstream target, xFANCD2, or whether xATRIP controlled chromatin recruitment of xFANCD2 independently of xFANCA. Testing chromatin binding of xATRIP in xFANCA-depleted extracts demonstrated that it is recruited to chromatin independently of xFANCA. In addition, xATRIP-chromatin binding was unaffected in extracts depleted of xFANCD2. These data support a model where recruitment of xFANCD2 to DNA lesions encountered by the replication machinery is under control of two entities: the xFA core complex and the xATR/xATRIP complex. The fact that absence of xATRIP blocked xFANCD2-chromatin binding even in the absence of exogenous DNA damage suggests that the xATR-xATRIP complex regulates FANCD2 not only during checkpoint activation but also as part of the DNA repair response that deals with basal levels of DNA damage during normal replication.
The current FA model suggests that modification and chromatin recruitment of FANCD2 are dependent on a functional FA core complex (1, 24, 36, 59, 62, 98). In support of this model, we found that the replication-associated chromatin binding of xFANCD2 is completely abrogated in egg extracts depleted of xFANCA. In contrast, depletion of xFANCD2 from S-phase extracts did not affect association of xFANCA with replicating chromatin. We conclude that the xFA proteins associate with chromatin in a coordinated manner. One possibility is that the xFA core complex binds to chromatin to recruit and activate xFANCD2 at specific types of DNA lesions encountered by the replication machinery.
Since our results demonstrated that accumulation of xFA proteins on chromatin is dependent on replication origin unwinding, we also investigated a potential requirement of xFA proteins in the replication process itself. As predicted by the absence of gross defects in genomic duplication in cells from FA patients, the rate and timing of replicative DNA synthesis were not affected in the absence of either xFANCA or xFANCD2. However, we found strikingly increased levels of DNA breaks in replication products from xFANCA- or xFANCD2-depleted extracts.
These results provide first proof that even in the absence of exogenous DNA damage, the xFA proteins are required to prevent accumulation of DNA DSBs that are known to arise during unperturbed replication. A similar function has been described for the xBlm and xMre11 proteins (15, 54), both of which are functionally dependent on the FA core complex (76, 78). Blm and the FA core complex proteins are part of a larger complex (named BRAFT), hinting at interrelated functions for the Blm and FA pathways (61). Rescue of the observed accumulation of replication-associated DNA DSBs will require adding back the respective recombinant wild-type xFA protein to the depleted egg extracts. However, rescue experiments are hampered by the lack of in vitro assays to assess the function of recombinant FANCD2 or any of the FA core complex proteins. In addition, rescue may not be possible with a single recombinant monomeric protein because the FA core complex contains at least eight proteins that act as an entity to mediate FANCD2 monoubiquitination (24, 34, 36, 58). On the other hand, the finding that depletion of xFANCA or xFANCD2 results in the same phenotype, i.e., accumulation of DSBs during replication, and the observation that xBlm or xMre11 protein levels are not reduced in extracts depleted of either xFA protein (data not shown) strongly suggest that the observed phenotype results from a defect in the FA pathway rather than from codepletion of other non-FA proteins such as xMre11 or the BRAFT complex components.
Since Mre11 and Blm have both been implicated in resolution and restart of stalled replication forks (29, 31-33, 77, 78, 99), our results support a model where concerted function among FA, Blm, and Mre11 pathways is required to prevent accumulation of DNA DSBs as a result of replication fork stalling.
In summary, we show new evidence that association of FA proteins with chromatin occurs specifically when replication forks encounter certain DNA lesions and that they are crucial to prevent chromosomal DNA breaks during normal replication. To our knowledge, this is the first report of a cell-free assay for FA protein function that introduces a powerful system for dissecting the function of the FA proteins in context with replication and within the network of proteins collaborating to ensure genomic stability in vertebrates.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Larry Thompson, Markus Grompe, Alan D’Andrea, Henri van de Vrugt, and Mathew Thayer for advice and many helpful discussions. We also thank James C. Bonner, Dietmar Gradl, and Doris Wedlich for providing us with XTC-2 cells. We are indebted to W. Dunphy for generously sharing antibodies for several Xenopus proteins and Jeff Parvin for sharing recombinant FA proteins.
