Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation (original) (raw)
Abstract
Chromatin-modifying factors play key roles in transcription, DNA replication and DNA repair. Post-translational modification of these proteins is largely responsible for regulating their activity. The FACT (facilitates chromatin transcription) complex, a heterodimer of hSpt16 and SSRP1, is a chromatin structure modulator whose involvement in transcription and DNA replication has been reported. Here we show that nucleosome binding activity of FACT complex is regulated by poly(ADP-ribosyl)ation. hSpt16, the large subunit of FACT, is poly(ADP-ribosyl)ated by poly(ADP-ribose) polymerase-1 (PARP-1) resulting from physical interaction between these two proteins. The level of hSpt16 poly(ADP-ribosyl)ation is elevated after genotoxic treatment and coincides with the activation of PARP-1. The enhanced hSpt16 poly(ADP-ribosyl)ation level correlates with the dissociation of FACT from chromatin in response to DNA damage. Our findings suggest that poly(ADP-ribosyl)ation of hSpt16 by PARP-1 play regulatory roles for FACT-mediated chromatin remodeling.
INTRODUCTION
Post-translational modification of proteins by phosphorylation, acetylation, methylation or poly(ADP-ribosyl)ation is known to modulate the chromatin-template activities. Poly(ADP-ribosyl)ation of proteins may affect protein–protein and protein–DNA interactions (1). In v_itro_ poly(ADP-ribosyl)ation has been described for many nuclear proteins that results in a down-regulation of their functions.
Poly(ADP-ribose) polymerases (PARPs) make up a family of enzymes with conserved catalytic domain at the C-terminal region (2). These enzymes catalyze the transfer of ADP-ribose from NAD+ onto acceptor proteins, including themselves (2). The most abundant and best-characterized member of PARP family is PARP-1. Formation of poly(ADP-ribose) polymers occurs rapidly after DNA damage and appears to be primarily due to PARP-1 activation. The sites of poly(ADP-ribose) accumulations often coincide with DNA damage foci or actively transcribed gene loci (3,4). Although not absolutely required for repair, loss of PARP-1 reduces the ability of cells to deal with DNA damage. Recent studies have established that poly(ADP-ribosyl)ation involves not only in the regulation of cellular response to genotoxic stress, but also ensures accurate transmission of genetic information during cell division (5). Additionally, studies in recent years suggest the involvement of poly(ADP-ribosyl)ation of proteins in chromatin organization. Many targets of PARP-1 are involved in establishing chromatin architecture (2). Histones, especially H1 and H2B, are the major poly(ADP-ribosyl)ated chromatin proteins (6). Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure (7). Genetic studies in Drosophila also showed that PARP-1 is required for proper chromatin organization throughout the life cycle (8).
The eukaryotic genome is packaged into chromatin, which present a constant barrier to transcription and other cellular processes that require access to DNA. The accessibility to DNA is modulated by protein complexes that remodel chromatin structure. Two main enzyme activities that regulate chromatin accessibility could be distinguished. One class of chromatin remodeling complex consists of chromatin modifying complex which modifies the histone tail (9). Another class, ATP-dependent chromatin remodeling complex, uses energy of ATP hydrolysis to alter the chromatin structure (10). A recent report using biochemical approach has identified a new chromatin remodeling factor, FACT (facilitates chromatin transcription), that allows RNA polymerase II (pol II) to proceed along the chromatin template (11). FACT, a heterodimer of hSpt16 and SSRP1 (12), binds and reorganizes the nucleosome to facilitate transcription on chromatin by pol II (13,14). FACT has been found to be chromatin-associated, consistent with its roles in modulating chromatin organization (15,16). Genetic studies in yeast have also demonstrated that both genes are essential for cell viability, global transcription and cell cycle progression (15,17,18).
Both PARP-1 and FACT are involved in the global regulation of chromatin architecture. Recently, two sets of experiments strongly indicate that there is functional link between PARP-1 and FACT. A study of Drosophila polytene chromosomes showed that FACT is recruited to actively transcribed loci after heat shock (19). The kinetics of FACT movement along the hsp70 gene is similar to that of pol II and elongation factors Spt5 and Spt6. Another study demonstrated that Drosophila PARP is required for loosening chromatin structure and normal induction of Hsp70 after heat-shock (3). Significant amount of poly(ADP-ribose) are observed on the hsp70 foci. These observations suggest that there is a possible functional link between poly(ADP-ribosyl)ation and FACT. We have sought to investigate the possible functional interaction between PARP-1 and FACT. We analyzed the modification of FACT immunoprecipitated from human cells. Our results showed that hSpt16 but not SSRP1 is poly(ADP-ribosyl)ated in vivo especially following genotoxic stress. Additionally, we showed that there is a direct interaction between hSpt16 and PARP-1. In vitro poly(ADP-ribosyl)ation assay demonstrated that hSpt16 is a substrate of PARP-1. We also showed that the nucleosome-binding activity of hSpt16 and FACT is decreased after poly(ADP-ribosyl)ation. These results suggest that transcription, DNA replication or repair may be regulated through the modulation of chromatin-binding property of FACT by PARP-1.
MATERIALS AND METHODS
Cell culture, plasmids and transient transfection
HeLa and 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. For inhibition of PARP activity, PARP inhibitor 3-aminobenzamide (3AB) was added to the culture 1 h prior to irradiation or H2O2 treatment. Cell cultures in mid-log phase of growth were treated with 500 µM H2O2 for 20 min at 37°C or irradiated in a 137Cs IBL irradiator at room temperature. Transfection of 293T cells were performed using a standard calcium-phosphate protocol. The cDNA encoding the full-length hSpt16, SSRP1 and PARP-1 were cloned into mammalian expression vector pCMV-Tag2 or recombinant baculovirus.
Antibodies
The primary antibodies used were as follows: mouse monoclonal antibodies anti-FLAG (M2/Sigma), anti-poly(ADP-ribose) polymer (10H-2/Riken; Japan), anti-tubulin α (Sigma) and rabbit anti-poly(ADP-ribose) polymer (Roche) were obtained commercially; monoclonal antibodies anti-PARP-1 (clone 5A5), anti-hSpt16 (clone 8D2) and anti-SSRP1 (clone 3E4) and rabbit anti-histone H3 serum were generated by our lab (Supplementary Figure S1).
