Diplotype Trend Regression Analysis of the ADH Gene Cluster and the ALDH2 Gene: Multiple Significant Associations with Alcohol Dependence (original) (raw)
Abstract
The set of alcohol-metabolizing enzymes has considerable genetic and functional complexity. The relationships between some alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) genes and alcohol dependence (AD) have long been studied in many populations, but not comprehensively. In the present study, we genotyped 16 markers within the ADH gene cluster (including the ADH1A, ADH1B, ADH1C, ADH5, ADH6, and ADH7 genes), 4 markers within the ALDH2 gene, and 38 unlinked ancestry-informative markers in a case-control sample of 801 individuals. Associations between markers and disease were analyzed by a Hardy-Weinberg equilibrium (HWE) test, a conventional case-control comparison, a structured association analysis, and a novel diplotype trend regression (DTR) analysis. Finally, the disease alleles were fine mapped by a Hardy-Weinberg disequilibrium (HWD) measure (J). All markers were found to be in HWE in controls, but some markers showed HWD in cases. Genotypes of many markers were associated with AD. DTR analysis showed that ADH5 genotypes and diplotypes of ADH1A, ADH1B, ADH7, and ALDH2 were associated with AD in European Americans and/or African Americans. The risk-influencing alleles were fine mapped from among the markers studied and were found to coincide with some well-known functional variants. We demonstrated that DTR was more powerful than many other conventional association methods. We also found that several ADH genes and the ALDH2 gene were susceptibility loci for AD, and the associations were best explained by several independent risk genes.
Several linkage studies, including the Collaborative Study on the Genetics of Alcoholism,1–4 a study by investigators at the National Institute on Alcohol Abuse and Alcoholism,5 and a study involving Mission Indians,6 have provided evidence supporting the localization of a risk locus or loci for alcohol dependence (AD [MIM 103780]) to a region harboring the alcohol dehydrogenase (ADH) gene cluster at chromosome 4q21-25 (reviewed by Luo et al.7). One or more risk alleles at the ADH gene cluster may directly predispose to AD. To identify these risk alleles, association studies using linkage disequilibrium (LD) mapping methods are most commonly used, which include case-only association designs,7 case-control association designs, and family-based association designs.
Both case-only designs (using a Hardy-Weinberg disequilibrium [HWD] test) and case-control designs can be valid association and fine-mapping methods. However, both designs are vulnerable to population stratification that could result in spurious findings. We therefore used a structured association (SA) method based on a case-control design, a novel method developed by Pritchard et al.,8 to exclude population stratification and admixture effects on associations. This method and related methods have been applied in several previous studies.e.g.,9–12 However, this method also has its limitations: (1) it does not take gene-gene interactions into account, and (2) it cannot accurately analyze haplotype data when some individuals have uncertain haplotype pairs (which are always observed when statistical inference is used to reconstruct haplotypes). The present study aims to extend this SA approach and to overcome its limitations by developing a novel method, which we call “diplotype trend regression” (DTR) analysis, a method similar to haplotype trend regression,13 that extends our previous application.11
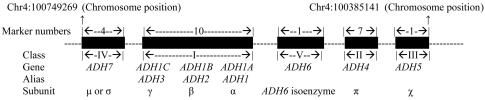
Certain ADH variants are among the best-known AD-vulnerability genes (table 1). This set of genes with partially redundant function may have created a situation relatively tolerant of functional variation in individual genes. Seven ADH genes at the ADH gene cluster are located so close together within an ∼364-kb region (fig. 1) that the LD between them cannot be neglected. Different markers within the same ADH gene could also, of course, be in strong LD. Furthermore, the expression products of different ADH genes—that is, the ADH isoenzymes—have similar amino acid sequences, structures, and properties, co-contributing to liver or stomach ADH activity, with only minor differences in preferred substrates.37–41 Therefore, theoretically, there may be interactions among different ADH genes that cause epistasis. For example, ADH1B (MIM 103720) and ADH1C (MIM 103730) have long been considered to be independent genes influencing risk of alcohol dependence, but Chen et al.29 and Osier et al.14 claimed that, on the basis of stratification analysis or regression analysis, the contribution to risk of alcoholism represented by ADH1C ∧ SNP17 (Ile/Val) might actually be attributable to LD with ADH1B ∧ SNP16 (Arg/His). Additionally, there may be strong physiological interactions between ADH genes and aldehyde dehydrogenase (ALDH) genes, because they appear to have the potential to exert multiplicative effects during the metabolism of alcohol: the ADHs convert alcohol to acetaldehyde, and then the ALDHs quickly convert acetaldehyde into acetate. Acetate is then oxidized via the tricarboxylic acid cycle to yield CO2 and H2O.
Table 1.
Positive Associations between the ADH and ALDH Genes and AD in Different Populations
| Gene or Allele | Positive Finding | Population(s) | Reference(s) |
|---|---|---|---|
| SNP16*Arg (ADH2*1) allele | Increases risk for AD | Japanese, Chinese | 6, 14–26 |
| SNP16*His (ADH2*2) or SNP14*Cys (ADH2*3) allele | Protects against AD | Taiwan Atayal natives, Chinese, Europeans, Jews, AAs | 6, 14–26 |
| SNP17*Ile (ADH3*1) allele | Protects against AD | Chinese, Europeans | 16, 20, 21, 23, 27, 28 |
| SNP17*Val (ADH3*2) allele | Increases risk for AD | Chinese, Mexican Americans, American Indians | 16, 20, 21, 23, 27, 28 |
| ALDH2*1 allele | Increases risk for AD | Chinese, Japanese | 20, 29–32 |
| ALDH2*2 allele | Protects against AD | East Asians, Asian Americans | 20, 29–32 |
| ADH5 gene | Two markers related to AD | … | 33 |
| ADH4 gene | Several variants associated with AD | EAs, Brazilians | 34, 35 |
| ADH7 gene | Epistatic role in protecting against AD | Asians (majority) | 36 |
Figure 1.
ADH gene cluster
Detection of gene-gene interactions among different ADH and ALDH genes is important for two main reasons. (1) Identifying an interaction will increase our understanding of the mechanisms through which the genes act to control expression of the trait; ignoring a true gene-gene interaction in an analysis can, erroneously, make the main effects of the genes appear nonsignificant.42 (2) Failing to model a gene-gene interaction in an analysis can lead to incorrect conclusions with respect to determination of the mode of inheritance and estimation of the magnitude of genetic effects.43,44 Thus, these marker-marker or gene-gene interactions should not be neglected. When gene-gene interactions are detected, we evaluate the strength of these interactions and study the effect of each gene by controlling for the interaction effects on the trait. One common analytic method to study gene-gene interaction effects is called “stratification analysis” (discussed by Luo et al.11). However, stratification analysis, through subsetting the sample, reduces statistical power for the identification of interactions. Another common analytic method to study gene-gene interaction effects is regression analysis, which directly models all the variables in a single analysis, thereby increasing the statistical power.11,45–48 DTR is one such regression model (see the “Material and Methods” section).
Because a multilocus haplotype incorporates the LD information from single markers and also might reflect additional information from unknown neighboring markers, it has the potential to provide more information in association analysis than any single marker. But inevitably, unambiguous haplotype pairs will often be unavailable if statistical inference is used to reconstruct haplotypes. In the analysis, if we use the most likely pair (i.e., the “best pair”) of haplotypes (“reduced mode”)—which has the highest probability among all the inferred uncertain haplotype pairs in each individual—so that we can use an existing analytic method such as SA (which requires that each individual’s haplotype be identified), the bias may become significant, including LD overestimation and biased estimates of haplotype effects. If we use all possible haplotype pairs inferred (“full mode”), which may have different probabilities in one individual, the bias will be maximally reduced, and the results will therefore be a better approximation of the truth. We are not aware of any previously existing analytic method that can use this “full mode” of haplotype pairs.
Disease is a natural-selection factor; this can be reflected in HWD at a disease locus, or in markers in LD with the disease locus. One may observe HWD at a locus when an association exists between that locus and disease.7 Under HWD, alleles at a locus are not independent of each other, and this may invalidate allelewise analysis of that locus.7,49 A multilocus haplotype is actually the subset of every single-locus allele; both allele and haplotype reflect the features of chromosomes in the population. Thus, under HWD, haplotypewise analysis may also be invalid. In this situation, genotypewise analysis may be the only way to draw fully valid conclusions. A diplotype (i.e., a haplotype pair) is the subset of every single-locus genotype; both genotype and diplotype represent the types of chromosome pairs in each individual. Therefore, under HWD, diplotypewise analysis may be a valid and maximally informative method. We also note that, under a recessive mode of inheritance, genotypewise and diplotypewise analyses should be considerably more powerful than allelewise and haplotypewise analyses.7,12 DTR is a diplotypewise analytic method (see the “Material and Methods” section).
In summary, in the present study, we used a DTR method that controls for any population stratification and admixture effects, allows for unknown haplotype phase, takes marker-marker and gene-gene interactions into account, obviates the need for Hardy-Weinberg equilibrium (HWE), and avoids multiple testing due to consideration of multiple populations, multiple markers, and multiple genes.
Many studies have shown positive associations between the ADH1B, ADH1C, ADH5 (MIM 103710), ADH4 (MIM 103740), ADH7 (MIM 600086), and ALDH2 (MIM 100650) loci and AD within specific populations or have shown consistent positive findings across different populations (e.g., see table 1). For the present study, we investigated associations between AD and all ADH genes (except ADH4, which we studied previously and reported elsewhere7,12) and ALDH2 in European Americans (EAs) and African Americans (AAs), the two most common distinct populations in the United States, and tested the population specificity of any detected associations, using DTR.
Material and Methods
Subjects
A total of 801 unrelated subjects were included in this study, as described elsewhere.11 This sample includes two different populations (651 EAs and 150 AAs; the populations were classified by statistical determination of ancestry proportions, as discussed below), comprising 365 healthy controls (317 EAs and 48 AAs) and 436 cases (334 EAs and 102 AAs) and including both females (_n_=324) and males (_n_=477). The cases met lifetime DSM-IIIR or DSM-IV criteria50,51 for AD. The control subjects were screened to exclude major axis I mental disorders, including substance-use disorders, psychotic disorders (including schizophrenia or schizophrenia-like disorders), mood disorders, and major anxiety disorders. Males constituted 75.9% of the cases and 40.0% of the controls. The average ages were 28.1±9.1 years for controls and 40.3±9.2 years for cases. The subjects were recruited at the University of Connecticut Health Center or at the VA Connecticut Healthcare System, West Haven Campus. All subjects gave informed consent before participating in the study, which was approved by the institutional review board at each institution.