A.S. received funding from the American Heart Association (0520117Z). V.C. and J.G. received funding from the National Institutes of Health (CA092245). K.A.C. received funding from the National Institutes of Health (GM62193). M.E.H. received funding from the Fanconi Anemia Research Fund, the Medical Research Foundation of Oregon, and the National Institutes of Health (CA112775).
Footnotes
REFERENCES
- 1.Andreassen, P. R., A. D. D'Andrea, and T. Taniguchi. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18**:**1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, H. L., and D. Cortez. 2005. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J. Biol. Chem. 280**:**31390-31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, H. L., J. S. Myers, and D. Cortez. 2005. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 16**:**2372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartek, J., and J. Lukas. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13**:**738-747. [DOI] [PubMed] [Google Scholar]
- 5.Bessho, T. 2003. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J. Biol. Chem. 278**:**5250-5254. [DOI] [PubMed] [Google Scholar]
- 6.Bomgarden, R. D., D. Yean, M. C. Yee, and K. A. Cimprich. 2004. A novel protein activity mediates DNA binding of an ATR-ATRIP complex. J. Biol. Chem. 279**:**13346-13353. [DOI] [PubMed] [Google Scholar]
- 7.Bridge, W. L., C. J. Vandenberg, R. J. Franklin, and K. Hiom. 2005. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37**:**953-957. [DOI] [PubMed] [Google Scholar]
- 8.Byun, T. S., M. Pacek, M. C. Yee, J. C. Walter, and K. A. Cimprich. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19**:**1040-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canman, C. E. 2003. Checkpoint mediators: relaying signals from DNA strand breaks. Curr. Biol. 13**:**R488-R490. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. J., D. Silver, S. Cantor, D. M. Livingston, and R. Scully. 1999. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 59(Suppl. 7)**:**1752s-1756s. [PubMed] [Google Scholar]
- 11.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294**:**1713-1716. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo, V., E. V. Avvedimento, M. E. Gottesman, J. Gautier, and D. Grieco. 1999. Protein kinase A is required for chromosomal DNA replication. Curr. Biol. 9**:**903-906. [DOI] [PubMed] [Google Scholar]
- 13.Costanzo, V., and J. Gautier. 2003. Single-strand DNA gaps trigger an ATR- and Cdc7-dependent checkpoint. Cell Cycle 2**:**17. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo, V., and J. Gautier. 2004. Xenopus cell-free extracts to study DNA damage checkpoints. Methods Mol. Biol. 241**:**255-267. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8**:**137-147. [DOI] [PubMed] [Google Scholar]
- 16.Costanzo, V., K. Robertson, C. Y. Ying, E. Kim, E. Avvedimento, M. Gottesman, D. Grieco, and J. Gautier. 2000. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol. Cell 6**:**649-659. [DOI] [PubMed] [Google Scholar]
- 17.Costanzo, V., D. Shechter, P. J. Lupardus, K. A. Cimprich, M. Gottesman, and J. Gautier. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11**:**203-213. [DOI] [PubMed] [Google Scholar]
- 18.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3**:**23-34. [DOI] [PubMed] [Google Scholar]
- 19.D'Andrea, A. D., and M. Grompe. 1997. Molecular biology of Fanconi anemia: implications for diagnosis and therapy. Blood 90**:**1725-1736. [PubMed] [Google Scholar]
- 20.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7**:**273-282. [DOI] [PubMed] [Google Scholar]
- 21.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20**:**7980-7990.11027268 [Google Scholar]