Protein purification
Bulk HeLa cells polynucleosomes were purified as described previously (20). Mononucleosomes were then created from these polynucleosomes by digestion with micrococcal nuclease and further purified by Sephacryl S-400 gel filtration chromatography. DNA of purified mononucleosome was analyzed on agarose gel. Recombinant human PARP-1, FLAG-tagged hSpt16 and His-tagged SSRP1 proteins were expressed in insect cells (Sf9) by recombinant baculoviruses. PARP-1 was purified to homogeneity by the procedure of Zahradka and Ebisuzaki (21). Briefly, insect cells were extracted with extraction buffer followed by two sequential ammonium sulfate precipitations of 30 and 70%. The pellet was suspended in buffer A (50 mM Tris–HCl, pH 8.0, 0.2 M NaCl, 10 mM 2-mercaptoethanol, 50 mM NaHSO3, 1 mM DTT and 1 mM EDTA) and desalted by passage through a Sephadex G-50 column. PARP-1 was concentrated by DNA-cellulose chromatography. Fractions containing purified enzyme were pooled and store at −80°C in aliquots. Analysis of the purified enzyme by SDS–PAGE revealed a single protein of 110 kDa by Coomassie blue staining. Recombinant FLAG-hSpt16 and His-SSRP1 were purified by standard M2 anti-FLAG agarose or nickel-NTA affinity chromatography. For purification of FACT heterodimer (14), the mixture of insect cell lysates containing FLAG-hSpt16 and His-SSRP1 was incubated with nickel-NTA. The bound proteins were eluted from the beads and subjected to M2 affinity purification. After extensive washing, the FACT heterodimers were eluted by FLAG peptide. The purities of these proteins were >98%, as judged by SDS–PAGE and Coomassie brilliant blue staining (Supplementary Figure S2).
Immunoprecipitation and protein interaction assay
HeLa cells were harvested by centrifugation, washed in phosphate-buffered saline (PBS) and directly lysed with lysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.1% Triton X-100) containing protease inhibitors [1 µg/ml leupeptin, 1 µg/ml pepstatin and 1 mM phenylmethlysulfonyl fluoride (PMSF)]. Antibodies pre-bound to protein G–Sepharose were incubated with lysates for 1 h at 4°C rotating continuously. Beads were washed three times with lysis buffer, boiled for 5 min followed by analysis on SDS–PAGE. For mononucleosome-binding assay, immunoprecipitated FLAG-hSpt16 was washed twice with binding buffer (10 mM Tris–HCl, pH 7.5, 100 mM KCl, 1 mM MgCl2 and 1 mM DTT). The precipitates were then resuspended in 100 µl of binding buffer containing 100 ng PARP-1 plus 1 µg mononucleosome and incubated at 37°C for 10 min. The reactions were started by adding NAD+ to 500 µM and incubated at 37°C for 40 min. Beads were washed three times with binding buffer, boiled for 5 min followed by analysis by SDS–PAGE. Binding of mononucleosome was detected by rabbit anti-histone H3 antibody. Endogenous FACT precipitated from HeLa cells was washed with lysis buffer containing 0.5 M NaCl. After poly(ADP-ribosyl)ation, PARP-1, DNA and NAD+ were washed away from the modified FACT using lysis buffer containing 0.5 M NaCl. The poly(ADP-ribosyl)ated FACT and control FACT (incubated with PARP-1 and DNA in the absence of NAD+) were incubated with mononucleosomes at 4°C for 2 h, washed three times with binding buffer and boiled for 5 min followed by western blot analysis. For pull-down assay, purified recombinant proteins (FLAG-hSpt16, His-SSRP1 or FACT heterodimer) were immobilized on M2 beads or nickel-resin and then incubated with purified PARP-1 or mononucleosome in lysis buffer.
PARP-1 poly(ADP-ribosyl)ation assay
Purified recombinant protein (1.5 µg) was incubated with 100 ng PARP-1 plus various activator (0.1 µg DNA, 6 µg core histone or 1 µg mononucleosome) in reaction buffer (10 mM Tris–HCl, pH 7.5, 1 mM MgCl2 and 1 mM DTT) and incubated at 37°C for 10 min. The reactions were started by adding NAD+ to 500 µM and incubated at 37°C for 10 more minutes. SDS sample buffer was added to stop the reaction. The samples were boiled and analyzed by SDS–PAGE. Poly(ADP-ribose) were detected by immunoblot analysis using 10H-2 (1:1000) or rabbit serum (1:10 000). To study the interaction between automodified PARP-1 and hSpt16, recombinant PARP-1 was bound to protein G beads (15 µl) with 4 µg rabbit anti-PARP-1 antibody and incubated with 0.1 µg DNA and FLAG-hSpt16 in the presence or absence of 500 µM NAD+ at 37°C for 30 min. The unbound proteins were washed out with binding buffer.
Biochemical fractionation
Biochemical fractionation of HeLa cells was performed as described previously with some modifications (22). Briefly, HeLa cells were harvested and washed with PBS, and resuspended in buffer A (10 mM Tris–HCl, pH 7.5, 10 mM NaCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 µg/ml leupeptin, 1 µg/ml pepstatin and 1 mM PMSF). Triton X-100 was added to final concentration of 0.1%, and the cells were incubated for 5 min on ice. Nuclei were washed once in buffer A, and then extracted sequentially with increasing concentrations of NaCl in buffer A. The final pellet was resuspended in sample buffer. Because sucrose and glycerol interferes the immunoprecipitation efficiency of hSpt16, 2 vol of buffer A without glycerol and sucrose were added to lysates before immunoprecipitation.