Marker Selection
The present study aimed to create a basis for a future fine-mapping study with denser sets of markers at each potential risk gene. These markers were selected because (1) they were available from and validated by Applied Biosystems (ABI) or were studied in a prior publication (e.g., four ALDH2 markers were selected from the study by Peterson et al.52) or (2) they had previously been reported to be associated with AD. After validation by PCR and allele-frequency evaluation in our sample, one ADH5 marker (located in a haplotype block that covers 80% of the full length of ADH5 [according to the ABI SNP and haplotype database]), one ADH6 (MIM 103735) marker (located in a haplotype block that covers 100% of the full length of ADH6), three ADH1A (MIM 103700) markers, four ADH1B markers, three ADH1C markers, four ADH7 markers, and four ALDH2 markers were ultimately included (table 2). Seven ADH4 markers were studied previously.7,12 Although the results with respect to phenotype have been reported elsewhere, these data were included in this study for LD analysis. All the rs numbers for these markers were available from the SNP database (dbSNP).
Table 2.
Information and Genotyping Methods for ADH and ALDH2 Gene Markers
| Marker | Alias | rsNumber | Chromosome | Position | Distancea(bp) | Substitution | AminoAcid | Location | GenotypingTechnique |
|---|---|---|---|---|---|---|---|---|---|
| ADH5 ∧ SNP1 | … | rs1154400 | 4 | 100468404 | 0 | C/T | … | Exon 1 | Assays-on-Demand |
| ADH4 ∧ SNP2 | … | rs6532795 | 4 | 100500615 | 32,211 | T/C | … | 3′ | Assays-on-Demand |
| ADH4 ∧ SNP3 | … | rs1042364 | 4 | 100503968 | 3,353 | G/A | Gly/Arg | Exon 10 | Assays-on-Demand |
| ADH4 ∧ SNP4 | … | rs1126671 | 4 | 100506808 | 2,840 | G/A | Val/Ile | Exon 8 | Assays-by-Design |
| ADH4 ∧ SNP5 | … | rs1126670 | 4 | 100511127 | 4,319 | T/G | Pro/Pro | Exon 7 | Assays-on-Demand |
| ADH4 ∧ SNP6 | … | rs7694646 | 4 | 100518126 | 6,999 | A/T | … | Intron 5 | Assays-on-Demand |
| ADH4 ∧ SNP7 | A−75C | rs1800759 | 4 | 100523903 | 5,777 | A/C | … | Promoter | Assays-on-Demand |
| ADH4 ∧ SNP8 | … | rs1984362 | 4 | 100529367 | 5,464 | C/T | … | 5′ | Assays-on-Demand |
| ADH6 ∧ SNP9 | … | rs13104485 | 4 | 100599217 | 69,850 | A/T | … | 3′ | Assays-on-Demand |
| ADH1A ∧ SNP10 | … | rs6837311 | 4 | 100653667 | 54,450 | A/T | … | 5′ | Assays-on-Demand |
| ADH1A ∧ SNP11 | … | rs975833 | 4 | 100660133 | 6,466 | C/G | … | Intron 7 | Assays-on-Demand |
| ADH1A ∧ SNP12 | … | rs1229966 | 4 | 100671827 | 11,694 | A/G | … | 3′ | Assays-on-Demand |
| ADH1B ∧ SNP13 | … | rs1042026 | 4 | 100686860 | 15,033 | C/T | … | Exon 11 | Assays-on-Demand |
| ADH1B ∧ SNP14 | ADH2*1/3 | rs2066702 | 4 | 100687411 | 551 | C/T | Arg/Cys | Exon 10 | PCR-RFLPb |
| ADH1B ∧ SNP15 | C96T | rs2066701 | 4 | 100696807 | 9,396 | C/T | … | Intron 3 | PCR-RFLPc |
| ADH1B ∧ SNP16 | ADH2*1/2 | rs1229984 | 4 | 100697713 | 906 | G/A | Arg/His | Exon 4 | PCR-RFLPd |
| ADH1C ∧ SNP17 | ADH3*1/2 | rs698 | 4 | 100719183 | 21,470 | A/G | Ile/Val | Exon 9 | PCR-RFLPe |
| ADH1C ∧ SNP18 | … | rs1693482 | 4 | 100722359 | 3,176 | A/G | Gln/Arg | Exon 7 | Assays-by-Design |
| ADH1C ∧ SNP19 | … | rs1693427 | 4 | 100725221 | 2,862 | C/T | … | Intron 4 | Assays-on-Demand |
| ADH7 ∧ SNP20 | … | rs284786 | 4 | 100792371 | 67,150 | A/T | … | Exon 11 | Assays-on-Demand |
| ADH7 ∧ SNP21 | … | rs971074 | 4 | 100800255 | 7,884 | C/T | Arg/Arg | Exon 7 | Assays-on-Demand |
| ADH7 ∧ SNP22 | … | rs1573496 | 4 | 100808063 | 7,808 | C/G | Ala/Gly | Exon 4 | Assays-by-Design |
| ADH7 ∧ SNP23 | … | rs1154470 | 4 | 100814731 | 6,668 | A/G | … | Intron 2 | Assays-on-Demand |
| ALDH2 ∧ SNP24 | G−355A | rs886205 | 12 | 110667147 | …f | G/A | … | 5′ | PCR-RFLPg |
| ALDH2 ∧ SNP25 | T348C | rs440 | 12 | 110691434 | 24,287 | T/C | … | Intron 6 | PCR-RFLPg |
| ALDH2 ∧ SNP26 | T483C | rs11613351 | 12 | 110691512 | 78 | T/C | … | Intron 6 | PCR-RFLPg |
| ALDH2 ∧ SNP27 | G69A | rs4646777 | 12 | 110692756 | 1,244 | G/A | … | Intron 8 | PCR-RFLPg |
Thirty-eight ancestry-informative markers (AIMs) unlinked to the ADH and ALDH genes, including 37 STRs and one Duffy antigen gene (FY) marker (rs2814778), were genotyped to detect the population structure of our sample. These marker sets were employed in many previous studies,9–12 and their characteristics have been described elsewhere53 in a report that included many of the subjects in the present study.
Genotyping
By TaqMan technique
Genomic DNA was extracted from peripheral blood by standard methods. Most SNPs were genotyped with a fluorogenic 5′-nuclease assay method: the TaqMan technique.54 PCR conditions were described elsewhere.7 All genotyping was performed in duplicate, and results were compared to ensure validity of the data. Mismatched genotypes, which constituted <0.5% of the total number of duplicate genotypes performed, were discarded.
By PCR-RFLP technique
Three ADH1B markers, one ADH1C marker, four ALDH2 markers, and one FY marker were genotyped by PCR-RFLP. The FY marker (rs2814778), highly informative for the ethnic ancestry of the subject, was genotyped by a PCR-RFLP technique as described elsewhere.55 Approximately 8% of genotypes on each plate cohort were genotyped again for quality control, with complete concordance.
By fluorescence capillary electrophoresis technique
The 37 STR markers were genotyped by a fluorescence capillary electrophoresis technique with the ABI PRISM 3100 semiautomated capillary fluorescence sequencer, as described elsewhere.53
Statistical Analysis
LD analysis
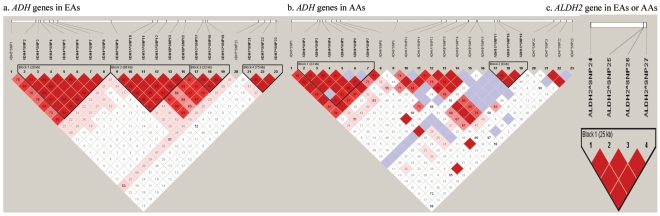
Pairwise LD between any two ADH or ALDH2 gene markers was analyzed separately by population (EAs and AAs). The _D_′ value for each LD pair was calculated and visualized using the program Haploview56 (fig. 2).
Figure 2.
LD analysis for ADH and ALDH2 markers in EAs and AAs. a, ADH genes in EAs. b, ADH genes in AAs. c, ALDH2 gene in EAs or AAs.
HWE test
HWE was tested within populations and separately in cases and controls, by use of an exact test of goodness of fit that is implemented in the program PowerMarker, version 3.0; P values are shown in table 3. Deviation from HWE expectations (i.e., HWD) in cases can indicate a valid disease-gene association.
Table 3.
P Values for HWE Tests in Cases[Note]
| P | ||
|---|---|---|
| Marker | EAs | AAs |
| ADH5 ∧ SNP1 | .037 | .058 |
| ADH1B ∧ SNP16 | .0001 | >.10 |
| ADH1C ∧ SNP17 | >.10 | .012 |
| ADH1C ∧ SNP18 | .055 | .035 |
| ADH1C ∧ SNP19 | .061 | .056 |
| ADH7 ∧ SNP22 | >.10 | .016 |
| ADH7 ∧ SNP23 | >.10 | .091 |
Genotype frequency analysis
Allele and genotype frequencies of the ADH and ALDH2 markers among EAs and AAs are shown in table 4. Genotype-phenotype associations were tested using exact tests (2 df) in the program PowerMarker; P values are listed in table 5.
Table 4.
