Mutually Exclusive T-Cell Receptor Induction and Differential Susceptibility to Human Immunodeficiency Virus Type 1 Mutational Escape Associated with a Two-Amino-Acid Difference between HLA Class I Subtypes (original) (raw)
Abstract
The relative contributions of HLA alleles and T-cell receptors (TCRs) to the prevention of mutational viral escape are unclear. Here, we examined human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses restricted by two closely related HLA class I alleles, B*5701 and B*5703, that differ by two amino acids but are both associated with a dominant response to the same HIV-1 Gag epitope KF11 (KAFSPEVIPMF). When this epitope is presented by HLA-B*5701, it induces a TCR repertoire that is highly conserved among individuals, cross-recognizes viral epitope variants, and is rarely associated with mutational escape. In contrast, KF11 presented by HLA-B*5703 induces an entirely different, more heterogeneous TCR β-chain repertoire that fails to recognize specific KF11 escape variants which frequently arise in clade C-infected HLA-B*5703+ individuals. These data show the influence of HLA allele subtypes on TCR selection and indicate that extensive TCR diversity is not a prerequisite to prevention of allowable viral mutations.
Sequence mutations resulting in loss of recognition by CD8+ T cells represent a key mechanism used by viruses to escape from host immune responses and a major challenge for vaccine development (2, 5, 6, 16, 20, 28, 29). Such sequence alterations can occur at anchor positions of the epitope and reduce or abrogate peptide binding to the restricting major histocompatibility complex (MHC) class I molecule. Alternatively, amino acid changes within or immediately adjacent to CD8+ T-cell epitopes can interfere with intracellular antigen processing (1, 9) or directly alter the structural interaction between the epitope of MHC class I complex and the T-cell receptor (TCR) of the corresponding CD8+ T cells (30).
Identification of the factors that influence mutational escape is likely to be critical for guiding human immunodeficiency virus type 1 (HIV-1) vaccine immunogen design, but obtaining insight into the individual parameters that contribute has been difficult due to host and viral heterogeneity. The fact that certain HIV-1 epitopes are presented in the context of multiple closely related HLA class I alleles or allele subtypes provides an opportunity to analyze the relative contributions of HLA class I molecules to TCR diversity and viral sequence evolution, independent of the presented epitope. One such example is the Gag epitope KF11 (KAFSPEVIPMF, p2430-40), which can be presented by HLA-B*5701 (13-15, 24, 25) and HLA-B*5703 (8, 18, 33). In the context of HLA-B*5701, this epitope is associated with a limited tendency for viral escape mutations (24), which might be due to structural viral constraints that preclude the virus from acquiring amino acid substitutions; however, KF11 sequence variants have clearly been described in naturally circulating viral strains, and emerging data from Africa (8, 13) suggest that this epitope is associated with greater sequence variation in geographic regions where the dominant HLA-B*57 subtype is B*5703. Since HLA-B*5701 and -B*5703 differ only by two amino acids, these data raise the possibility that the same viral epitope may be under differential mutational constraints that are influenced by minor differences in HLA amino acid sequences.
In this study, we examined HIV-1-specific immune responses to the immunodominant KF11 Gag epitopes as well as viral sequence mutations within this epitope in a total of 48 persons expressing HLA-B*5701 and 69 expressing HLA-B*5703. Subjects were recruited from cohorts in India, Africa, Europe, and North America and infected with either clade B or clade C viruses. Our data indicate that the observed paucity of KF11 sequence variations in HLA-B*5701-expressing individuals is associated with a highly conserved KF11-specific TCR repertoire that is able to cross-recognize the major naturally occurring KF11 peptide variants. In contrast, individuals expressing HLA-B*5703, which differs from HLA-B*5701 only in two amino acid residues and presents the identical epitope, recruit an entirely different KF11-specific TCR repertoire that frequently fails to cross-recognize specific KF11 escape variants that arise commonly in vivo in clade C-infected HLA-B*5703-expressing individuals. These results therefore provide clear evidence that the restricting HLA allele itself, together with the induced TCR repertoire, can have a dominant influence on the prevention of mutational escape.