Chromatin immunoprecipitation assay
HeLa cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M, and the incubation was continued for 5 min. Cross-linked cells were washed with PBS and lysed in 1.5 ml lysis buffer (10 mM Tris–HCl, pH 7.5, 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA and protease inhibitors) on ice for 5 min. Cells were centrifuged at 600 g for 5 min at 4°C. Nuclei were washed with wash buffer (10 mM Tris–HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and protease inhibitor) and centrifuged again under the same condition. Nuclear pellet was resuspended in 400 µl sonication buffer (10 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and 1% SDS), sonicated with a MSE soniprep 150 sonicator (eight bursts of 5 s each) on ice, and centrifugation at 10 000 g for 10 min. The soluble chromatin was adjusted to RIPA buffer (10 mM Tris, pH 7.5, 0.3% Triton X-100, 0.1% SDS, 1 mM EDTA, 1 mM EGTA and 150 mM NaCl). For immunoprecipitation, chromatin was pre-cleared with 30 µl of protein A–Sepharose beads and immunoprecipitated with 10 µl protein G beads coupled with antibodies 8D2, 3E4 or rabbit anti-histone H3 antiserum for 2 h at 4°C.Immunoprecipitates were washed three times with wash buffer A (20 mM Tris–HCl, pH 7.5, 0.5% Triton X-100, 0.1% SDS and 0.5 N NaCl), once with wash buffer B (20 mM Tris–HCl, pH 7.5, 0.5% Triton X-100, 0.1% SDS and 0.9 N NaCl) and once with TE (20 mM Tris–HCl, pH 7.5 and 1 mM EDTA). Immunocomplexes were eluted twice with 100 µl of elution buffer (TE with 30 mM NaCl and 1% SDS). The input and pooled eluates were incubated for 6 h at 65°C to reverse the formaldehyde cross-linkage, followed by dilution with 100 µl of water containing 0.16 µg/µl of proteinase K and incubated for 1 h at 50°C. DNA was purified with phenol/chloroform and a fraction was used as PCR template to detect the presence of actin gene (25–30 cycles of 45 s melting at 95°C, 45 s annealing at 52°C, 45 s extension at 72°C) using the primers listed below: Act-F GCTGTTCCAGGCTCTGTTCC; and Act-R ATGCTCACACGCCACAACATGC. The PCR products were resolved in 1.9% agarose gels and visualized by ethidium bromide staining.
RESULTS
hSpt16 but not SSRP1 is poly(ADP-ribosyl)ated in vivo
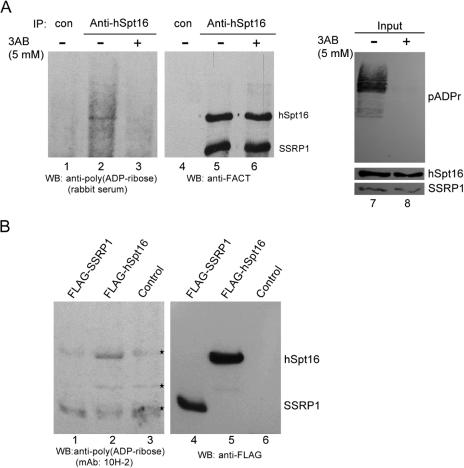
To gain insight into the regulation of chromatin organization by poly(ADP-ribosyl)ation of proteins, we tested whether FACT heterodimer is poly(ADP-ribosyl)ated in vivo. The poly(ADP-ribosyl)ation status of endogenous FACT immunoprecipitated from HeLa cells by hSpt16 specific antibody was determined using rabbit anti-poly(ADP-ribose) antibody (Figure 1A). FACT heterodimer remained intact under this immunoprecipitation condition, as determined by Coomassie blue staining (data not shown). A specific, albeit relatively weak, poly(ADP-ribose) signal corresponding to hSpt16 was detected in the immunoprecipitated FACT (Figure 1A, lane 2). The results showed that hSpt16 but not SSRP1 is poly(ADP-ribosyl)ated in vivo. A similar result was obtained when anti-SSRP1 monoclonal antibody was used to immunoprecipitate FACT heterodimer (data not shown and Figure 2B).
Figure 1.
hSpt16 is poly(ADP-ribosyl)ated in vivo. (A) Poly(ADP-ribosyl)ation status of endogenous FACT. HeLa cells were cultured in the absence or presence of 5 mM 3AB for 24 h. FACT heterodimer was immunoprecipitated from lysates and the status of poly(ADP-ribosyl)ation was determined using rabbit anti-poly(ADP-ribose) antibody (lanes 1–3). The same blot was probed sequentially with anti-FACT antibody (8D2 and 3E4, lanes 4–6). The levels of poly(ADP-ribose) (pADPr) of cell extracts were shown (lanes 7 and 8). The protein levels of hSpt16 and SSRP1 in lysates were also shown (lower panel, lanes 7 and 8). (B) FLAG-hSpt16 but not FLAG-SSRP1 is poly(ADP-ribosyl)ated in vivo. Whole cell extracts from 293T cells overexpressing FLAG-hSpt16 or FLAG-SSRP1 were immunoprecipitated with anti-FLAG M2 agarose followed by immunoblotting with monoclonal anti-poly(ADP-ribose) antibody (10H-2) (lanes 1–3). The same blot was reprobed with anti-FLAG antibody (lanes 4–6). The asterisk indicates bands that were recognized by 10H-2 antibody non-specifically.
Figure 2.
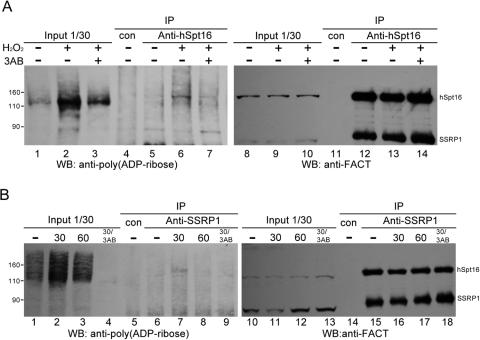
Enhanced poly(ADP-ribosyl)ation of hSpt16 in response to DNA damage. (A) HeLa cells were mock exposed or exposed to 500 µM H2O2 in the presence or absence of PARP inhibitor 3AB as indicated. FACT immunoprecipitated from extracts derived from equal cell numbers by anti-hSpt16 antibody were analyzed by immunoblot analysis. Same blot was probed sequentially with rabbit anti-poly(ADP-ribose) antibody (lanes 1–7) and anti-FACT antibodies (8D2 and 3E4, lanes 8–14). Numbers on the left side indicate positions of protein molecular weight markers. Signals of poly(ADP-ribosyl)ated polypeptides (∼110 kDa) in lanes 1–3 should be poly(ADP-ribosyl)ated PARP-1. (B) HeLa cells were subjected to γ-irradiation (8 Gy) in the presence or absence of PARP inhibitor 3AB. FACT was immunoprecipitated with anti-SSRP1 antibody (3E4) from lysates prepared before or after irradiation at indicated time and the immunoprecipitates were analyzed by immunoblot analysis. Same blot was probed sequentially with anti-poly(ADP-ribose) antibody (lanes 1–9) and anti-FACT antibodies (lanes 10–18). The input lanes showed the loading of HeLa extracts from equal cells numbers (lanes 1–4 and 10–13). Numbers on the left side indicate positions of protein molecular weight markers.