- 22.de Winter, J. P., F. Leveille, C. G. van Berkel, M. A. Rooimans, L. van Der Weel, J. Steltenpool, I. Demuth, N. V. Morgan, N. Alon, L. Bosnoyan-Collins, J. Lightfoot, P. A. Leegwater, Q. Waisfisz, K. Komatsu, F. Arwert, J. C. Pronk, C. G. Mathew, M. Digweed, M. Buchwald, and H. Joenje. 2000. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am. J. Hum. Genet. 67**:**1306-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Winter, J. P., M. A. Rooimans, L. van Der Weel, C. G. van Berkel, N. Alon, L. Bosnoyan-Collins, J. de Groot, Y. Zhi, Q. Waisfisz, J. C. Pronk, F. Arwert, C. G. Mathew, R. J. Scheper, M. E. Hoatlin, M. Buchwald, and H. Joenje. 2000. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat. Genet. 24**:**15-16. [DOI] [PubMed] [Google Scholar]
- 24.de Winter, J. P., L. van Der Weel, J. de Groot, S. Stone, Q. Waisfisz, F. Arwert, R. J. Scheper, F. A. Kruyt, M. E. Hoatlin, and H. Joenje. 2000. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum. Mol. Genet. 9**:**2665-2674. [DOI] [PubMed] [Google Scholar]
- 25.de Winter, J. P., Q. Waisfisz, M. A. Rooimans, C. G. van Berkel, L. Bosnoyan-Collins, N. Alon, M. Carreau, O. Bender, I. Demuth, D. Schindler, J. C. Pronk, F. Arwert, H. Hoehn, M. Digweed, M. Buchwald, and H. Joenje. 1998. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat. Genet. 20**:**281-283. [DOI] [PubMed] [Google Scholar]
- 26.Donahue, S. L., and C. Campbell. 2002. A DNA double strand break repair defect in Fanconi anemia fibroblasts. J. Biol. Chem. 277**:**46243-46247. [DOI] [PubMed] [Google Scholar]
- 27.Donahue, S. L., R. Lundberg, R. Saplis, and C. Campbell. 2003. Deficient regulation of DNA double-strand break repair in Fanconi anemia fibroblasts. J. Biol. Chem. 278**:**29487-29495. [DOI] [PubMed] [Google Scholar]
- 28.Dronkert, M. L., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486**:**217-247. [DOI] [PubMed] [Google Scholar]
- 29.Enomoto, T. 2001. Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication. J. Biochem. (Tokyo) 129**:**501-507. [DOI] [PubMed] [Google Scholar]
- 30.Falck, J., J. Coates, and S. P. Jackson. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434**:**605-611. [DOI] [PubMed] [Google Scholar]
- 31.Franchitto, A., and P. Pichierri. 2002. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 157**:**19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franchitto, A., and P. Pichierri. 2002. Protecting genomic integrity during DNA replication: correlation between Werner's and Bloom's syndrome gene products and the MRE11 complex. Hum. Mol. Genet. 11**:**2447-2453. [DOI] [PubMed] [Google Scholar]
- 33.Franchitto, A., and P. Pichierri. 2004. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle 3**:**1331-1339. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Higuera, I., Y. Kuang, D. Näf, J. Wasik, and A. D. D'Andrea. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19**:**4866-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Higuera, I., Y. Kuang, J. Denham, and A. D. D'Andrea. 2000. The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 96**:**3224-3230. [PubMed] [Google Scholar]
- 36.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7**:**249-262. [DOI] [PubMed] [Google Scholar]
- 37.Grompe, M., and A. D'Andrea. 2001. Fanconi anemia and DNA repair. Hum. Mol. Genet. 10**:**2253-2259. [DOI] [PubMed] [Google Scholar]
- 38.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14**:**2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hekmat-Nejad, M., Z. You, M. C. Yee, J. W. Newport, and K. A. Cimprich. 2000. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 10**:**1565-1573. [DOI] [PubMed] [Google Scholar]
- 40.Hirano, S., K. Yamamoto, M. Ishiai, M. Yamazoe, M. Seki, N. Matsushita, M. Ohzeki, Y. M. Yamashita, H. Arakawa, J. M. Buerstedde, T. Enomoto, S. Takeda, L. H. Thompson, and M. Takata. 2005. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 24**:**418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holzel, M., P. J. Van Diest, P. Bier, M. Wallisch, M. E. Hoatlin, H. Joenje, and J. P. de Winter. 2003. FANCD2 protein is expressed in proliferating cells of human tissues that are cancer-prone in Fanconi anaemia. J. Pathol. 201**:**198-203. [DOI] [PubMed] [Google Scholar]