Genotype and Allele Frequencies in EAs and AAs[Note]
| EAs | AAs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| With AD | Controls | With AD | Controls | ||||||
| Marker | Genotypeor Allele | n | Frequency | n | Frequency | n | Frequency | n | Frequency |
| ADH5 ∧ SNP1 | TT | 153 | .475 | 136 | .459 | 46 | .455 | 20 | .417 |
| ADH5 ∧ SNP1 | TC | 127 | .394 | 135 | .456 | 50 | .495 | 24 | .500 |
| ADH5 ∧ SNP1 | CC | 42 | .130 | 25 | .084 | 5 | .050 | 4 | .083 |
| ADH5 ∧ SNP1 | T | 433 | .672 | 407 | .688 | 142 | .703 | 64 | .667 |
| ADH5 ∧ SNP1 | C | 211 | .328 | 185 | .313 | 60 | .297 | 32 | .333 |
| ADH6 ∧ SNP9 | TT | 94 | .296 | 83 | .278 | 73 | .730 | 35 | .729 |
| ADH6 ∧ SNP9 | AT | 156 | .491 | 140 | .468 | 24 | .240 | 13 | .271 |
| ADH6 ∧ SNP9 | AA | 68 | .214 | 76 | .254 | 3 | .030 | 0 | .000 |
| ADH6 ∧ SNP9 | T | 344 | .541 | 306 | .512 | 170 | .850 | 83 | .865 |
| ADH6 ∧ SNP9 | A | 292 | .459 | 292 | .488 | 30 | .150 | 13 | .135 |
| ADH1A ∧ SNP10 | TT | 126 | .387 | 106 | .353 | 80 | .808 | 37 | .787 |
| ADH1A ∧ SNP10 | AT | 152 | .466 | 142 | .473 | 19 | .192 | 10 | .213 |
| ADH1A ∧ SNP10 | AA | 48 | .147 | 52 | .173 | 0 | .000 | 0 | .000 |
| ADH1A ∧ SNP10 | T | 404 | .620 | 354 | .590 | 179 | .904 | 84 | .894 |
| ADH1A ∧ SNP10 | A | 248 | .380 | 246 | .410 | 19 | .096 | 10 | .106 |
| ADH1A ∧ SNP11 | GG | 198 | .623 | 169 | .569 | 59 | .596 | 21 | .438 |
| ADH1A ∧ SNP11 | CG | 105 | .330 | 108 | .364 | 36 | .364 | 26 | .542 |
| ADH1A ∧ SNP11 | CC | 15 | .047 | 20 | .067 | 4 | .040 | 1 | .021 |
| ADH1A ∧ SNP11 | G | 501 | .788 | 446 | .751 | 154 | .778 | 68 | .708 |
| ADH1A ∧ SNP11 | C | 135 | .212 | 148 | .249 | 44 | .222 | 28 | .292 |
| ADH1A ∧ SNP12 | AA | 136 | .421 | 123 | .423 | 13 | .134 | 3 | .063 |
| ADH1A ∧ SNP12 | AG | 141 | .437 | 127 | .436 | 44 | .454 | 24 | .500 |
| ADH1A ∧ SNP12 | GG | 46 | .142 | 41 | .141 | 40 | .412 | 21 | .438 |
| ADH1A ∧ SNP12 | A | 413 | .639 | 373 | .641 | 70 | .361 | 30 | .313 |
| ADH1A ∧ SNP12 | G | 233 | .361 | 209 | .359 | 124 | .639 | 66 | .688 |
| ADH1B ∧ SNP13 | TT | 174 | .537 | 154 | .520 | 83 | .847 | 43 | .915 |
| ADH1B ∧ SNP13 | TC | 119 | .367 | 115 | .389 | 14 | .143 | 4 | .085 |
| ADH1B ∧ SNP13 | CC | 31 | .096 | 27 | .091 | 1 | .010 | 0 | .000 |
| ADH1B ∧ SNP13 | T | 467 | .721 | 423 | .715 | 180 | .918 | 90 | .957 |
| ADH1B ∧ SNP13 | C | 181 | .279 | 169 | .285 | 16 | .082 | 4 | .043 |
| ADH1B ∧ SNP14 | CC | 316 | 1.000 | 280 | 1.000 | 70 | .745 | 24 | .511 |
| ADH1B ∧ SNP14 | TC | 0 | .000 | 0 | .000 | 22 | .234 | 22 | .468 |
| ADH1B ∧ SNP14 | TT | 0 | .000 | 0 | .000 | 2 | .021 | 1 | .021 |
| ADH1B ∧ SNP14 | C | 632 | 1.000 | 560 | 1.000 | 162 | .862 | 70 | .745 |
| ADH1B ∧ SNP14 | T | 0 | .000 | 0 | .000 | 26 | .138 | 24 | .255 |
| ADH1B ∧ SNP15 | CC | 131 | .524 | 116 | .487 | 78 | .839 | 43 | .915 |
| ADH1B ∧ SNP15 | TC | 101 | .404 | 101 | .424 | 14 | .151 | 4 | .085 |
| ADH1B ∧ SNP15 | TT | 18 | .072 | 21 | .088 | 1 | .011 | 0 | .000 |
| ADH1B ∧ SNP15 | C | 363 | .726 | 333 | .700 | 170 | .914 | 90 | .957 |
| ADH1B ∧ SNP15 | T | 137 | .274 | 143 | .300 | 16 | .086 | 4 | .043 |
| ADH1B ∧ SNP16 | GG | 218 | .956 | 215 | .888 | 83 | .988 | 41 | .953 |
| ADH1B ∧ SNP16 | AG | 8 | .035 | 27 | .112 | 1 | .012 | 2 | .047 |
| ADH1B ∧ SNP16 | AA | 2 | .009 | 0 | .000 | 0 | .000 | 0 | .000 |
| ADH1B ∧ SNP16 | G | 444 | .974 | 457 | .944 | 167 | .994 | 84 | .977 |
| ADH1B ∧ SNP16 | A | 12 | .026 | 27 | .056 | 1 | .006 | 2 | .023 |
| ADH1C ∧ SNP17 | AA | 108 | .359 | 105 | .376 | 57 | .626 | 35 | .761 |
| ADH1C ∧ SNP17 | AG | 134 | .445 | 126 | .452 | 34 | .374 | 10 | .217 |
| ADH1C ∧ SNP17 | GG | 59 | .196 | 48 | .172 | 0 | .000 | 1 | .022 |
| ADH1C ∧ SNP17 | A | 350 | .581 | 336 | .602 | 148 | .813 | 80 | .870 |
| ADH1C ∧ SNP17 | G | 252 | .419 | 222 | .398 | 34 | .187 | 12 | .130 |
| ADH1C ∧ SNP18 | GG | 123 | .381 | 113 | .382 | 64 | .640 | 38 | .792 |
| ADH1C ∧ SNP18 | AG | 139 | .430 | 130 | .439 | 36 | .360 | 9 | .188 |
| ADH1C ∧ SNP18 | AA | 61 | .189 | 53 | .179 | 0 | .000 | 1 | .021 |
| ADH1C ∧ SNP18 | G | 385 | .596 | 356 | .601 | 164 | .820 | 85 | .885 |
| ADH1C ∧ SNP18 | A | 261 | .404 | 236 | .399 | 36 | .180 | 11 | .115 |
| ADH1C ∧ SNP19 | TT | 115 | .365 | 111 | .370 | 62 | .674 | 38 | .792 |
| ADH1C ∧ SNP19 | TC | 138 | .438 | 132 | .440 | 30 | .326 | 9 | .188 |
| ADH1C ∧ SNP19 | CC | 62 | .197 | 57 | .190 | 0 | .000 | 1 | .021 |
| ADH1C ∧ SNP19 | T | 368 | .584 | 354 | .590 | 154 | .837 | 85 | .885 |
| ADH1C ∧ SNP19 | C | 262 | .416 | 246 | .410 | 30 | .163 | 11 | .115 |
| ADH7 ∧ SNP20 | TT | 166 | .519 | 147 | .497 | 26 | .260 | 9 | .188 |
| ADH7 ∧ SNP20 | AT | 127 | .397 | 116 | .392 | 56 | .560 | 22 | .458 |
| ADH7 ∧ SNP20 | AA | 27 | .084 | 33 | .111 | 18 | .180 | 17 | .354 |
| ADH7 ∧ SNP20 | T | 459 | .717 | 410 | .693 | 108 | .540 | 40 | .417 |
| ADH7 ∧ SNP20 | A | 181 | .283 | 182 | .307 | 92 | .460 | 56 | .583 |
| ADH7 ∧ SNP21 | CC | 241 | .746 | 225 | .771 | 61 | .610 | 24 | .511 |
| ADH7 ∧ SNP21 | TC | 75 | .232 | 62 | .212 | 35 | .350 | 20 | .426 |
| ADH7 ∧ SNP21 | TT | 7 | .022 | 5 | .017 | 4 | .040 | 3 | .064 |
| ADH7 ∧ SNP21 | C | 557 | .862 | 512 | .877 | 157 | .785 | 68 | .723 |
| ADH7 ∧ SNP21 | T | 89 | .138 | 72 | .123 | 43 | .215 | 26 | .277 |
| ADH7 ∧ SNP22 | GG | 249 | .764 | 242 | .796 | 96 | .970 | 44 | .936 |
| ADH7 ∧ SNP22 | CG | 70 | .215 | 56 | .184 | 3 | .030 | 3 | .064 |
| ADH7 ∧ SNP22 | CC | 7 | .021 | 6 | .020 | 0 | .000 | 0 | .000 |
| ADH7 ∧ SNP22 | G | 568 | .871 | 540 | .888 | 195 | .985 | 91 | .968 |
| ADH7 ∧ SNP22 | C | 84 | .129 | 68 | .112 | 3 | .015 | 3 | .032 |
| ADH7 ∧ SNP23 | GG | 133 | .430 | 133 | .455 | 65 | .707 | 36 | .750 |
| ADH7 ∧ SNP23 | AG | 138 | .447 | 118 | .404 | 27 | .293 | 11 | .229 |
| ADH7 ∧ SNP23 | AA | 38 | .123 | 41 | .140 | 0 | .000 | 1 | .021 |
| ADH7 ∧ SNP23 | G | 404 | .654 | 384 | .658 | 157 | .853 | 83 | .865 |
| ADH7 ∧ SNP23 | A | 214 | .346 | 200 | .342 | 27 | .147 | 13 | .135 |
| ALDH2 ∧ SNP24 | AA | 213 | .705 | 129 | .686 | 11 | .118 | 5 | .114 |
| ALDH2 ∧ SNP24 | AG | 83 | .275 | 52 | .277 | 42 | .452 | 22 | .500 |
| ALDH2 ∧ SNP24 | GG | 6 | .020 | 7 | .037 | 40 | .430 | 17 | .386 |
| ALDH2 ∧ SNP24 | A | 509 | .843 | 310 | .824 | 64 | .344 | 32 | .364 |
| ALDH2 ∧ SNP24 | G | 95 | .157 | 66 | .176 | 122 | .656 | 56 | .636 |
| ALDH2 ∧ SNP25 | TT | 173 | .671 | 123 | .715 | 62 | .689 | 25 | .581 |
| ALDH2 ∧ SNP25 | TC | 80 | .310 | 42 | .244 | 27 | .300 | 17 | .395 |
| ALDH2 ∧ SNP25 | CC | 5 | .019 | 7 | .041 | 1 | .011 | 1 | .023 |
| ALDH2 ∧ SNP25 | T | 426 | .826 | 288 | .837 | 151 | .839 | 67 | .779 |
| ALDH2 ∧ SNP25 | C | 90 | .174 | 56 | .163 | 29 | .161 | 19 | .221 |
| ALDH2 ∧ SNP26 | TT | 178 | .685 | 117 | .718 | 61 | .678 | 26 | .591 |
| ALDH2 ∧ SNP26 | TC | 77 | .296 | 39 | .239 | 28 | .311 | 16 | .364 |
| ALDH2 ∧ SNP26 | CC | 5 | .019 | 7 | .043 | 1 | .011 | 2 | .045 |
| ALDH2 ∧ SNP26 | T | 433 | .833 | 273 | .837 | 150 | .833 | 68 | .773 |
| ALDH2 ∧ SNP26 | C | 87 | .167 | 53 | .163 | 30 | .167 | 20 | .227 |
| ALDH2 ∧ SNP27 | GG | 146 | .658 | 117 | .701 | 57 | .663 | 26 | .619 |
| ALDH2 ∧ SNP27 | AG | 71 | .320 | 44 | .263 | 28 | .326 | 16 | .381 |
| ALDH2 ∧ SNP27 | AA | 5 | .023 | 6 | .036 | 1 | .012 | 0 | .000 |
| ALDH2 ∧ SNP27 | G | 363 | .818 | 278 | .832 | 142 | .826 | 68 | .810 |
| ALDH2 ∧ SNP27 | A | 81 | .182 | 56 | .168 | 30 | .174 | 16 | .190 |
Table 5.