To demonstrate the specificity of hSpt16 poly(ADP-ribosyl)ation, we examined the effects of a general PARP inhibitor, 3AB, on the poly(ADP-ribosyl)ation patterns of FACT. The signal of poly(ADP-ribose) polymers in the 3AB-treated HeLa cells was significantly decreased (Figure 1A, compare lanes 7 and 8) while no obvious cytotoxic effects were evident. The inhibition of poly(ADP-ribosyl)ation of hSpt16 by 3AB suggests that PARP activity is involved (Figure 1A, compare lanes 2 and 3). To exclude the possibility that the observed signal of poly(ADP-ribose) is due to associated proteins which co-migrate with hSpt16 on SDS–PAGE, recombinant FLAG-tagged hSpt16 or FLAG-tagged SSRP1 was immunoprecipitated from transfected 293T cells with anti-FLAG M2-agarose and followed by extensive high salt (0.8 M NaCl) wash to remove possible associated proteins (Figure 1B). Western blot analysis using anti-poly (ADP-ribose) antibody (10H-2) revealed that poly(ADP-ribosyl)ated hSpt16 could still be seen (Figure 1B, lane 2). The level of precipitated proteins were shown in Figure 1B (lanes 4–6). This result confirms that hSpt16, but not SSRP1, can bepoly(ADP-ribosyl)ated.
A number of proteins are known to be poly(ADP-ribosyl)ated during DNA damage response. We investigated the level of poly(ADP-ribosyl)ation of hSpt16 when cells are exposed to genotoxic stress. As shown in Figure 2A (lanes 5 and 6), the level of poly(ADP-ribosyl)ation of immunoprecipitated hSpt16 increased significantly after exposure of the cells to H2O2. Pre-incubation of the cells with 3AB decreased the poly(ADP-ribosyl)ated hSpt16 level (Figure 2A, compares lanes 2–3 and 6–7). We have also analyzed the poly(ADP-ribosyl)ation of the FACT immunoprecipitated from the extracts of γ-irradiated cells by SSRP1-specific antibody (Figure 2B). The amount of poly(ADP-ribosyl)ated hSpt16 increased 30 min post-irradiation. The kinetics of hSpt16 poly(ADP-ribosyl)ation correlates with the poly(ADP-ribose) synthesis of cell extracts following γ-irradiation (Figure 2B, compare lanes 1–3 and 6–8). 3AB treatment also abolished the incorporation of poly(ADP-ribose) into hSpt16 (Figure 2B, lane 9). The same membrane was re-probed with anti-FACT antibodies to demonstrate equal loading of proteins (Figure 2B, lanes 10–18). Taken together, these observations suggest that FACT is ploy(ADP-ribosyl)ated under genotoxic stress conditions. These data further show that poly(ADP-ribosyl)ated hSpt16 remains heterodimerized with SSRP1.
Association of FACT with PARP-1
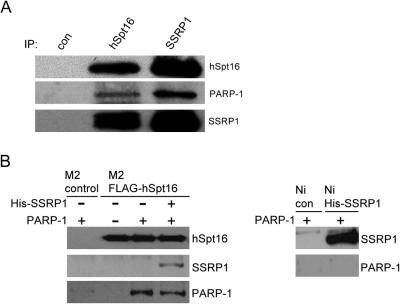
PARP-1 is the most abundant of PARPs in the cell, accounting for the synthesis of >90% of the poly(ADP-ribose) following DNA damage. To examine the possible interaction between FACT and PARP-1, we performed co-immunoprecipitation experiments by using whole cell extracts from HeLa cells. Both monoclonal antibodies against hSpt16 and SSRP1 were used to immunoprecipitate FACT heterodimer from the cell extracts. As shown in Figure 3A, PARP-1 could be detected in both anti-hSpt16 and anti-SSRP1 but not in control antibody immunoprecipitates. These results demonstrate that FACT heterodimer and PARP1 co-exist in a complex. We further examined the possible direct interaction between FACT and PARP-1 by performing in vitro pull-down assay using purified recombinant proteins. Purified PARP-1 could be co-precipitated by recombinant hSpt16 or FACT heterodimer, while the binding to M2 gel control, Ni-resin control or His-SSRP1 was close to background level (Figure 3B). Taken together, these data show that PARP-1 associates with FACT through direct physical interaction with hSpt16. In vitro pull-down assay using serial deletion mutants of hSpt16 showed that the central region of hSpt16 is responsible for its interaction with PARP-1 (Supplementary Figure S3).
Figure 3.