- 42.Howlett, N. G., T. Taniguchi, S. G. Durkin, A. D. D'Andrea, and T. W. Glover. 2005. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 14**:**693-701. [DOI] [PubMed] [Google Scholar]
- 43.Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz, C. De Die-Smulders, N. Persky, M. Grompe, H. Joenje, G. Pals, H. Ikeda, E. Fox, and A. D'Andrea. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297**:**606-609. [DOI] [PubMed] [Google Scholar]
- 44.Hussain, S., J. B. Wilson, A. L. Medhurst, J. Hejna, E. Witt, S. Ananth, A. Davies, J. Y. Masson, R. Moses, S. C. West, J. P. de Winter, A. Ashworth, N. J. Jones, and C. G. Mathew. 2004. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 13**:**1241-1248. [DOI] [PubMed] [Google Scholar]
- 45.Itakura, E., K. K. Takai, K. Umeda, M. Kimura, M. Ohsumi, K. Tamai, and A. Matsuura. 2004. Amino-terminal domain of ATRIP contributes to intranuclear relocation of the ATR-ATRIP complex following DNA damage. FEBS Lett. 577**:**289-293. [DOI] [PubMed] [Google Scholar]
- 46.Itakura, E., K. Umeda, E. Sekoguchi, H. Takata, M. Ohsumi, and A. Matsuura. 2004. ATR-dependent phosphorylation of ATRIP in response to genotoxic stress. Biochem. Biophys. Res. Commun. 323**:**1197-1202. [DOI] [PubMed] [Google Scholar]
- 47.Jasin, M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21**:**8981-8993. [DOI] [PubMed] [Google Scholar]
- 48.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2**:**446-457. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi, T., S. Tada, T. Tsuyama, H. Murofushi, M. Seki, and T. Enomoto. 2002. Focus-formation of replication protein A, activation of checkpoint system and DNA repair synthesis induced by DNA double-strand breaks in Xenopus egg extract. J. Cell Sci. 115**:**3159-3169. [DOI] [PubMed] [Google Scholar]
- 50.Komori, K., M. Hidaka, T. Horiuchi, R. Fujikane, H. Shinagawa, and Y. Ishino. 2004. Cooperation of the N-terminal helicase and C-terminal endonuclease activities of archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 279**:**53175-53185. [DOI] [PubMed] [Google Scholar]
- 51.Lee, M., M. J. Daniels, and A. R. Venkitaraman. 2004. Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene 23**:**865-872. [DOI] [PubMed] [Google Scholar]
- 52.Levitus, M., Q. Waisfisz, B. C. Godthelp, Y. de Vries, S. Hussain, W. W. Wiegant, E. Elghalbzouri-Maghrani, J. Steltenpool, M. A. Rooimans, G. Pals, F. Arwert, C. G. Mathew, M. Z. Zdzienicka, K. Hiom, J. P. de Winter, and H. Joenje. 2005. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 37**:**934-935. [DOI] [PubMed] [Google Scholar]
- 53.Levran, O., C. Attwooll, R. T. Henry, K. L. Milton, K. Neveling, P. Rio, S. D. Batish, R. Kalb, E. Velleuer, S. Barral, J. Ott, J. Petrini, D. Schindler, H. Hanenberg, and A. D. Auerbach. 2005. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 37**:**931-933. [DOI] [PubMed] [Google Scholar]
- 54.Li, W., S. M. Kim, J. Lee, and W. G. Dunphy. 2004. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J. Cell Biol. 165**:**801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo Ten Foe, J. R., M. A. Rooimans, L. Bosnoyan-Collins, N. Alon, M. Wijker, L. Parker, J. Lightfoot, M. Carreau, D. F. Callen, A. Savoia, N. C. Cheng, C. G. van Berkel, M. H. Strunk, J. J. Gille, G. Pals, F. A. Kruyt, J. C. Pronk, F. Arwert, M. Buchwald, and H. Joenje. 1996. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat. Genet. 14**:**320-323. [DOI] [PubMed] [Google Scholar]
- 56.Luciani, M. G., M. Oehlmann, and J. J. Blow. 2004. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 117**:**6019-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lupardus, P. J., T. Byun, M. C. Yee, M. Hekmat-Nejad, and K. A. Cimprich. 2002. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 16**:**2327-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medhurst, A. L., P. A. Huber, Q. Waisfisz, J. P. de Winter, and C. G. Mathew. 2001. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol. Genet. 10**:**423-429. [DOI] [PubMed] [Google Scholar]