P Values of Comparisons for Genotype Frequency Distributions between Cases and Controls in EAs and AAs[Note]
| P Beforea | P Afterb | |||
|---|---|---|---|---|
| Marker | EAs | AAs | EAs | AAs |
| ADH1A ∧ SNP11 | >.10 | >.10 | >.10 | .075 |
| ADH1B ∧ SNP14 | NA | .012 | NA | .004 |
| ADH1B ∧ SNP16 | .001 | >.10 | .007 | >.10 |
| ADH1C ∧ SNP17 | >.10 | .040 | >.10 | >.10 |
| ADH1C ∧ SNP18 | >.10 | .025 | >.10 | .056 |
| ADH1C ∧ SNP19 | >.10 | .068 | >.10 | >.10 |
| ADH7 ∧ SNP20 | >.10 | .068 | >.10 | .068 |
Fine mapping the risk alleles
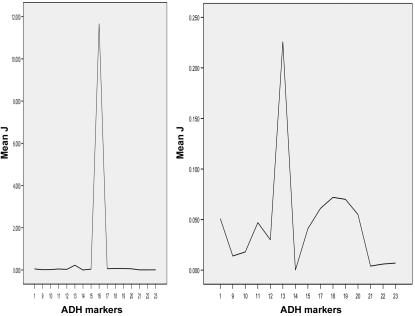
HWD of a marker in cases sometimes indicates a valid gene-phenotype association, especially when the marker is in HWE in controls.7,11 Thus, HWD measures can be used for fine mapping a risk locus—ideally, in the situation where markers are in HWD in cases but in HWE in controls, as was often the case in the present study (table 3) and in the study by Luo et al.7 Many measures of HWD in case-only samples have been advanced for this purpose, including F, F_′, J,_ and _J_′.57,58 Among these, J is the preferred disequilibrium measure for fine mapping, because it is a direct decreasing function of the recombination fraction between the disease and the marker loci and does not depend on allele frequencies of the disease and marker loci. J can be derived from the genotype frequency data but not from the allele frequency data.7,58 If there are several peak J values in the ADH gene cluster, this might suggest that there are several risk alleles for disease within that cluster (fig. 3). Therefore, among the many HWD measures, this statistic is best suited for fine mapping in the present application.
Figure 3.
Fine mapping the risk alleles at the ADH gene cluster in EA cases on the basis of J values. The _X_-axis represents the marker names; the _Y_-axis represents the J values. Marker numbers (which do not include markers mapped to the ADH4 gene) correspond to the order presented in table 2. The marker ADH1B ∧ SNP16 (i.e., ADH2*Arg/His, with the highest J value) is included in the left figure but excluded in the right figure (to enlarge the scale of the _Y_-axis).
Population structure analysis
The two most common genetically distinguishable populations in the United States—EAs and AAs—have their origins in ancestral populations that migrated from multiple geographic locations in Europe and Africa, respectively. Both populations have admixture histories in recent generations in the United States, although the admixture rate for EAs is much lower than that for AAs. As reported elsewhere,59,60 AAs are admixed primarily with EAs, and some EA individuals have (usually small) proportions of African ancestry. Thus, both of these populations were treated as potentially admixed populations in the present study.
Even when the statistical analysis is conducted separately for EAs and AAs, population stratification could still have an effect on the analysis, because admixture within these two populations could still produce spurious LD block size, confuse HWD tests, or cause spurious associations. Pritchard et al.8 and Falush et al.61 developed a software program, STRUCTURE, based on a model-based clustering method, that can infer ancestry proportions of an admixed sample to detect its underlying population structure by use of information from unlinked AIMs. For this purpose, we selected 38 AIMs, including 37 STR markers and 1 FY marker. The suitability of these AIMs for detecting the presence of population structure, their adequacy for providing information for assigning all individuals into different genetic ancestral populations, and the feasibility of validly analyzing them with the program STRUCTURE have already been demonstrated by many previous studies.9–12,53 These 38 AIMs are unlinked to each other and to the ADH and ALDH2 genes. All AIMs were in HWE, and there was no LD among these AIMs, nor was there association between the AIMs and any phenotype. These AIMs are appropriate for detection of population structure without significant bias. More details of the features of this set of AIMs are provided elsewhere.12,53
To estimate the ancestry proportions of the subjects more accurately, all subjects were studied together as a single “admixed” sample. Parameter settings for running STRUCTURE are reported elsewhere.12
SA analysis
In admixed populations, each individual may have ancestries from different populations, and the ancestry proportions may vary among individuals, which can cause spurious findings in association analysis. By stratifying the admixed population to nonadmixed subpopulations and then performing the association analysis within these subpopulations, spurious findings can be avoided; or, by conditioning the association analysis on the ancestry proportions of each subject, the admixture effects can be accounted for statistically and thereby eliminated. Conversely, correction of the spurious associations—for example, elimination of the associations between the 38 unlinked AIMs and any phenotypes—also indicates that the admixed populations have been successfully structured or that the admixture effects have been successfully controlled. This can be achieved by an SA analysis performed using the program STRAT.62 (Parameter settings for running STRAT are described elsewhere.12) It should be noted that the association analysis was limited to the genotypewise level, not the allelewise level, because of HWD existing among the ADH and ALDH2 markers. This SA method is also not suitable for the unphased diplotype data.
Haplotype reconstruction
The expectation-maximization (EM) algorithm, as employed by many programs that reconstruct estimated haplotypes, assumes HWE. But, in our study, the genotype frequency distributions of many markers were in HWD in the cases (table 3), which violates the assumption of the EM algorithm. This may increase the error of EM estimates, especially when the HWD is attributable to an excess of the expected heterozygote frequency over that observed.63 The Bayesian approach and the partition-ligation algorithm that the program PHASE is based on have been claimed to be more accurate in reconstructing haplotypes than the EM algorithm and are valid even under HWD.64–66 Consequently, we applied PHASE to reconstruct haplotypes and to estimate the diplotype (haplotype pair) probabilities for each subject in the present study. Parameter settings for running PHASE are presented elsewhere.12 Haplotypes were reconstructed for “genetic” EAs (European ancestry proportion >0.5) and AAs (African ancestry proportion >0.5) rather than self-reported EAs and AAs. In the present study, all analyses conducted separately by population were performed using “genetic” EAs and AAs rather than self-reported EAs and AAs.
Alleles at the ADH gene markers that map to the cluster on chromosome 4, especially those within the same haplotype block (e.g., alleles at ADH6, ADH1A, and ADH1B) (fig. 2), can be “put” in the same haplotype, but we constructed haplotypes only within single genes because we wanted to differentiate haplotype effects among different genes. (Alternatively, interactions between different genes were considered via the regression methods described below.)
Gene-gene interaction analysis
Pairwise LD analysis between markers can direct us to the observation of marker-marker correlation. However, single markers usually cannot fully reflect the information for an entire gene. Haplotype-haplotype or diplotype-diplotype interactions might be more representative of gene-gene interaction. Haplotypes or diplotypes themselves incorporate the marker-marker LD information. A multilocus haplotype or diplotype is actually the subset of an allele or a genotype of a single marker, so haplotype or diplotype analysis is actually equivalent to stratification analysis of every single marker,67 with the correlations among single markers already incorporated. Thus, the use of haplotype or diplotype data obviates the analysis of marker-marker interaction effects. Haplotypes or diplotypes are mutually exclusive in structure (i.e., no two haplotypes can be located on the same chromosome), and interactions among them may reflect their joint effects on the trait. To study correlations among diplotypes at different genes, a Pearson correlation analysis can be performed between any two diplotypes (a similar procedure was used by Dong et al.68). Correlation analysis on single markers can be used as a valid LD measure.69 Strong correlation between two intergene diplotypes suggests that these two diplotypes may have additive, or multiplicative, effects on the trait. Strong correlation between two within-gene diplotypes suggests that these two diplotypes may have similar effects on trait. Any two diplotypes within the same gene that are highly correlated can be combined as a single variable in the DTR model (if the variance inflation factor is >10),70 or the interactions between them should be considered if they are not combined as a single variable in DTR. Only the interactions between those diplotypes having correlations with _r_>0.9 and P<.01 were considered in DTR.
Determined by statistical inference but not molecular experimentation, the inferred haplotype probability in each individual is usually not equal to 1.0; uncertainty remains. Thus, most individuals have several possible diplotypes even within one gene, which can be described as follows (“full mode”): the individual has a% of diplotype A (i.e., the probability is a% that A is the correct diplotype), b% of diplotype B, and [100-(a+b)]% of diplotype C (if there are three possible diplotypes). Supposing this individual’s true diplotype is A, we can look at it as a special case of the “full mode”—that is, the individual has 100% of diplotype A, 0% of diplotype B, and 0% of diplotype C. Thus, this method of analysis fits for any certain or uncertain diplotype data.
DTR analysis
A backward stepwise logistic regression analysis implemented in SPSS, version 13.0, was used to test associations between genes and diseases within “genetic” EAs and AAs (see the regression model elsewhere11,12). Backward regression variable selection was applied. In the regression model, phenotypes served as the dependent variables, and the covariates included ancestry proportion, age, sex, genotype probabilities at ADH5 and ADH6 (we only genotyped one SNP at each of these two genes), diplotype probabilities at other genes, and interactions among genotypes or diplotypes. Age and sex were included because they were highly asymmetrically distributed between cases and controls and therefore could potentially confound the association analysis. Ancestry proportions were included in the model to control for population stratification and admixture effects. Genotype and diplotype probabilities were included, but allele and haplotype probabilities were excluded because of HWD.7 Genotypes at ADH5 and ADH6 and diplotypes at other genes can be entered into a single DTR model, because genotypes can be taken as supersets of diplotypes.