MATERIALS AND METHODS
Subjects.
A total of 608 chronically HIV-1 infected individuals (426 clade C infected and 182 clade B infected) were enrolled in this study, all of whom gave informed consent to participate. Subjects were recruited from the Massachusetts General Hospital (Boston, MA), the University of KwaZulu-Natal (Durban, South Africa), Oxford University (Oxford, United Kingdom), the University of Alabama (Birmingham, AL), the All India Institute for Medical Sciences (New Delhi, India) and from the Adolescent Medicine Trials Network (ATN) for HIV/AIDS Intervention (United States) as part of protocol ATN 026.
High-resolution HLA-B*57 typing.
Genomic DNA was extracted from samples typed as HLA-B*57, exons 1 and 2 of the HLA-B alleles were amplified with oligonucleotide primers specific for the HLA-B locus, and the HLA-B PCR products were directly DNA sequenced (10). HLA-B types were determined with Assign software (version 3.2.7; Conexio Genomics, Australia). Samples typed as ambiguous due to polymorphisms outside of exons 2 and 3 were resolved by sequence-specific primer typing (Invitrogen) while HLA-B ambiguities within exons 2 and 3 were resolved by the cloning and DNA sequencing of individual HLA-B*57 clones throughout exons 2 and 3.
Synthetic peptides.
Peptides corresponding to described optimal HIV-1 CD8+ T-cell epitopes and their variants (http://hiv-web.lanl.gov) were synthesized at the Massachusetts General Hospital Peptide Core Facility on an automated peptide synthesizer using F-moc technology.
ELISPOT assay.
Enzyme-linked immunospot (ELISPOT) assays were carried out as described previously (22). Briefly, peripheral blood mononuclear cells (PBMCs) were plated in 96-well polyvinylidene plates that had been precoated with 0.5 μg/ml of an anti-human gamma interferon (IFN-γ) monoclonal antibody (Mabtech, Stockholm, Sweden). PBMCs were added at a concentration of 100,000 cells per well in a volume of 100 μl of RPMI 1640 medium supplemented with fetal calf serum (10%), HEPES buffer (10 mM), l-glutamine (2 mM), and penicillin-streptomycin (50U/ml). Plates were incubated overnight at 37°C in 5% CO2 and developed on the next day as described elsewhere (22). Wells containing PBMCs and medium with phytohemagglutinin or without any peptide were used as positive or negative controls, respectively, and run in triplicate on each plate. To calculate the number of specific T cells, the number of spots in the negative control wells was subtracted from the counted number of spots in each well. Responses were considered positive if there were >50 spot-forming cells/106 PBMCs and at least three times the mean number of spot-forming cells of the three control wells.
Sorting of tetramer-positive and HIV-1-specific CD8+ T-cell populations.
Fresh or frozen PBMC samples were stained with phycoerythrin-labeled B5701/KF11 tetramer, which has been previously shown to bind equally to B5701- and B5703-restricted KF11-specific CD8+ T cells (13) refolded with epitopic HIV-1 peptides (Beckman Coulter, Fullerton, CA) and fluorophore-labeled CD8+ antibodies, followed by decontamination with fixation solution A (1:100 dilution; Caltag). Tetramer-positive CD8+ cells were sorted on a fluorescence-activated cell sorting Aria instrument (BD Biosciences) at 70 lb/in2. Electronic compensation was performed with antibody capture beads (BD Biosciences, San Jose, CA) stained separately with individual antibodies used in the test samples. The purity of sorted cell populations was consistently greater than 98%.