- 59.Meetei, A. R., J. P. de Winter, A. L. Medhurst, M. Wallisch, Q. Waisfisz, H. J. Van De Vrugt, A. B. Oostra, Z. Yan, C. Ling, C. E. Bishop, M. E. Hoatlin, H. Joenje, and W. Wang. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35**:**165-170. [DOI] [PubMed] [Google Scholar]
- 60.Meetei, A. R., A. L. Medhurst, C. Ling, Y. Xue, T. R. Singh, P. Bier, J. Steltenpool, S. Stone, I. Dokal, C. G. Mathew, M. Hoatlin, H. Joenje, J. P. de Winter, and W. Wang. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37**:**958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23**:**3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meetei, A. R., Z. Yan, and W. Wang. 2004. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle 3**:**179-181. [PubMed] [Google Scholar]
- 63.Menut, S., J. M. Lemaitre, A. Hair, and M. Mechali. 1999. DNA replication and chromatin assembly using Xenopus egg extract, p. 196-226. In J. D. Richter (ed.), Advances in molecular biology: a comparative methods approach to the study of oocytes and embryos. Oxford University Press, New York, N.Y.
- 64.Mi, J., and G. M. Kupfer. 2005. The Fanconi anemia core complex associates with chromatin during S phase. Blood 105**:**759-766. [DOI] [PubMed] [Google Scholar]
- 65.Montes de Oca, R., P. R. Andreassen, S. P. Margossian, R. C. Gregory, T. Taniguchi, X. Wang, S. Houghtaling, M. Grompe, and A. D. D'Andrea. 2005. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 105**:**1003-1009. [DOI] [PubMed] [Google Scholar]
- 66.Mosedale, G., W. Niedzwiedz, A. Alpi, F. Perrina, J. B. Pereira-Leal, M. Johnson, F. Langevin, P. Pace, and K. J. Patel. 2005. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 12**:**763-771. [DOI] [PubMed] [Google Scholar]
- 67.Murray, A. 1991. Cell cycle extracts. Methods Cell Biol. 36**:**581-605. [PubMed] [Google Scholar]
- 68.Nakanishi, K., T. Taniguchi, V. Ranganathan, H. V. New, L. A. Moreau, M. Stotsky, C. G. Mathew, M. B. Kastan, D. T. Weaver, and A. D. D'Andrea. 2002. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 4**:**913-920. [DOI] [PubMed] [Google Scholar]
- 69.Nakanishi, K., Y. G. Yang, A. J. Pierce, T. Taniguchi, M. Digweed, A. D. D'Andrea, Z. Q. Wang, and M. Jasin. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA 102**:**1110-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niedzwiedz, W., G. Mosedale, M. Johnson, C. Y. Ong, P. Pace, and K. J. Patel. 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15**:**607-620. [DOI] [PubMed] [Google Scholar]
- 71.Nishino, T., K. Komori, Y. Ishino, and K. Morikawa. 2005. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure (Cambridge) 13**:**1183-1192. [DOI] [PubMed] [Google Scholar]
- 72.Nishino, T., K. Komori, D. Tsuchiya, Y. Ishino, and K. Morikawa. 2005. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure (Cambridge) 13**:**143-153. [DOI] [PubMed] [Google Scholar]
- 73.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36**:**617-656. [DOI] [PubMed] [Google Scholar]
- 74.Pacek, M., and J. C. Walter. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23**:**3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pasero, P., K. Shimada, and B. P. Duncker. 2003. Multiple roles of replication forks in S phase checkpoints: sensors, effectors and targets. Cell Cycle 2**:**568-572. [PubMed] [Google Scholar]
- 76.Pichierri, P., D. Averbeck, and F. Rosselli. 2002. DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum. Mol. Genet. 11**:**2531-2546. [DOI] [PubMed] [Google Scholar]
- 77.Pichierri, P., and A. Franchitto. 2004. Werner syndrome protein, the MRE11 complex and ATR: menage-a-trois in guarding genome stability during DNA replication? Bioessays 26**:**306-313. [DOI] [PubMed] [Google Scholar]
- 78.Pichierri, P., and F. Rosselli. 2004. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 23**:**1178-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Powell, S. N., and L. A. Kachnic. 2003. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 22**:**5784-5791. [DOI] [PubMed] [Google Scholar]
- 80.Qiao, F., A. Moss, and G. M. Kupfer. 2001. Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem. 276**:**23391-23396. [DOI] [PubMed] [Google Scholar]