In the regression model, phenotype and sex are categorical variables, whereas ancestry proportion, age, genotype probability, and diplotype probability are continuous variables. The use of continuous variables, such as proportions and probabilities, preserves more information than does the use of categorical variables, such as population categories, genotype categories, and diplotype categories. We named this regression analysis that uses diplotype probability as the predictor variable “diplotype trend regression” (DTR) analysis, analogous to haplotype trend regression.13
(As an alternative to this DTR analysis, an even more complete analysis of “full mode” would involve the use of a true complete mixture model,71,72 in which the probabilities of various diplotypes for each person are considered in the analysis. This was beyond the scope of the present study.)
Results
ADH markers were located in several haplotype blocks, whereas ALDH2 markers were in one haplotype block (fig. 2). Twenty-three ADH markers span 346,327 bp, covering 95% of the full length of the ADH gene cluster (364,128 bp) on chromosome 4, with an average intermarker distance of 15 kb (table 2). LD between ADH markers differs substantially between EAs and AAs (fig. 2a and 2b). Pairwise LD analysis showed that three ADH1C markers belong to one haplotype block (_D_′>0.9) in both EAs and AAs. The seven ADH4 markers also belong to one haplotype block in both EAs and AAs (as described by Luo et al.7). The sets of markers at ADH6, ADH1A, and ADH1B belong to one haplotype block in EAs, and three markers at ADH7 belong to another haplotype block in EAs, but these markers do not define any haplotype blocks in AAs. (Markers were in much weaker LD in AAs than in EAs, possibly because AAs are an older population in which recombination may have had more time to reduce haplotype block size.) In both EAs and AAs, there were no significant differences in LD between cases and controls for these markers (data not shown).
Four ALDH2 markers, spanning 25,609 bp of the gene on chromosome 12, cover 60% of the full length of ALDH2 (table 2). LD analysis showed that these four markers were in one haplotype block in both EAs and AAs (fig. 2). Two markers, T348C and T483C, are in complete LD (_D_′=1). In both EAs and AAs, there were no significant differences in LD between cases and controls for these markers (data not shown).
The genotype frequency distributions of all markers were in HWE in both EA and AA controls, but some markers were in HWD in either EA or AA cases (table 3). In EAs, all ADH and ALDH markers were in HWE in controls. However, many ADH markers were nominally in significant (P<.03), modest (.03⩽_P_⩽.05), or suggestive (.05<P<.09) HWD in cases (table 3), including ADH5 ∧ SNP1, ADH1B ∧ SNP16 (Arg/His), ADH1C ∧ SNP18 (Gln/Arg), and ADH1C ∧ SNP19. Seven ADH4 markers were also in significant HWD in cases, as reported elsewhere.7 After correction for multiple testing by use of SNPSpD (an effective Bonferroni-type correction that takes marker correlation into account),73 ADH1B ∧ SNP16 remained in significant HWD (_P_=.0001).
In AAs, all ADH and ALDH markers were in HWE in controls (except ADH1A ∧ SNP11 [_P_=.044], which we presume is because of its rare genotype frequency and the small sample size). However, many ADH markers were nominally in significant (P<.03), modest (.03⩽_P_⩽.05), or suggestive (.05<P<.09) HWD in cases (table 3), including ADH5 ∧ SNP1, ADH1C ∧ SNP17 (Ile/Val), ADH1C ∧ SNP18 (Gln/Arg), ADH1C ∧ SNP19, and ADH7 ∧ SNP22 (Ala/Gly). After correction by SNPSpD, no markers remained in significant HWD.
Genotypes of some ADH markers were associated with AD (table 5). In EAs, the genotypes of ADH1B ∧ SNP16 were nominally associated with AD. (Genotypes of seven ADH4 markers were also significantly associated with AD, as reported elsewhere.7) After correction by SNPSpD, ADH1B ∧ SNP16 remained significantly associated with AD (_P_=.0013).
In AAs, the genotypes of many markers were nominally significantly (P<.03), modestly (.03⩽_P_⩽.05), or suggestively (.05<P<.09) associated with AD, including ADH1B ∧ SNP14 (Arg/Cys), ADH1C ∧ SNP17, ADH1C ∧ SNP18, ADH1C ∧ SNP19, and ADH7 ∧ SNP20. After multiple-comparison correction by SNPSpD, no association remained significant.
There are several peak J values among markers within the ADH gene cluster and the ALDH2 gene for AD in EAs and AAs (fig. 3). In both EAs (fig. 3) and AAs (not shown), there are several peak J values among the ADH markers that might indicate proximity of the risk alleles. The highest J peak in the ADH gene cluster is at a functional variant, ADH1B ∧ SNP16 (Arg/His) (|J|=11.667 in EAs and 1.000 in AAs). Other J peaks are at the following markers (grouped by gene): (1) ADH5 ∧ SNP1 (|J|=0.051 in EAs and 0.439 in AAs), (2) ADH1A ∧ SNP10 (|J|=1.000 in AAs) and ADH1A ∧ SNP11 (6.5 kb to SNP10) (|J|=0.047 in EAs), (3) ADH1B ∧ SNP13 (|J|=0.226 in EAs) and ADH1B ∧ SNP14 (551 bp to ADH1B ∧ SNP13) (|J|=0.112 in AAs), (4) ADH1C ∧ SNP17 (|J|=0.053 in AAs) and ADH1C ∧ SNP18 (3.2 kb to SNP17) (|J|=0.072 in EAs), and (5) ADH7 ∧ SNP20 (|J|=0.055 in EAs) and ADH7 ∧ SNP22 (Ala→Gly) (|J|=1.000 in AAs).
Peak J values among the ALDH2 markers were at SNP24 (|J|=0.197 in EAs) and SNP27 (|J| = 0.618 in AAs). We note that every gene had at least one marker with a J peak.
Two ancestries were detected in our sample. The genotypes of some ADH markers were associated with AD after admixture effects were controlled for. These results are almost completely consistent with, although less statistically significant than, those from the aforementioned case-control genotypewise analysis (table 5).
All subjects were assigned to two ancestral populations, Europeans and Africans; therefore, each subject has two complementary ancestry proportions. According to the ancestry proportions, the mixed sample can be separated into two distinct subpopulations: “genetic” EAs (European ancestry proportion >0.5) and “genetic” AAs (African ancestry proportion >0.5). The concordances between the “genetic” status and the self-reported ethnicity are 100% for EAs and 99.1% for AAs. Among the “genetic” EA subjects, the admixture degree is 1.7%; among the “genetic” AA subjects, the admixture degree is 4.0% (more details given elsewhere12). These two groups are quite distinct, not only in their asymmetric ancestry proportions, but also in the greatly different results from LD analysis, HWE tests, and case-control association analysis.
SA analysis based on this structured sample showed that, in “genetic” EAs, genotypes of ADH1B ∧ SNP16 were significantly associated with AD (_P_=.007). In “genetic” AAs, genotypes of many markers were nominally significantly (P<.03), modestly (.03⩽_P_⩽.05), or suggestively (.05<P<.09) associated with AD, including ADH1A ∧ SNP11, ADH1B ∧ SNP14, ADH1C ∧ SNP18 (Gln/Arg), and ADH7 ∧ SNP20. After correction by SNPSpD, no association remained significant (table 5).
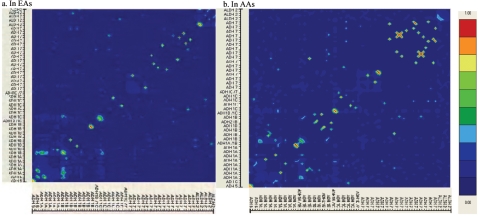
There were correlations between different diplotypes, mainly within genes (fig. 4). Within each population, the results from correlation analyses in cases and controls were similar. However, the correlations were quite different between populations. In EAs, there were significant diplotype-diplotype correlations within the ADH1B, ADH1C, ADH7, and ALDH2 genes (_r_>0.9; P<.01) but weak correlations between genes. In AAs, there were significant diplotype-diplotype correlations within the _ADH1A, ADH1B, ADH1C,_ and _ADH7_ genes (_r_>0.9; P<.01). There were also diplotype-diplotype correlations between ADH1B and ADH7 in AA cases.
Figure 4.
Pairwise correlations between different genotypes (at the ADH5 and ADH6 genes), diplotypes (at other genes) in EAs (a) and AAs (b). The gene names corresponding to the genotypes and diplotypes are shown on the axes, but the detailed names of genotypes and diplotypes are not shown (the names of parts of the risk genotypes and diplotypes can be found in table 6). The colored scale denotes the correlation coefficient (r). This figure was generated using the program GOLD.74
DTR analysis demonstrated that several genes studied were risk genes for AD (table 6). In both EAs and AAs, the genotypes of ADH5 ∧ SNP1 and some diplotypes at the ADH1A, ADH1B, ADH7, and ALDH2 genes were associated with AD. Some of these risk diplotypes exerted consistent effects on phenotype across EAs and AAs. For example, the diplotype TCCG/CCTG at the ADH1B gene protected against disease in both populations (β<0). Some of the risk genotypes or diplotypes exerted opposite effects on phenotype in EAs and AAs. For example, genotype C/C of _ADH5_ _∧_ _SNP1_ and all of the diplotypes at _ADH1A_ increased risk for disease in EAs (β>0) but protected against disease in AAs (β<0). Some of the risk diplotypes exerted effects on phenotype in EAs only. For example, the diplotype CCTG/CCTG at _ADH1B_ and the diplotype ACGG/TCGA at _ADH7_ increased risk for disease in EAs (β>0). The diplotype-diplotype interaction effects occurred mainly in EAs. For example, the diplotype ATTG/ATTG and the diplotype ATTG/GCCA at ALDH2 have interaction effects on phenotype in EAs. Some of the risk diplotypes exerted effects on phenotype in AAs only—for example, the diplotypes TCCG/TCCG and TTCG/TCCG at ADH1B and the diplotype TTGG/TCGG at ADH7 protected against disease (β<0) in AAs, whereas the diplotype TCGG/TCGG at _ADH7_ increased risk for disease in AAs (β>0). Table 6 lists only those variables that remained in the last step of the DTR equations.
Table 6.