TCR α- and β-chain sequencing.
mRNA was extracted from tetramer-positive CD8+ T cells using an RNeasy mini kit (QIAGEN, Valencia, CA). Anchored reverse transcription-PCR was then performed using a modified version of the SMART (switching mechanism at 5′ end of RNA transcript) procedure and a TCR α- or β-chain constant region 3′ primer to obtain PCR products containing the variable (V) α or β chain in addition to the CDR3 region, the joining (J) α/β region, and the beginning of the constant (C) α/β region. Briefly, reverse transcription was carried out at 42°C for 90 min with primers provided for the 5′ rapid amplification of cDNA ends reaction in a SMART-rapid amplification of cDNA ends PCR kit (BD Biosciences). First- and second-round PCR were then performed using a universal 5′ end primer (5′-CTAATACGACTCACTATAGGGC-3′) and nested gene-specific 3′ end primers annealing to the constant region of the TCR α or β chain (Cα outer, GTCCATAGACCTCATGTCTAGCACAG; Cα inner, ATACACATCAGAATCCTTACTTTG; Cβ outer, 5′-TGTGGCCAGGCACACCAGTGTGGCC-3′; Cβ inner, 5′-GGTGTGGGAGATCTCTGCTTCTGA-3′). PCR conditions were as follows: for the first run, 5 cycles of 95° for 30 s and 72° for 2 min; 5 cycles of 95° for 30 s, 70° for 30 s, and 72°for 2 min; and 25 cycles of 95° for 30 s, 60° for 30 s, and 72° for 1 min. For the second run, the program was 30 cycles of 95° for 30 s, 60° for 30 s, and 72° for 1 min. The PCR product was ligated into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) and used to transform E_s_cherichia coli (Mach 1; Invitrogen). Colonies were selected, amplified by PCR with M13 primers, and sequenced by T7 or T3 primers on an ABI 3100 PRISM automated sequencer. Sequences were edited and aligned using Sequencher (Gene Codes Corp., Ann Arbor, MI) and Se-Al (University of Oxford, Oxford, United Kingdom) and compared to the human TCR genes database (http://imgt.cines.fr:8104/home.html). The TCR Vα/β chain classification system used is that of the international ImMunoGeneTics database (19).
Generation of CTL clones.
Cytotoxic T-cell (CTL) clones were isolated by limiting dilution as previously described (39), using the CD3-specific monoclonal antibody 12F6 as a stimulus for T-cell proliferation. Developing clones were screened for HIV-1-specific CTL activity by a chromium-51 release assay against autologous B lymphoblastoid cell lines (B-LCL) pulsed with the target peptide. HIV-1-specific clones were maintained by stimulation every 14 to 21 days with an anti-CD3 monoclonal antibody and irradiated allogeneic PBMCs.
Chromium release assay.
B-LCL target cells were labeled with chromium-51 by incubation with 50 μCi of Na2CrO4 for 1 h at 37°C in 5% CO2 in the presence or absence of epitopic peptides (20 μg/ml). After three washes, target cells were mixed with effector cells at the indicated effector-to-target concentration. The supernatants were harvested following 4 h of further incubation at 37°C in 5% CO2.
Flow cytometric detection of antigen-induced intracellular IFN-γ.
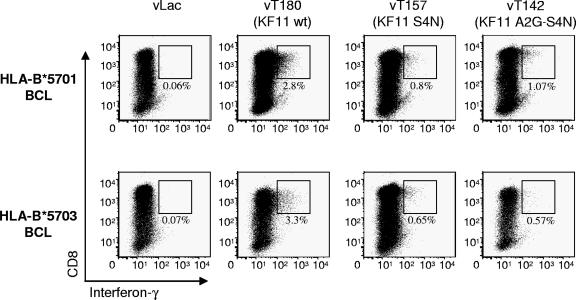
Intracellular cytokine staining assays were performed as described previously (39). Cells were analyzed on a LSR II flow cytometer (Becton Dickinson). Vaccinia virus experiments, utilizing constructs vLac (control), vT142, v180, and vT157 (NIH AIDS Research and Reference Reagent Program), were performed by infecting B-LCL target cells overnight with a multiplicity of infection of 3:1, as described previously (1). Target cells were then washed three times and used in the intracellular cytokine staining assay at an effector-to-target ratio of 1:5.