Interaction between FACT and PARP-1. (A) Co-immunoprecipitation of FACT and PARP-1 from HeLa cells extracts. HeLa whole-cell extracts were immunoprecipitated with control (non-specific), hSpt16 or SSRP1 antibody. The immunoprecipitates were examined for the presence of PARP-1 (middle panel) or FACT heterodimer (upper and lower panels) by immunoblotting. (B) FACT interacts with PARP-1 via hSpt16. M2-agarose bound FLAG-hSpt16 (left panel) or Ni-resin bound His-SSRP1 (right panel) was incubated with different set of purified proteins as indicated. After unbound proteins were removed by washing, the immunoprecipitates were subjected to immunoblot analysis using the indicated antibodies. M2-agarose beads or Ni-resin were used as controls.
hSpt16 is poly(ADP-ribosyl)ated by PARP-1 in vitro
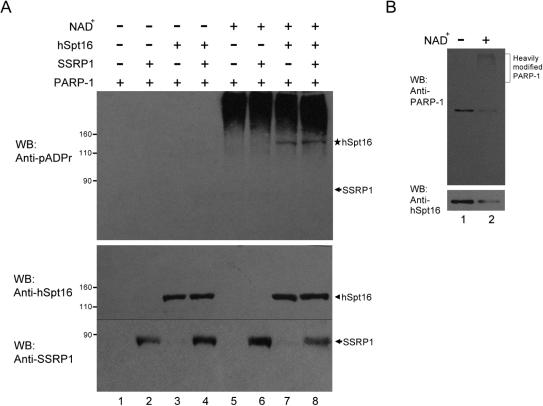
To test the poly(ADP-ribosyl)ation of hSpt16 by PARP-1, we performed in vitro poly(ADP-ribosyl)ation assays using purified recombinant proteins. Poly(ADP-ribosyl)ation depends on the presence of NAD+ (Figure 4A, upper panel, lanes 1–4). In the presence of NAD+, the slowly migrating smearing bands detected by the poly(ADP-ribose) antibody are the result of auto-poly(ADP-ribosyl)ation of PARP-1 (Figure 4A, upper panel, lane 5). When either FLAG-hSpt16 or FACT heterodimer was included in the reaction, an additional band of poly(ADP-ribose) was observed (Figure 4A, lanes 7 and 8, shown as asterisk on the right side of upper panel). This band corresponds to the position of FLAG-hSpt16 (Figure 4A, lower panel, arrowhead) suggesting that the observed signal is due to poly(ADP-ribosyl)ation of FLAG-hSpt16. No signal of poly(ADP-ribose) was observed in association with SSRP1 even in the presence of hSpt16 (Figure 4A, upper panel, lanes 6 and 8). Therefore, these results suggest that hSpt16 but not SSRP1 is a substrate of PARP-1.
Figure 4.
In vitro poly(ADP-ribosyl)ation of hSpt16 by PARP-1. (A) hSpt16 is an in vitro substrate of PARP-1. In vitro poly(ADP-ribosyl)ation reactions were performed using the indicated sets of recombinant proteins. Proteins were separated on SDS–PAGE followed by immunoblot. The same blot was probed sequentially with monoclonal anti-poly(ADP-ribose) (pADPr) antibody (upper panel) and anti-FACT antibodies (hSpt16: 8D2 and SSRP-1: 3E4, lower panel). The band indicated by asterisk represent FLAG-hSpt16 that had been poly(ADP-ribosyl)ated. Arrows indicate the position of SSRP-1. PARP-1 was activated by adding fragmented DNA and NAD+. (B) Poly(ADP-ribosyl)ation disrupts the interaction between hSpt16 and PARP-1. In vitro poly(ADP-ribosyl)ation was carried out as described in (A). PARP-1 was precipitated by rabbit antiserum and the immunoprecipitates were subjected to immunoblot analysis as indicated.
Having demonstrated the poly(ADP-ribosyl)ation of hSpt16 by PARP-1, we then addressed the effect of poly(ADP-ribosyl)ation on the interaction between hSpt16 and PARP-1. In vitro poly(ADP-ribosyl)ation assays were carried out using solid-phase PARP-1 (prebound to protein G beads with PARP-1 specific antibody) and purified FLAG-hSpt16. The interaction between hSpt16 and PARP-1 before or after PARP-1 automodification was compared. As shown in Figure 4B, slow migrating PARP-1 was present on top portion of the gel, suggesting that PARP-1 was heavily automodified, while the level of hSpt16 remained in the immunoprecipitate was reduced. These results suggest that like many other PARP-1 substrates, hSpt16 does not interact with the automodified PARP-1.
Poly(ADP-ribosyl)ation of hSpt16 alters the nucleosome binding property of FACT
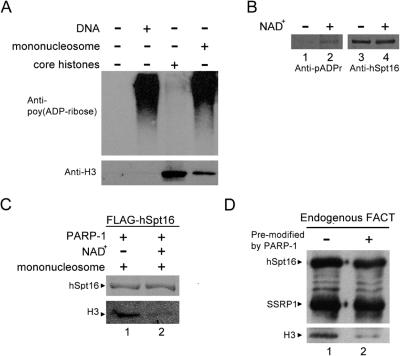
Previous studies demonstrated that FACT interacts with mononucleosome via hSpt16 subunit (14). To investigate the binding of poly(ADP-ribosyl)ated hSpt16 to nucleosomes, we performed in vitro binding assay using purified mononucleosomes, recombinant PARP-1 and FLAG-hSpt16. Mononucleosomes and core histones were purified from HeLa cells (Supplementary Figure S2B). We first tested whether mononucleosome could be activated by mononucleosome. As shown in Figure 5A, PARP-1 could be activated by DNA or mononucleosomes but not by core histones. Furthermore, in the presence of mononucleosomes, hSpt16 could be poly(ADP-ribosyl)ated by PARP-1 (Figure 5B). These data suggest that mononucleosome could serve as PARP-1 activator in the following in vitro mononucleosome-binding assay. FLAG-hSpt16 was incubated with purified PARP-1 and mononucleosome in the presence or absence of NAD+. After incubation, unbound proteins were removed by washing and the bound proteins were subjected to immunoblot analysis (Figure 5C). FLAG-hSpt16 could bind to mononucleosome as reported previously (Figure 5C, lane 1 and data not shown). However, reduced binding of mononucleosome to hSpt16 was observed after poly(ADP-ribosyl)ation reaction (Figure 5C, compare lanes 1 and 2). The effect of poly(ADP-ribosyl)ation of hSpt16 on the nucleosome binding of FACT was also studied. To exclude the possibility that the decreased interaction of FACT with mononucleosomes is due to modification of nucleosomal histones, endogenous FACT was purified from HeLa cells, and modified with PARP-1 in the presence of absence of NAD+ (as described in Figure 4A). PARP-1 and DNA were removed from FACT after poly(ADP-ribosyl)ation of hSpt16. The nucleosome-binding activity of pre-modified or control FACT was compared (Figure 5D). Consistent with earlier results, disruption of interaction of poly(ADP-ribosyl)ated FACT with mononucleosome could be observed. These results indicate that PARP-1-dependent poly(ADP-ribosyl)ation of hSpt16 diminishes FACT's ability to associate with nucleosome in vitro.