- 81.Robertson, K., C. Hensey, and J. Gautier. 1999. Isolation and characterization of Xenopus ATM (X-ATM): expression, localization, and complex formation during oogenesis and early development. Oncogene 18**:**7070-7079. [DOI] [PubMed] [Google Scholar]
- 82.Rothfuss, A., and M. Grompe. 2004. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24**:**123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saleh-Gohari, N., and T. Helleday. 2004. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 32**:**3683-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarkaria, J. N. 2003. Identifying inhibitors of ATM and ATR kinase activities. Methods Mol. Med. 85**:**49-56. [DOI] [PubMed] [Google Scholar]
- 85.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6**:**648-655. [DOI] [PubMed] [Google Scholar]
- 86.Stokes, M. P., R. Van Hatten, H. D. Lindsay, and W. M. Michael. 2002. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 158**:**863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strathdee, C. A., H. Gavish, W. R. Shannon, and M. Buchwald. 1992. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature 356**:**763-767. (Erratum, **358:**434.) [DOI] [PubMed] [Google Scholar]
- 88.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3**:**107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100**:**2414-2420. [DOI] [PubMed] [Google Scholar]
- 90.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. Andreassen, R. Gregory, S. Kim, W. Lane, M. Kastan, and A. D'Andrea. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109**:**459-472. [DOI] [PubMed] [Google Scholar]
- 91.Tercero, J. A., M. P. Longhese, and J. F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11**:**1323-1336. [DOI] [PubMed] [Google Scholar]
- 92.Thompson, L. H., J. M. Hinz, N. A. Yamada, and N. J. Jones. 2005. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ. Mol. Mutagen. 45**:**128-142. [DOI] [PubMed] [Google Scholar]
- 93.Timmers, C., T. Taniguchi, J. Hejna, C. Reifsteck, L. Lucas, D. Bruun, M. Thayer, B. Cox, S. Olson, A. D. D'Andrea, R. Moses, and M. Grompe. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7**:**241-248. [DOI] [PubMed] [Google Scholar]
- 94.Unsal-Kacmaz, K., and A. Sancar. 2004. Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Mol. Cell. Biol. 24**:**1292-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venkitaraman, A. R. 2003. A growing network of cancer-susceptibility genes. N. Engl. J. Med. 348**:**1917-1919. [DOI] [PubMed] [Google Scholar]
- 96.Waisfisz, Q., J. P. de Winter, F. A. Kruyt, J. de Groot, L. van der Weel, L. M. Dijkmans, Y. Zhi, F. Arwert, R. J. Scheper, H. Youssoufian, M. E. Hoatlin, and H. Joenje. 1999. A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc. Natl. Acad. Sci. USA 96**:**10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5**:**617-627. [DOI] [PubMed] [Google Scholar]
- 98.Wang, X., P. R. Andreassen, and A. D. D'Andrea. 2004. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24**:**5850-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426**:**870-874. [DOI] [PubMed] [Google Scholar]
- 100.Yamamoto, K., S. Hirano, M. Ishiai, K. Morishima, H. Kitao, K. Namikoshi, M. Kimura, N. Matsushita, H. Arakawa, J. M. Buerstedde, K. Komatsu, L. H. Thompson, and M. Takata. 2005. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 25**:**34-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamamoto, K., M. Ishiai, N. Matsushita, H. Arakawa, J. E. Lamerdin, J. M. Buerstedde, M. Tanimoto, M. Harada, L. H. Thompson, and M. Takata. 2003. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 23**:**5421-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.You, Z., L. Kong, and J. Newport. 2002. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 277**:**27088-27093. [DOI] [PubMed] [Google Scholar]
- 103.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300**:**1542-1548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]