Proliferation assays.
The proliferative activity of HIV-1-specific CD8+ T cells was measured by a flow cytometric assay after carboxyfluorescein succinimidyl ester (CFSE) labeling (21). Briefly, PBMCs were labeled with 0.25 μM of CFSE (Invitrogen) and placed in 48-well plates in RPMI medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 10 mM HEPES, and 50 IU/ml interleukin-2 (IL-2; provided by the AIDS Research and Reference Reagent Repository, NIH). Antigenic peptides were added at a final concentration of 2μg/ml. On day 6, cells were harvested, washed with phosphate-buffered saline, and stained with allophycocyanin -labeled B57-KF11 tetramers and phycoerythrin-labeled CD8 antibodies. Cells were then washed, fixed in 1% paraformaldehyde, and subjected to flow cytometric analysis on an LSRII flow cytometer.
Sequencing of autologous virus.
Nested PCR for Gag on proviral DNA or plasma viral RNA was performed as previously described (1). PCR fragments were population sequenced to identify regions of sequence variation. All fragments were sequenced bidirectionally on an ABI 3100 PRISM automated sequencer (Applied Biosystems, Foster City, CA). If the height of the secondary peak at a given residue in the chromatogram was more than 25% of the dominant peak, a mixed base was considered present at that position. Sequencher (Gene Codes Corp., Ann Arbor, MI) and MacVector, version 4.1 (Oxford Molecular, Oxford, United Kingdom), were used to edit and align sequences. Clade determination of viruses was performed with phylogenetic trees using MacVector software.
Statistical analysis.
Amino acid variability in TCR CDR3 regions was determined using the Shannon entropy (H) calculation for protein sites as described previously (32) by the formula: H = −Σ pi log2 _p_i where _p_i is the fraction of residues at a site that is amino acid type i. For the 20 amino acids, H can range from 0 (site contains only one amino acid in all sequences) to 4.32 (all amino acids are represented equally at this site). Positions that contained >50% gaps were excluded from analysis. Statistical analysis was based on Fisher's exact tests, chi-square tests, or Mann-Whitney U tests as appropriate; a P value of <0.05 was considered significant.
RESULTS
Frequency of KF11 escape mutations.
The KF11 Gag epitope is a frequent and dominant target for CD8+ T cells in chronic HIV-1 infection in HLA-B*57-expressing individuals infected with clade B or clade C viruses. When presented by HLA-B*5701 in clade B-infected individuals (24), the amino acid sequence of this epitope is highly conserved; however, recent data suggest that this epitope shows considerable sequence variation in clade C viruses circulating in Southern Africa (8, 13). To better define sequence alterations within this epitope in both clade B and clade C infection, we sequenced the viral region encoding the KF11 epitope in a cohort of 426 HIV-1 clade C-infected individuals recruited from South Africa or India and in 182 HIV-1 clade B-infected persons from the United States or the United Kingdom.