DTR Analysis in EAs and AAs[Note]
| Population and Variable | f | P | β |
|---|---|---|---|
| EAs: | |||
| European ancestry | … | .0678 | − |
| Male | … | 1.7 × 10−7 | + |
| Age | … | 8.7 × 10−29 | + |
| ADH5: | |||
| C/C | … | .0124 | + |
| ADH1A: | |||
| AGA/TGA | .203 | .0109 | + |
| AGA/TCG | .181 | .0108 | + |
| AGA/AGA | .164 | .0110 | + |
| TCG/TGA | .109 | .0108 | + |
| AGA/TGG | .088 | .0109 | + |
| TGA/TGA | .060 | .0109 | + |
| TCG/TGG | .060 | .0110 | + |
| TGA/TGG | .057 | .0108 | + |
| TCG/TCG | .055 | .0112 | + |
| ADH1B: | |||
| TCCG/CCTG | .366 | .0071 | − |
| CCTG/CCTG | .075 | .0945 | + |
| TCCA/TCCG | .058 | .0005 | − |
| ADH7: | |||
| ACGG/TCGA | .036 | .0590 | + |
| ALDH2: | |||
| ATTG/ATTG × ATTG/GCCAa | … | 4.6 × 10−9 | − |
| AAs: | |||
| Male | … | .0012 | + |
| Age | … | .0035 | + |
| ADH5: | |||
| T/T | … | .0042 | + |
| ADH5: | |||
| C/T | … | .0073 | + |
| ADH1A: | |||
| TCG/TCG | .083 | .0083 | − |
| TGA/TGA | .059 | .0451 | − |
| ADH1B: | |||
| TCCG/TCCG | .425 | .0106 | − |
| TTCG/TCCG | .270 | .0089 | − |
| TCCG/CCTG | .113 | .0100 | − |
| ADH7: | |||
| ACGG/TCGG | .216 | .0259 | + |
| ACGG/ACGG | .124 | .0425 | − |
| TCGG/TCGG | .108 | .0381 | + |
| TTGG/TCGG | .057 | .0265 | − |
| ALDH2: | |||
| GTTG/GCCA | .197 | .0306 | + |
Discussion
Two main issues in this study warrant discussion: (1) the implications of the results in terms of the gene-phenotype relationships and (2) the properties and advantages of the DTR method. Some ADH and ALDH genes have been shown by other studies to be important risk factors for AD, mainly in Asians (table 1), but we show that they are also important in EAs and AAs, and we are the first to show that other ADH and ALDH genes are important for risk of AD in these two populations. In the present study, we found, using DTR, associations between AD and the ADH5, ADH1A, ADH1B, ADH7, and ALDH2 genes, findings that are consistent with the roles of ADH and ALDH isoenzymes in the metabolism of alcohol. We expected to find evidence of association between ADH loci and AD, but the association was surprisingly comprehensive.
These associations constitute an important part of the genetic risk for AD. This is reflected both in the overall attributable risk for this set of genes, each of which has an independent contribution to disease, and in the fact that this genomic region has consistently been identified as one that harbors AD risk-affecting loci in linkage studies.
DTR is a powerful method, and, in using it, we detected associations that were not seen using many other association methods, such as the HWD test, case-control comparison, and SA. Several features make DTR more powerful than other conventional association methods. First, DTR allows use of a case-control sample, which is easier than a family sample to collect and to expand to reach sufficient statistical power. Second, cases and controls, and even different populations, can be combined in a single DTR model, thereby increasing sample size and statistical power. Third, an unmatched case-control design has been demonstrated to be more powerful than a matched case-control design or a family-based association design in detecting gene-gene interactions, especially when the disease prevalence is moderate (such as with AD).75 Fourth, different variables, including different genotypes and diplotypes from different genes, can be entered into a single DTR model, which avoids the multiple testing that leads to loss of information. Fifth, DTR allows analysis even in the presence of deviation from HWE. Sixth, DTR allows diplotype phase to be uncertain (in the present study, the maximal proportion of individuals with unambiguous diplotypes [i.e., probability = 1] in a single gene was only 37%; the proportion of individuals with unambiguous diplotypes across all the genes studied was only 15%). Seventh, DTR can control for population stratification and admixture effects on association analysis (assuming, of course, that ancestry coefficients are available), and it allows for the control of other potential confounders of association analysis, such as age and sex. Eighth, DTR takes into account gene-gene interactions, an approach that has been demonstrated to be more powerful than single-locus analysis (despite correction for multiple comparisons).76 Finally, DTR is able to account for LD effects and, additionally, _cis_-acting functional effects. There is reason to believe that, in some cases, _cis_-acting elements are mediating phenotypic expression (e.g., there are variants in the promoter of a gene that influence the way other variants impact that gene’s function, so that it is necessary to know, from a functional standpoint, what specific variants are on each chromosome). These may be detected by using diplotype-based (or haplotype-based) analytic approaches but not by using other methods that employ multilocus genotype data. On the basis of these considerations, findings obtained through application of DTR have a high likelihood of being valid.
In our sample, the genotypes of all markers were in HWE in controls, but some were in HWD in cases, indicating the existence of associations between genes and disease.7,57,58,77–81 Comparing the results of these HWD tests with the case-control comparisons, we found two things. First, the results from these methods are largely consistent, which supports the notion that the HWD test can be a valid association method, equivalent to a case-control approach. Second, more markers were found to be associated with phenotypes by the HWD test than by case-control comparison, and P values generally were lower by the HWD test than by case-control comparison. Some P values greater than but close to .05 in the case-control study were <.05 by the HWD test; thus, the HWD test sometimes appears to be more powerful than a case-control approach, which supports the conclusions of Nielsen et al.77 and Luo et al.7 This may reflect a recessive mode of inheritance.
Case-only studies and case-control studies are potentially vulnerable to population stratification, so all association analyses were performed separately for EAs and AAs. To control for admixture effects, SA was applied via the program STRAT, which gave results similar to those obtained using a case-control comparison, indicating that admixture effects were not strong in our sample. We noted that many associations from the HWD test, case-control comparison, and SA method became nonsignificant after correction for multiple tests, which indicates that these association methods often led to information loss. However, this information is preserved using DTR, which does not require adjustment of significance level for multiple tests.
Under HWD, alleles and haplotypes are not independent of one another. The effects of disease-predisposing alleles and haplotypes may be “masked” by other non–disease-predisposing alleles and haplotypes (i.e., epistatic interactions).82 This may be particularly true for recessive diseases, in which the non–disease-associated allele obscures an effect of the disease-associated allele. Therefore, allelewise and haplotypewise analyses might lose power or otherwise be invalid.7,49 Since some of our markers were in HWD, exploratory allelewise and haplotypewise analyses were performed and showed fewer and less significant positive results than genotypewise and diplotypewise analyses for our sample (authors' unpublished data), which is consistent with conclusions from our other studies7,11,12 about the relative power of these methods in an HWD situation. Genotypewise and diplotypewise analyses may be valid even under HWD, and therefore they served as the primary analyses in the present study.
The HWD test, case-control comparison, and SA analyses cannot correct for interaction effects between markers and between genes. Diplotypes incorporate the LD information from different markers, and the interactions between diplotypes can be considered in the DTR model. A diplotype is more representative of gene background than is a single genotype, and diplotype-diplotype interactions from different genes are more representative of gene-gene interactions than are marker-marker interactions. Therefore, DTR works well with respect to the evaluation of gene-gene interactions.
Under HWD, the EM algorithm is not suitable for reconstructing diplotypes. However, in the DTR model, we used the diplotype probabilities predicted by the program PHASE that waived the HWE assumption. When the PHASE approach to haplotype reconstruction is used, DTR is thus also independent of the HWE assumption.
In summary, our findings by DTR analysis include the following points. (1) In EAs and/or AAs, the genotypes of ADH5 ∧ SNP1 and the diplotypes at the ADH1A, ADH1B, ADH7, and ALDH2 genes are associated with AD. Some associations are universal across both populations. Some associations have opposite effects in different populations (suggesting that the actual risk-influencing variant is in a different phase in the two populations, or, alternatively, that there are differing epistatic effects). Some associations are population specific—that is, some associations appear only in EAs or in AAs (table 6). (2) Most associations from DTR analysis are much more significant than those from other association methods. DTR detected strong associations between ALDH2 and disease that were not observed at all by use of other association methods—including a multilocus genotype data analysis with a regression method (the results of which were similar to the single-locus genotype frequency analysis in table 5; data not shown), which may reflect a _cis_-acting functional effect in this gene. (3) The correlations between the genes are weak. But within the genes, diplotype-diplotype correlations are strong, which include those within ADH1A in AAs, ALDH2 in EAs, and ADH1B, ADH1C, and ADH7 in both populations (data not shown). Considering these correlations by DTR, only a significant interaction effect between two diplotypes within ALDH2 was detected in EAs (table 6). Additionally, we found that ADH1C diplotypes were significantly associated with drug dependence, one of the disorders most commonly comorbid with AD (authors' unpublished data).
Markers can be dependent on one another (i.e., correlated) without being in complete LD, or their dependence may not be statistically significant, so that the effects of markers on traits can be decomposed into main effects and interaction effects. If an interaction effect is strong, one marker can “mask” the main effect of another marker.82 The interaction effect depends on the correlation between markers and is related to the trait of interest. Correlation between markers per se (such as LD) depends on the physical distance between markers, the allele frequencies of markers, population history, and the nature of the traits, including the definition of phenotypes (e.g., mutation-related disease), sample size, and ethnicity. Several of these factors—notably, allele frequencies and population history—also vary between populations. Therefore, the interaction effects of markers are affected by many factors. Such effects may also be population-specific. In the present study, ADH1B ∧ SNP16 (Arg/His) was associated with AD in EAs (_P_=.001), and ADH1C ∧ SNP17 (Ile/Val) was associated with AD in AAs (_P_=.040) (table 5). Our EA sample size was relatively large, and the correlation between ADH1B ∧ SNP16 and ADH1C ∧ SNP17 was weak (_D_′=0.758; _r_2=0.019; _P_=.463>.05). In our AA sample, the correlation between these two markers was also weak (_D_′=0.900; _r_2=0.004; _P_=.231>.05; here, we interpret the high _D_′ as being reflective of the different allele frequencies for the two markers). Thus, the interaction effect of these two markers was weak, but the main effect was strong in both populations (by use of regression analysis). Even with this interaction effect taken into account via stratification analysis, as per Osier et al.,14 the main effects of these two markers did not change significantly (data not shown), and the effect of _ADH1B_∧Arg/His and that of _ADH1C_∧Ile/Val did not modify each other significantly in our samples. These findings are not consistent with those reported by Osier et al.,14 who claimed that the contribution of _ADH1C_∧Ile/Val to risk for AD was actually attributable to LD with _ADH1B_∧Arg/His in the Taiwanese Chinese population. This inconsistency may result from the population specificity of the interaction effects; in other words, this effect could be weak in EAs and AAs but strong in Taiwanese Chinese.14 However, the conclusion by Osier et al.14 may simply be incorrect, given the following points. (1) Their sample size (_n_=135) was small. Such a sample size might result in type I error in analysis of interaction effects. (2) The use of a stratification analytic method, and not a regression method, to consider the marker-marker interaction effects could reduce power, because dividing the sample (i.e., into nine subgroups based on three genotypes for each marker) further reduces the sample size. Moreover, this should have occasioned correction for multiple comparisons. (3) The reported _D_′ of 0.77 between the _ADH1B_∧Arg/His and _ADH1C_∧Ile/Val variants14 does not constitute strong enough disequilibrium for the markers to be in the same haplotype block (as defined by Gabriel et al.83); markers should usually show higher LD to exert interaction effects on traits through that mechanism. Increasing the sample size may help to clarify whether this _D_′ value was accurate and whether such LD can result in as strong an interaction effect in Taiwanese Chinese as that reported by Osier et al.14 (4) Finally, two markers represent only two points or two haplotype blocks in genes; a marker-marker interaction effect is not sufficient to represent a gene-gene interaction effect (that is, additional markers at these two loci might not have any interaction effects at all). It may therefore have been excessive to state that the ADH1C gene exerted its effect via the ADH1B gene in all populations, especially because the authors tested only two markers in a small sample of a specific population. Our design overcomes these particular limitations, and we were able to demonstrate that these two genes exert independent main effects on phenotype—at least in EA and AA populations.