Figure 5.
Poly(ADP-ribosyl)ation of hSpt16 reduced its binding to nucleosome. (A) Stimulation of PARP-1 activity by purified HeLa mononucleosomes. Auto poly(ADP-ribosyl)ation of PARP-1 were performed using DNA, mononucleosome, and core histones activators as indicated. Poly(ADP-ribose) polymers were detected by monoclonal antibody 10H-2 (upper panel). The input of core histone and mononucleosome was shown using anti-H3 antibody (lower panel). (B) hSpt16 could be poly(ADP-ribosyl)ated when using mononucleosome to activate PARP-1. In vitro poly(ADP-ribosyl)ation was carried out as described in Figure 4A except that mononucleosome (not DNA) was used to activate PARP-1. The same blot was probed sequentially with monoclonal anti-poly(ADP-ribose) antibody (lanes 1 and 2) and anti-hSpt16 antibodies (lanes 3 and 4). (C) M2 agarose bound FLAG-hSpt16 was incubated with PARP-1 and mononucleosome in the presence or absence of NAD+. After incubation for 30 min at 37°C, unbound proteins were removed by washing and bound proteins were subject to immunoblot analysis using antibodies as indicated by the left side of each panel. (D) Endogenous FACT was immobilized on protein G beads using anti-hSpt16 antibody. FACT was either pre-modified or not by PARP-1 as described in Figure 4A. Mononucleosome binding assay was performed as in (C).
Release of FACT from chromatin during DNA damage response
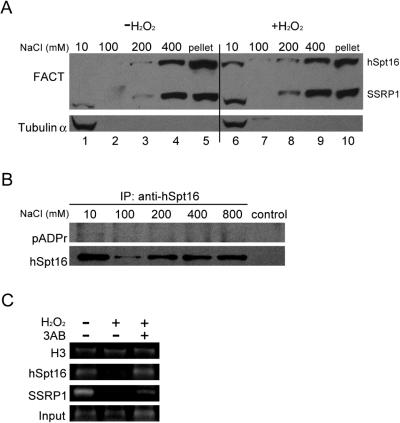
Finally we investigated whether the altered nucleosome binding of poly(ADP-ribosyl)ated FACT could be observed in cells. Previous reports demonstrated that most endogenous FACT is associated with chromatin (15,16). To study the possible dissociation of FACT from chromatin in response to DNA damage, we analyzed the chromatin-bound FACT by extracting the nuclear preparations from control or H2O2-treated HeLa cells with increasing concentrations of NaCl. In the absence of DNA damage, the majority of FACT was associated with chromatin and only insignificant amount of FACT was released between 10 and 100 mM NaCl. The release of hSpt16 and SSRP1 was observed at 200 mM NaCl indicating that it is chromatin associated (23). However, following H2O2 treatment, significant portions of FACT were detected in the soluble fraction (i.e. 10–100 mM NaCl) suggesting that a population of FACT is released from the chromatin (Figure 6A, compare lanes 1 and 6). These results suggest that poly(ADP-ribosyl)ated FACT has weaker chromatin binding ability. We further studied the poly(ADP-ribosyl)ation status of FACT immunoprecipitated from different salt extracted fractions. As shown in Figure 6B, higher level of poly(ADP-ribose) signal was detected in hSpt16 precipitated from low salt (100 mM NaCl) than high salt (i.e. 200 mM) fractions. Taken together, these data suggest that DNA damage-induced poly(ADP-ribosyl)ated Spt16 is released from the chromatin.
Figure 6.
FACT is released from chromatin after poly(ADP-ribosyl)ation. (A) Differential release of FACT from chromatin by increasing concentration of NaCl. Control or H2O2-treated HeLa cells were sequentially extracted with extraction buffer containing the indicated concentration of NaCl. The released proteins and the final pellet were subject to immunoblot analysis using anti-FACT antibodies (upper panel). Tubulin α was used as a marker of soluble fraction (lower panel). (B) Poly(ADP-ribosyl)ation status of hSpt16 extracted with different salt concentrations. HeLa cells were treated with 500 µM H2O2 for 20 min and sequentially extracted with extraction buffer containing the indicated concentrations of NaCl. hSpt16 precipitated from different fractions were subject to immunoblot analysis using monoclonal anti-poly(ADP-ribose) (pADPr) antibody (upper panel). The same blot was reprobed with anti-hSpt16 antibody (lower panel). (C) ChIP analysis of chromatin binding of FACT. The associations of FACT or histone H3 with γ-actin gene under different conditions were analyzed. HeLa cells were subjected to different treatment before crosslinking with formaldehyde. Crosslinked chromatin was immunoprecipitated with the indicated antibodies (on the left side of each panel). Input, 1% of input chromatin was amplified as control.
To further confirm these observations, we performed chromatin immunoprecipitation (ChIP) assays which can monitor the interaction between protein and chromatin more directly. We analyzed the binding of FACT and histone H3 on the coding region of constitutively expressed γ-actin gene (Figure 6C). The association of histone H3 with actin gene showed no significant changes under control or DNA damage conditions. Consistent with the results in Figure 6A, the level of hSpt16 or SSRP1 associated with actin gene was greatly reduced after H2O2 treatment. Furthermore, the reduction in actin gene association of hSpt16 was alleviated by pre-treatment of the cells with 3AB. The binding of SSRP1 to the actin gene was only partially attenuated by pre-treatment of the cells with 3AB. These results further support the concept that poly(ADP-ribosyl)ation modulates the nucleosome binding activity of FACT.