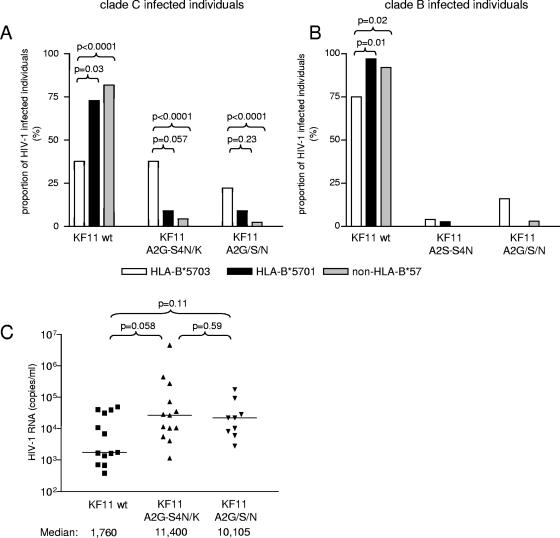
Overall, we found that the KF11 wild-type sequence was found in 328/426 (76.9%) of clade C-infected individuals and in 165/182 (90.6%) of clade B-infected individuals (P < 0.001) (Table 1), indicating a significant interclade difference between the genetic conservation of this epitope. In clade B-infected individuals, there was no significant difference in the frequency of any of KF11 mutations between HLA-B*57-positive and HLA-B*57-negative individuals, and the KF11 wild-type sequence was observed in 54/61 (88.5%) of the tested HLA-B*57-positive study subjects. In contrast, in clade C-infected individuals, the KF11 wild-type sequence was only detected in 44.6% of HLA-B*57-positive patients. Two types of mutations were significantly more frequently encountered in HLA-B*57-expressing clade C-infected individuals compared to the clade C-infected HLA-B*57-negative background population. These involved single amino acid alterations at position 2 [A162(G/S/N), called A2(G/S/N) from here on; 19.6% versus 2.4%; P < 0.0001] and combined amino acid changes at positions 2 and 4 [A162G and S164(N/K), called A2G-S4(N/K) from here on; 32.1% versus 4.3%; P < 0.0001]. High-resolution HLA class I subtyping revealed that within clade C-infected study persons, the frequency of these mutations critically depended on the HLA-B*57 subtype. This was particularly striking for the KF11 A2G-S4(N/K) mutations, which represented the most frequent KF11 epitope variants and occurred in 37.7% of all HLA-B*5703-expressing individuals, compared to 9.1% and 4.3% in HLA-B*5701- and HLA-B*57-negative persons, respectively (Fig. 1A and B). A similar, yet less pronounced, difference was observed for KF11 A2(G/S/N) mutations, which occurred in 22.2% of all HLA-B*5703-expressing persons but in only 9.1% of HLA-B*5701-expressing individuals and 2.4% of HLA-B*57-negative study subjects (Fig. 1A and B). These KF11 sequence mutations may be of clinical significance, as trends towards higher viral loads were found in carriers of the KF11 A2G-S4(N/K) (P = 0.058) or the KF11 A2(G/S/N) mutations (P = 0.11), compared to individuals harboring the KF11 wild-type (wt) sequence (Fig. 1C). Overall, these data indicate that HLA-B*57-associated KF11 escape mutations are more frequent in clade C than in clade B viruses and, within clade C-infected individuals, are preferentially selected in the context of HLA-B*5703.
TABLE 1.
KF11 sequence variations in HIV-1 clade B and C strains in populations of HLA-B*5701- or HLA-B*5703-expressing individuals
| Study subject infection type | KF11 sequence | Sequence name | HLA-B*57 | HLA-B*57 negative | Total | ||
|---|---|---|---|---|---|---|---|
| Total B*57 | HLA-B*5703 | HLA-B*5701 | |||||
| Clade C | KAFSPEVIPMF | wt | 25 | 17 | 8 | 303 | 328 |
| -G-N------- | A2G-S4(N/K) | 15 | 14 | 1 | 16 | 31 | |
| -G-K------- | A2G-S4(N/K) | 3 | 3 | 0 | 0 | 3 | |
| -G--------- | A2G | 2 | 2 | 0 | 3 | 5 | |
| -N--------- | A2N | 7 | 6 | 1 | 4 | 11 | |
| -S--------- | A2S | 2 | 2 | 0 | 2 | 4 | |
| -S----T---- | A2S-V7T | 1 | 1 | 0 | 0 | 1 | |
| ------I---- | V7I | 1 | 0 | 1 | 29 | 30 | |
| ---N----I---- | S4N-V7I | 0 | 0 | 0 | 6 | 6 | |
| ---N------- | S4N | 0 | 0 | 0 | 5 | 5 | |
| R---------- | K1R | 0 | 0 | 0 | 1 | 1 | |
| -----K----- | E6K | 0 | 0 | 0 | 1 | 1 | |