The multiplicity of gene effects that we observed (several ADH genes and the ALDH2 gene were associated with AD) confirms that these disorders are multigenic—minor effects from different genes produced additive effects on risk for AD. This is consistent with the roles of different ADH and ALDH isoenzymes in contributing to alcohol metabolism (although different isoenzymes have minor differences in the preferred substrates). Although the activity of ADH1 enzyme (α subunit) is weak in adults, the ADH1A gene still has effects on risk for AD.
Replacing multilocus diplotypes with single-locus genotypes in the DTR model can be done to fine map the risk locus (data not shown). One advantage of DTR as a fine-mapping method is that it allows for marker-marker interactions, so that the confounding effects of these interactions can be accounted for. However, DTR fine mapping is limited by the fact that it does not control for the influence of the allele frequency of markers.12 The best approach for fine mapping would be to combine DTR with HWD measures—that is, use DTR to screen potential susceptibility genes and then use an HWD measure, such as the J value, to fine map the risk alleles within those genes. In the present study, the results from fine mapping with a J value (fig. 3) are basically consistent with those from HWD tests (table 3) and case-control comparisons (table 5).
We noted that every gene had at least one marker with a J peak. This suggests that, despite the fact that LD is sometimes present between markers at different genes, association signals are actually originating within the genes that show J peaks, which is consistent with the DTR results. Interestingly, we localized some risk alleles close to well-known functional variants, such as ADH1B ∧ SNP16 (Arg/His; previously called “_ADH2*1/2_”), ADH1B ∧ SNP14 (Arg/Cys; previously called “_ADH2*1/3_”), ADH1C ∧ SNP17 (Ile/Val; previously called “_ADH3*1/2_”), and ADH7 ∧ SNP22 (Ala→Gly), which is consistent with findings from the existing literature (listed in table 1) and supports the validity of our findings. Among these peaks, the J value at ADH1B ∧ SNP16 for AD in EAs is extremely high (11.667) and is consistent with the significance levels from HWD tests and case-control comparisons, suggesting either that this marker is extremely close to the disease locus at the ADH1B gene or that the marker might be the disease locus itself. In future studies aimed at fine mapping the risk alleles, a denser set of markers at each risk gene will be required. This is a necessary next step in understanding the complex association between the genes encoding multiple alcohol-metabolizing enzymes and AD in a variety of populations.
Acknowledgments
This work was supported in part by National Institutes of Health grants R01-DA12849, R01-DA12690, K24-DA15105, R01-AA11330, P50-AA12870, K08-AA13732, K24-AA13736, K02-MH01387, and M01-RR06192 (to University of Connecticut General Clinical Research Center), by funds from the U.S. Department of Veterans Affairs (to the VA Medical Research Program; the VA Connecticut–Massachusetts Mental Illness Research, Education and Clinical Center; and the VA Research Enhancement Award Program research center), a National Alliance for Research on Schizophrenia and Depression Young Investigator Award (to X.L.), and Alcoholic Beverage Medical Research Foundation grant award R06932 (to X.L.). Ann Marie Lacobelle provided excellent technical assistance. Dr. Jaakko Lappalainen gave helpful comments. The constructive, knowledgeable, and sagacious comments of the two anonymous reviewers are highly appreciated.
Web Resources
The URLs for data presented herein are as follows:
- Applied Biosystems (ABI), http://www.appliedbiosystems.com/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD, ADH1B, ADH1C, ADH5, ADH4, ADH7, ALDH2, ADH6, and ADH1A)
- PowerMarker, http://www.powermarker.net/ (for genetic data analysis software)
References
- 1.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H (1998) Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81:207–221 [DOI] [PubMed] [Google Scholar]
- 2.Zinn-Justin A, Abel L (1999) Genome search for alcohol dependence using the weighted pairwise correlation linkage method: interesting findings on chromosome 4. Genet Epidemiol Suppl 1 17:S421–S426 [DOI] [PubMed] [Google Scholar]
- 3.Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J (1999) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet 65:1148–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP (2000) A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet 96:632–637 [DOI] [PubMed] [Google Scholar]
- 5.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed] [Google Scholar]
- 6.Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC (2004) Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet 129:110–115 10.1002/ajmg.b.30057 [DOI] [PubMed] [Google Scholar]
- 7.Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J (2005) ADH4 gene variation is associated with alcohol and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology (http://www.nature.com/npp/journal/vaop/ncurrent/abs/1300925a.html) (electronically published October 12, 2005; accessed April 6, 2006) [DOI] [PubMed] [Google Scholar]
- 8.Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein MB, Schork MJ, Gelernter J (2004) A polymorphism of the β1-adrenergic receptor is associated with shyness and low extraversion. Biol Psychiatry 56:217–224 10.1016/j.biopsych.2004.05.020 [DOI] [PubMed] [Google Scholar]
- 10.Kaufman J, Yang BZ, Douglas-Palumberi H, Houshyar S, Lipschitz D, Krystal JH, Gelernter J (2004) Social supports and serotonin transporter gene moderate depression in maltreated children. Proc Natl Acad Sci USA 101:17316–17321 10.1073/pnas.0404376101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J (2005) CHRM2 gene predisposes to alcohol dependence, drug dependence, and affective disorders: results from an extended case-control structured association study. Hum Mol Genet 14:2421–2434 10.1093/hmg/ddi244 [DOI] [PubMed] [Google Scholar]
- 12.Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J (2005) ADH4 gene variation is associated with alcohol and drug dependence in European Americans: results from family-controlled and population-structured association studies. Pharmacogenet Genomics 15:755–768 [DOI] [PubMed] [Google Scholar]
- 13.Zaykin DV, Westfall PH, Young SS, Karnoub MC, Wagner MJ, Ehm MG (2002) Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53:79–91 10.1159/000057986 [DOI] [PubMed] [Google Scholar]
- 14.Osier M, Pakstis AJ, Kidd JR, Lee JF, Yin SJ, Ko HC, Edenberg HJ, Lu RB, Kidd KK (1999) Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 64:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomasson HR, Crabb DW, Edenberg HJ, Li TK, Hwu HG, Chen CC, Yeh EK, Yin SJ (1994) Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res 18:640–643 10.1111/j.1530-0277.1994.tb00923.x [DOI] [PubMed] [Google Scholar]
- 16.Chao YC, Wang MF, Tang HS, Hsu CT, Yin SJ (1994) Genotyping of alcohol dehydrogenase at the ADH2 and ADH3 loci by using a polymerase chain reaction and restriction-fragment-length polymorphism in Chinese alcoholic cirrhotics and non-alcoholics. Proc Natl Sci Counc Repub China B 18:101–106 [PubMed] [Google Scholar]
- 17.Nakamura K, Suwaki H, Matsuo Y, Ichikawa Y, Miyatake R, Iwahashi K (1995) Association between alcoholics and the genotypes of ALDH2, ADH2, ADH3 as well as P-4502E1 [in Japanese]. Arukoru Kenkyuto Yakubutsu Ison 30:33–42 [PubMed] [Google Scholar]
- 18.Nakamura K, Iwahashi K, Matsuo Y, Miyatake R, Ichikawa Y, Suwaki H (1996) Characteristics of Japanese alcoholics with the atypical aldehyde dehydrogenase 2*2. I. A comparison of the genotypes of ALDH2, ADH2, ADH3, and cytochrome P-4502E1 between alcoholics and nonalcoholics. Alcohol Clin Exp Res 20:52–55 10.1111/j.1530-0277.1996.tb01043.x [DOI] [PubMed] [Google Scholar]
- 19.Tanaka F, Shiratori Y, Yokosuka O, Imazeki F, Tsukada Y, Omata M (1996) High incidence of ADH2*1/ALDH2*1 genes among Japanese alcohol dependents and patients with alcoholic liver disease. Hepatology 23:234–239 [DOI] [PubMed] [Google Scholar]
- 20.Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT (1996) Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry 168:762–767 [DOI] [PubMed] [Google Scholar]
- 21.Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J, Zhao ZL, Xia GY (1997) Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–1277 [PubMed] [Google Scholar]
- 22.Neumark YD, Friedlander Y, Thomasson HR, Li TK (1998) Association of the ADH2*2 allele with reduced ethanol consumption in Jewish men in Israel: a pilot study. J Stud Alcohol 59:133–139 [DOI] [PubMed] [Google Scholar]
- 23.Borras E, Coutelle C, Rosell A, Fernandez-Muixi F, Broch M, Crosas B, Hjelmqvist L, Lorenzo A, Gutierrez C, Santos M, Szczepanek M, Heilig M, Quattrocchi P, Farres J, Vidal F, Richart C, Mach T, Bogdal J, Jornvall H, Seitz HK, Couzigou P, Pares X (2000) Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology 31:984–989 10.1053/he.2000.5978 [DOI] [PubMed] [Google Scholar]
- 24.Shea SH, Wall TL, Carr LG, Li TK (2001) ADH2 and alcohol-related phenotypes in Ashkenazic Jewish American college students. Behav Genet 31:231–239 10.1023/A:1010261713092 [DOI] [PubMed] [Google Scholar]
- 25.Hasin D, Aharonovich E, Liu X, Mamman Z, Matseoane K, Carr L, Li TK (2002) Alcohol and ADH2 in Israel: Ashkenazis, Sephardics, and recent Russian immigrants. Am J Psychiatry 159:1432–1434 10.1176/appi.ajp.159.8.1432 [DOI] [PubMed] [Google Scholar]