DISCUSSION
In this report we demonstrated that hSpt16 is poly(ADP-ribosyl)ated by PARP-1. We also discovered that this modification is up-regulated in response to genotoxic stress. These results suggest that the biological functions of FACT may be regulated by poly(ADP-ribosyl)ation. This finding lends further support for the importance of PARP-1-mediated poly(ADP-ribosyl)ation of chromatin modulators in the regulation of DNA-dependent processes (24).
Under normal conditions, basal level of poly(ADP-ribosyl)ated proteins detected in the cells suggest that they participate not only in cellular response to DNA damage, but are also involved in the homeostasis of cellular functions. Previous studies have demonstrated that PARP-1 could be activated under normal conditions by certain undamaged DNA structures (25), polynucleosomes (26) and PARP-1 interacting proteins (27). Enzymatic involvement of PARP-1 in the regulation of pol II dependent transcription has been reported (28). In a number of model organisms, PARP-1 has been implicated in regulating gene expression. For example, activation of PARP-1 has been observed during transcription activation of genes in Drosophila salivary gland chromosome (3). Recent studies with PARP-1 null mice and PARP-1 inhibitors have also demonstrated that the activity of PARP-1 is essential for NF-κB-dependent gene expression induced by various inflammatory stimuli (29). Poly(ADP-ribosyl)ation is also known to induce local chromatin re-organization (28). The nucleosome-binding activity of hSpt16 and FACT are altered when poly(ADP-ribosyl)ated by PARP-1 (Figure 5). Our data indicate that PARP-1 may regulate FACT activity in the pol II dependent transcription. Belotserkovskaya et al. (14) showed that FACT helps to remove histones H2B/H2A dimer from a nucleosome during pol II transcription. Their model suggests that FACT facilitates pol II transcription by altering nucleosome structure in front of pol II and then reestablish chromatin architecture after pol II pass. But how FACT is switched off and on in vivo remains a mystery. Although the detailed interaction between FACT and nucleosome is still unclear, there is a strong preference for hSpt16 to bind to H2B/H2A dimer (14). Whether poly(ADP-ribosyl)ated FACT dissociate from nucleosome through the disruption of its interaction with the histones remains to be determined.
Transcriptional arrest in response to DNA damage may be regulated by several mechanisms (30). For example, DNA damage lesion has been shown to block the progression of elongating RNA polymerase and the stalled pol II can be released from the DNA template (30–32). A recent study found that arrested pol II elongation complexes in response to DNA damage are the preferred substrates for ubiquitylation (33). This DNA damage-induced polyubiquitylation of pol II is believed to induce destruction of irreversibly stalled elongation complex, enabling DNA repair and subsequent rounds of transcription (34). Published results also supported that poly(ADP-ribosyl)ation acts as a switch between transcription and DNA repair (35). Following DNA damage, PARP-1 is highly activated leading to rapid and transient poly(ADP-ribosyl)ation of nuclear proteins and itself (2). Active PARP-1 has been suggested to silent transcription through poly(ADP-ribosyl)ation of transcription factors which interferes with their DNA-binding activity (36,37). Our results demonstrate that the narrow time frame of the increased modification on hSpt16 coincides with the activation of PARP-1 under these conditions (Figure 2). This immediate modification correlates with the decreased affinity of FACT to chromatin (Figure 6). These data suggest that when activated, PARP-1 may regulate pol II-dependent transcription through modification of the nucleosome binding subunit of FACT.
There are at least 18 known members of the PARP gene family (38). In the present study, we do not exclude the possibility that other PARPs may also be able to modify hSpt16. PARP-2, which shares some common properties with PARP-1, is a possible candidate. PARP-2 is the only other PARP reported to mediate poly(ADP-ribose) synthesis in response to DNA damage. PARP-1 and PARP-2 participate in overlapping DNA damage response (39). The possible involvement of PARP-2 in the regulation of FACT may be studied in the future.
In this report, poly(ADP-ribosyl)ation of SSRP1 was not observed. The reduced association of SSRP1 with chromatin was only partially preserved by pre-treatment with 3AB (Figure 6C). The interaction between hSpt16 and SSRP1 was not significantly influenced by poly(ADP-ribosyl)ation (Figures 2 and 5D). Although previous i_n vitro_ assays using purified nucleosome have shown that hSpt16, but not SSRP1, is responsible for nucleosome binding property of FACT (14), SSRP1 was known to bind the specific structures of DNA (40,41)—a likely scenario for the observed results in Figure 6C. In agreement with this possibility, the study of Li et al. (42) demonstrated that phosphorylation of SSRP1 by CK2 in response to UV inhibited the DNA-binding activity of SSRP1 and FACT in vitro. These observations suggest that FACT is a target of multiple enzymes that confine transcription of pol II following genotoxic stress. Indeed, the complex network of signaling pathways leads to various post-translational modifications of FACT or sub-population of FACT may determine its activity. Accordingly, poly(ADP-ribosyl)ation of FACT by PARP-1 during genotoxic stress may be confined to a sub-population of FACT. Other modifications of FACT such as phosphorylation–dephosphorylation and acetylation–deacetylation may act in conjunction with poly(ADP-ribosyl)ation to modulate its nucleosome-binding and functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
[Supplementary Data]
Acknowledgments
We thank Ming-Yue Lee for reading of the manuscript. This work was supported by grants from the National Science Council (NSC92-2320-B002-190 and NSC93-2320-B002-109) and the Institute of Biological Chemistry, Academia Sinica. Funding to pay the Open Access publication charges for this article was provided by National Science Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yung T.M., Sato S., Satoh M.S. Poly(ADP-ribosyl)ation as a DNA damage-induced post-translational modification regulating poly(ADP-ribose) polymerase-1-topoisomerase I interaction. J. Biol. Chem. 2004;279:39686–39696. doi: 10.1074/jbc.M402729200. [DOI] [PubMed] [Google Scholar]