| Total no. of variations | 56 | 45 | 11 | 370 | 426 | ||
| Clade B | KAFSPEVIPMF | wt | 54 | 18 | 36 | 111 | 165 |
| -G-N------- | A2G-S4(N/K) | 0 | 0 | 0 | 0 | 0 | |
| -G-K------- | A2G-S4(N/K) | 0 | 0 | 0 | 0 | 0 | |
| -G--------- | A2G | 1 | 1 | 0 | 1 | 2 | |
| -N--------- | A2N | 1 | 1 | 0 | 2 | 3 | |
| -S--------- | A2S | 1 | 1 | 0 | 1 | 2 | |
| -S-N------- | A2S-S4N | 2 | 1 | 1 | 0 | 2 | |
| -S----I---- | A2S-V7I | 2 | 2 | 0 | 0 | 2 | |
| ------I---- | V7I | 0 | 0 | 0 | 3 | 3 | |
| ---N------- | S4N | 0 | 0 | 0 | 1 | 1 | |
| R---------- | K1R | 0 | 0 | 0 | 1 | 1 | |
| ------A---- | V7A | 0 | 0 | 0 | 1 | 1 | |
| Total no. of variations | 61 | 24 | 37 | 121 | 182 |
FIG. 1.
Frequency of KF11 mutations in HIV-1 clade B- and clade C-infected individuals. Data are from clade C-infected individuals recruited from South Africa and India (A) and from clade B-infected persons from the United States and Europe (B). (C) HIV-1 RNA of clade C-infected HLA-B*5703-expressing individuals with KF11 wt, A2G-S4(N/K), or A2(G/S/N) sequences.
TCR repertoire of KF11-specific CD8+ T cells in HLA-B*5701- or HLA-B*5703-expressing individuals.
The above data indicate that in clade C-infected persons, there is a substantial difference in the frequency of KF11 A2G-S4(N/K) mutations in the context of HLA-B*5701 or -B*5703. These mutations involve a change in a putative TCR contact residue at position 4 (33), and thus the HLA-B*57-subtype-dependent frequency might reflect a divergent TCR usage of KF11-specific CD8+ T cells restricted by these closely related HLA alleles. We therefore analyzed the TCR sequences of sorted KF11-specific CD8+ T-cell populations from HLA-B*5701- and HLA-B*5703-expressing individuals. Persons included in the TCR repertoire analysis were either clade B or clade C infected and had an autologous wild-type KF11 sequence in order to analyze the TCR repertoire to the wild-type sequence and in the absence of detectable escape variants that might result in additional TCR alterations.
As indicated in Table 2, we observed an inter- and intra-individually highly conserved or “public” pattern of KF11-specific TCR β-chain sequences in individuals expressing the HLA-B*5701 subtype, with a consistent dominant or exclusive use of the TCR Vβ chain 19 (Vβ19) in conjunction with a conserved CDR3β motif, C-A-S-(S/T)-X-X-Y-G-Y-T. In contrast, in HLA-B*5703-expressing individuals, we found intra- and inter-individually heterogeneous repertoires of TCR β-chain sequences that were different from the KF11-specific TCR sequences discovered in individuals expressing HLA-B*5701 and were frequently dominated by recruitment of Vβ7.9, Vβ5.6, and Vβ28 (Table 2). In two of the HLA-B*5703-expressing persons, we found a monoclonal KF11-specific TCR β-chain usage that did not involve Vβ19, while in the remaining eight study subjects, the TCR β-chain repertoire was broadly diversified with simultaneous usage of multiple different Vβ chains, each with one or several different CDR3β binding motifs. Analysis of sequence variability at each amino acid residue within the respective CDR3β regions revealed a significantly lower CDR3β Shannon entropy score (less variability) among sequences obtained from those expressing HLA B*5701 compared to those expressing B*5703 (P = 0.03; data not shown).