- 26.Ehlers CL, Gilder DA, Harris L, Carr L (2001) Association of the ADH2*3 allele with a negative family history of alcoholism in African American young adults. Alcohol Clin Exp Res 25:1773–1777 10.1111/j.1530-0277.2001.tb02189.x [DOI] [PubMed] [Google Scholar]
- 27.Konishi T, Calvillo M, Leng AS, Feng J, Lee T, Lee H, Smith JL, Sial SH, Berman N, French S, Eysselein V, Lin KM, Wan YJ (2003) The ADH3*2 and CYP2E1 c2 alleles increase the risk of alcoholism in Mexican American men. Exp Mol Pathol 74:183–189 10.1016/S0014-4800(03)00006-6 [DOI] [PubMed] [Google Scholar]
- 28.Mulligan CJ, Robin RW, Osier MV, Sambuughin N, Goldfarb LG, Kittles RA, Hesselbrock D, Goldman D, Long JC (2003) Allelic variation at alcohol metabolism genes (ADH1B, ADH1C, ALDH2) and alcohol dependence in an American Indian population. Hum Genet 113:325–336 10.1007/s00439-003-0971-z [DOI] [PubMed] [Google Scholar]
- 29.Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ (1999) Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet 65:795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwahashi K, Matsuo Y, Suwaki H, Nakamura K, Ichikawa Y (1995) CYP2E1 and ALDH2 genotypes and alcohol dependence in Japanese. Alcohol Clin Exp Res 19:564–566 10.1111/j.1530-0277.1995.tb01549.x [DOI] [PubMed] [Google Scholar]
- 31.McCarthy DM, Wall TL, Brown SA, Carr LG (2000) Integrating biological and behavioral factors in alcohol use risk: the role of ALDH2 status and alcohol expectancies in a sample of Asian Americans. Exp Clin Psychopharmacol 8:168–175 10.1037/1064-1297.8.2.168 [DOI] [PubMed] [Google Scholar]
- 32.Hara K, Terasaki O, Okubo Y (2000) Dipole estimation of alpha EEG during alcohol ingestion in males genotyped for ALDH2. Life Sci 67:1163–1173 10.1016/S0024-3205(00)00707-4 [DOI] [PubMed] [Google Scholar]
- 33.Edman K, Maret W (1992) Alcohol dehydrogenase genes: restriction fragment length polymorphisms for ADH4 (π-ADH) and ADH5 (χ-ADH) and construction of haplotypes among different ADH classes. Hum Genet 90:395–401 [DOI] [PubMed] [Google Scholar]
- 34.Foroud T, Dick D, Xuei X, Goate A, Porjesz B, Begleiter H, Reich T, Edenberg HJ (2003) Association of SNPs in the ADH gene cluster with alcoholism. Am J Hum Genet Suppl 73:521 [Google Scholar]
- 35.Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M (2005) Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry 162:1005–1007 10.1176/appi.ajp.162.5.1005 [DOI] [PubMed] [Google Scholar]
- 36.Osier MV, Lu R-B, Pakstis AJ, Kidd JR, Huang S-Y, Kidd KK (2004) Possible epistatic role of ADH7 in the protection against alcoholism. Am J Med Genet B Neuropsychiatr Genet 126:19–22 10.1002/ajmg.b.20136 [DOI] [PubMed] [Google Scholar]
- 37.Smith M, Hopkinson DA, Harris H (1972) Alcohol dehydrogenase isozymes in adult human stomach and liver: evidence for activity of the ADH(3) locus. Ann Hum Genet 35:243–253 [DOI] [PubMed] [Google Scholar]
- 38.Smith M, Hopkinson DA, Harris H (1973) Studies on the subunit structure and molecular size of the human dehydrogenase isozymes determined by the different loci, ADH(1), ADH(2), and ADH(3). Ann Hum Genet 36:401–414 [DOI] [PubMed] [Google Scholar]
- 39.Li TK, Bosron WF, Dafeldecker WP, Lange LG, Vallee BL (1977) Isolation of Π-alcohol dehydrogenase of human liver: is it a determinant of alcoholism? Proc Nat Acad Sci 74:4378–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zgombic-Knight M, Foglio MH, Duester G (1995) Genomic structure and expression of the ADH7 gene encoding human class IV alcohol dehydrogenase, the form most efficient for retinol metabolism in vitro. J Biol Chem 270:4305–4311 10.1074/jbc.270.9.4305 [DOI] [PubMed] [Google Scholar]
- 41.Edenberg HJ (2000) Regulation of the mammalian alcohol dehydrogenase genes. Prog Nucleic Acid Res Mol Biol 64:295–341 [DOI] [PubMed] [Google Scholar]
- 42.Ottman R (1990) An epidemiologic approach to gene-environment interaction. Genet Epidemiol 7:177–185 10.1002/gepi.1370070302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiret L, Abel L, Rakotovao R (1993) Effect of ignoring genotype-environment interaction on segregation analysis of quantitative traits. Genet Epidemiol 10:581–586 10.1002/gepi.1370100642 [DOI] [PubMed] [Google Scholar]
- 44.Eaves LJ (1984) The resolution of genotype × environment interaction in segregation analysis of nuclear families. Genet Epidemiol 1:215–228 10.1002/gepi.1370010302 [DOI] [PubMed] [Google Scholar]
- 45.Breiman L, Friedman JH, Olshen RA, Stone CJ (1984) Classification and regression trees. Wadsworth, Belmont, CA [Google Scholar]
- 46.Friedman JH (1991) Multivariate adaptive regression splines. Ann Statist 19:1–67 [Google Scholar]
- 47.Gauderman WJ, Faucett CL (1997) Detection of gene-environment interactions in joint segregation and linkage analysis. Am J Hum Genet 61:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cook NR, Zee RYL, Ridker PM (2004) Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat Med 23:1439–1453 10.1002/sim.1749 [DOI] [PubMed] [Google Scholar]
- 49.Sasieni PD (1997) From genotypes to genes: doubling the sample size. Biometrics 53:1253–1261 [PubMed] [Google Scholar]
- 50.American Psychiatric Association (1987) Diagnostic and statistical manual of mental disorders, third edition, revised. American Psychiatric Press, Washington, DC [Google Scholar]
- 51.American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, fourth edition. American Psychiatric Press, Washington, DC [Google Scholar]
- 52.Peterson RJ, Goldman D, Long JC (1999) Nucleotide sequence diversity in non-coding regions of ALDH2 as revealed by restriction enzyme and SSCP analysis. Hum Genet 104:177–187 10.1007/s004390050932 [DOI] [PubMed] [Google Scholar]
- 53.Yang BZ, Zhao H, Kranzler HR, Gelernter J (2005) Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol 28:302–312 10.1002/gepi.20070 [DOI] [PubMed] [Google Scholar]
- 54.Shi MM, Myrand SP, Bleavins MR, de la Iglesia FA (1999) High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol 52:295–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo X, Klempan TA, Lappalainen J, Rosenheck RA, Charney DS, Erdos J, van Kammen DP, Kranzler HR, Kennedy JL, Gelernter J (2004) NOTCH4 gene haplotype is associated with schizophrenia in African-Americans. Biol Psychiatry 55:112–117 10.1016/S0006-3223(03)00588-2 [DOI] [PubMed] [Google Scholar]
- 56.Barrett J, Fry B, Maller J, Daly J (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 57.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, et al (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13:399–408 10.1038/ng0896-399 [DOI] [PubMed] [Google Scholar]
- 58.Jiang R, Dong J, Wang D, Sun FZ (2001) Fine-scale mapping using Hardy-Weinberg disequilibrium. Ann Hum Genet 65:207–219 10.1046/j.1469-1809.2001.6520207.x [DOI] [PubMed] [Google Scholar]
- 59.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fallin D, Schork NJ (2000) Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am J Hum Genet 67:947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niu T, Qin ZD, Xu X, Liu JS (2002) Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am J Hum Genet 70:157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valdes AM, Thomson G (1997) Detecting disease-predisposing variants: the haplotype method. Am J Hum Genet 60:703–716 [PMC free article] [PubMed] [Google Scholar]
- 68.Dong C, Wang S, Li WD, Li D, Zhao H, Price A (2003) Interacting genetic loci on chromosomes 20 and 10 influence extreme human obesity. Am J Hum Genet 72:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38:226–231 10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- 70.Haan CT (2002) Statistical methods in hydrology, second edition. Iowa State University Press, Ames [Google Scholar]
- 71.Self SG, Longton G, Kopecky KJ, Liang KY (1991) On estimating HLA/disease association with application to a study of aplastic anemia. Biometrics 47:53–61 [PubMed] [Google Scholar]
- 72.Chiano MN, Clayton DG (1998) Fine genetic mapping using haplotype analysis and the missing data problem. Ann Hum Genet 62:55–60 10.1017/S0003480098006678 [DOI] [PubMed] [Google Scholar]
- 73.Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- 75.Wang S, Zhao H (2003) Sample size needed to detect gene-gene interactions using association designs. Am J Epidemiol 158:899–914 10.1093/aje/kwg233 [DOI] [PubMed] [Google Scholar]
- 76.Marchini J, Donnelly P, Cardon LR (2005) Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet 37:413–417 10.1038/ng1537 [DOI] [PubMed] [Google Scholar]
- 77.Nielsen DM, Ehm MG, Weir BS (1999) Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet 63:1531–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoh J, Wille A, Ott J (2001) Trimming, weighting, and grouping SNPs in human case-control association studies. Genome Res 11:2115–2119 10.1101/gr.204001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee WC (2003) Searching for disease-susceptibility loci by testing for Hardy-Weinberg disequilibrium in a gene bank of affected individuals. Am J Epidemiol 158:397–400 10.1093/aje/kwg150 [DOI] [PubMed] [Google Scholar]
- 80.Hao K, Xu X, Laird N, Wang X, Xu X (2004) Power estimation of multiple SNP association test of case-control study and application. Genet Epidemiol 26:22–30 10.1002/gepi.10293 [DOI] [PubMed] [Google Scholar]
- 81.Wittke-Thompson JK, Pluzhnikov A, Cox NJ (2005) Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet 76:967–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cordell HJ (2002) Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet 11:2463–2468 10.1093/hmg/11.20.2463 [DOI] [PubMed] [Google Scholar]
- 83.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]