- 2.D'Amours D., Desnoyers S., D'Silva I., Poirier G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 3.Tulin A., Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 4.Tartier L., Spenlehauer C., Newman H.C., Folkard M., Prise K.M., Michael B.D., Menissier-de Murcia J., de Murcia G. Local DNA damage by proton microbeam irradiation induces poly(ADP-ribose) synthesis in mammalian cells. Mutagenesis. 2003;18:411–416. doi: 10.1093/mutage/geg015. [DOI] [PubMed] [Google Scholar]
- 5.Meyer-Ficca M.L., Meyer R.G., Jacobson E.L., Jacobson M.K. Poly(ADP-ribose) polymerases: managing genome stability. Int. J. Biochem. Cell Biol. 2005;37:920–926. doi: 10.1016/j.biocel.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Adamietz P., Rudolph A. ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J. Biol. Chem. 1984;259:6841–6846. [PubMed] [Google Scholar]
- 7.d'Erme M., Yang G., Sheagly E., Palitti F., Bustamante C. Effect of poly(ADP-ribosyl)ation and Mg2+ ions on chromatin structure revealed by scanning force microscopy. Biochemistry. 2001;40:10947–10955. doi: 10.1021/bi002742a. [DOI] [PubMed] [Google Scholar]
- 8.Tulin A., Stewart D., Spradling A.C. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaquero A., Loyola A., Reinberg D. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003;2003:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 10.Flaus A., Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 12.Orphanides G., Wu W.H., Lane W.S., Hampsey M., Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 13.Rhoades A.R., Ruone S., Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 15.Wittmeyer J., Joss L., Formosa T., Lycan D., Mikesell G., Bunger M., Breeden L. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- 16.Loyola A., He S., Oh S., McCafferty D.G., Reinberg D. Techniques used to study transcription on chromatin templates. Methods Enzymol. 2004;377:474–499. doi: 10.1016/S0076-6879(03)77031-1. [DOI] [PubMed] [Google Scholar]
- 17.Rowley A., Singer R.A., Johnston G.C. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol. 1991;11:5718–5726. doi: 10.1128/mcb.11.11.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lycan D., Mikesell G., Bunger M., Breeden L. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:7455–7465. doi: 10.1128/mcb.14.11.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 20.Utley R.T., Owen-Hughes T.A., Juan L.J., Cote J., Adams C.C., Workman J.L. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 21.Zahradka P., Ebisuzaki K. Poly(ADP-ribose) polymerase is a zinc metalloenzyme. Eur. J. Biochem. 1984;142:503–509. doi: 10.1111/j.1432-1033.1984.tb08314.x. [DOI] [PubMed] [Google Scholar]
- 22.Mendez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichota J., Grasser K.D. Differential chromatin association and nucleosome binding of the maize HMGA, HMGB, and SSRP1 proteins. Biochemistry. 2001;40:7860–7867. doi: 10.1021/bi010548y. [DOI] [PubMed] [Google Scholar]
- 24.Petermann E., Keil C., Oei S.L. Importance of poly(ADP-ribose) polymerases in the regulation of DNA-dependent processes. Cell Mol. Life Sci. 2005;62:731–738. doi: 10.1007/s00018-004-4504-2. [DOI] [PubMed] [Google Scholar]
- 25.Kun E., Kirsten E., Ordahl C.P. Coenzymatic activity of randomly broken or intact double-stranded DNAs in auto and histone H1 trans-poly(ADP-ribosylation), catalyzed by poly(ADP-ribose) polymerase (PARP I) J. Biol. Chem. 2002;277:39066–39069. doi: 10.1074/jbc.C200410200. [DOI] [PubMed] [Google Scholar]
- 26.Kim M.Y., Mauro S., Gevry N., Lis J.T., Kraus W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Oei S.L., Shi Y. Transcription factor Yin Yang 1 stimulates poly(ADP-ribosyl)ation and DNA repair. Biochem. Biophys. Res. Commun. 2001;284:450–454. doi: 10.1006/bbrc.2001.4985. [DOI] [PubMed] [Google Scholar]
- 28.Kraus W.L., Lis J.T. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 29.Oliver F.J., Menissier-de Murcia J., Nacci C., Decker P., Andriantsitohaina R., Muller S., de la Rubia G., Stoclet J.C., de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tornaletti S. Transcription arrest at DNA damage sites. Mutat Res. 2005;577:131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Hara R., Selby C.P., Liu M., Price D.H., Sancar A. Human transcription release factor 2 dissociates RNA polymerases I and II stalled at a cyclobutane thymine dimer. J. Biol. Chem. 1999;274:24779–24786. doi: 10.1074/jbc.274.35.24779. [DOI] [PubMed] [Google Scholar]
- 32.Tremeau-Bravard A., Riedl T., Egly J.M., Dahmus M.E. Fate of RNA polymerase II stalled at a cisplatin lesion. J. Biol. Chem. 2004;279:7751–7759. doi: 10.1074/jbc.M309853200. [DOI] [PubMed] [Google Scholar]
- 33.Somesh B.P., Reid J., Liu W.F., Sogaard T.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Multiple mechanisms confining RNA polymerase II Ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Muratani M., Tansey W.P. How the ubiquitin–proteasome system controls transcription. Nature Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 35.Ziegler M., Oei S.L. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays. 2001;23:543–548. doi: 10.1002/bies.1074. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza-Alvarez H., Alvarez-Gonzalez R. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J. Biol. Chem. 2001;276:36425–36430. doi: 10.1074/jbc.M105215200. [DOI] [PubMed] [Google Scholar]
- 37.Oei S.L., Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem. Biophys. Res. Commun. 2001;285:27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- 38.Ame J.C., Spenlehauer C., de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 39.Huber A., Bai P., de Murcia J.M., de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst.) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Gariglio M., Ying G.G., Hertel L., Gaboli M., Clerc R.G., Landolfo S. The high-mobility group protein T160 binds to both linear and cruciform DNA and mediates DNA bending as determined by ring closure. Exp. Cell Res. 1997;236:472–481. doi: 10.1006/excr.1997.3742. [DOI] [PubMed] [Google Scholar]
- 41.Bruhn S.L., Pil P.M., Essigmann J.M., Housman D.E., Lippard S.J. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl Acad. Sci. USA. 1992;89:2307–2311. doi: 10.1073/pnas.89.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Keller D.M., Scott J.D., Lu H. CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J. Biol. Chem. 2005;280:11869–11875. doi: 10.1074/jbc.M413944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]