TABLE 2.
TCR sequences recruited by KF11-specific CD8+ T cells in HLA-B*5701- or HLA-B*5703-expressing individuals
| HLA allele and study subject | TCR sequence (frequency)a | |
|---|---|---|
| β Chain | α Chain | |
| HLA-B*5701 | ||
| AC-121 | Vβ19-CASSGQGYGYA-J1.2 (5/46) | Vα5-CAASGGYQKVT-FGTGTKLQVIP (6/27) |
| Vβ19-CASTGGGYGYT-J1.2 (8/46) | Vα5-CAVSGGYQKVT-FGTGTKLQVIP (10/27) | |
| Vβ19-CASSGQGGYT-J1.2 (22/46) | Vα5-CAESGGYQKVT-J13 (9/27) | |
| Vβ19-CASSGQDYGYT-J1.2 (3/46) | Vα5-CAAYGGYQKVT-FGTGTKLQVIP (2/27) | |
| Vβ19-CASTGSGYGYT-J1.2 (1/46) | ||
| Vβ19-CASSGQEYGYT-J1.2 (1/46) | ||
| Vβ6.1-CASTDSYGYT-J1.2 (6/46) | ||
| Vβ19-CATTDTYGYT-J1.2 (35/36) | Vα5-CAVSGTYQKVT-FGTGTKLQVIP (8/22) | |
| Vβ19-CATTGTYGYT-J1.2 (1/36) | Vα5-CAASGGYQKVT-J13 (14/22) | |
| 013569H | Vβ19-CASSSRQNYGYT-J1.2 (19/32) | ND |
| Vβ19-CASSGQLYGYT-J1.2 (2/32) | ||
| Vβ24 CATSDLGAGTSGTGELF-J2.2 (11/32) | ||
| 010 | Vβ19-CASSATYGYT-J1.2 (39/39) | ND |
| AC-43 | Vβ19-CASTGTAYGYT-J1.2 (24/38) | Vα5-CAGSGGYQKVT-J13 (7/15) |
| Vβ19-CASTGTDYGYT-J1.2 (4/38) | Vα5-CALSGGYQKVT-J13 (4/15) | |
| Vβ19-CASSGQNYGYT-J1.2 (2/38) | Vα5-CAMSGGYQKVT-J13 (2/15) | |
| Vβ19-CASSGQSYGYA-J1.2 (1/38) | Vα5-CAESGGYQKTV-J13 (2/15) | |
| Vβ19-CASSGRNYGYT-J1.2 (3/38) | ||
| Vβ19-CAGTGTAYGYT-J1.2 (1/38) | ||
| Vβ30-CAWTGTNYGYT-J1.2 (3/38) | ||
| AC-167 | Vβ19-CASSREVYGYT-J1.2 (15/38) | Vα5-CAESPGGKLI-J23 (25/25) |
| Vβ19-CASSTSYGYT-J1.2 (4/38) | ||
| Vβ19-CASNSREVYGYT-J1.2 (1/38) | ||
| Vβ7.9-CAAGGQF YGYT-J1.2 (3/38) | ||
| Vβ5.4-CASSLTAPDTDAF-J1.1 (15/38) | Vα5-CAVSGGYQKVT-J13 (15/22) | |
| CTR28 | Vβ19-CASSGGSYGYT-J1.2 (6/32) | Vα5-CAGSGGYQKVT-J13 (2/22) |
| Vβ19-CATTGSYGYT-J1.2 (23/32) | Vα5-CAESGGYQKVT-J13 (3/22) | |
| Vβ19-CASSGQN YGYT-J1.2 (2/32) | Vα5-CAESTGNQFY-J49 (1/22) | |
| Vβ19-CASTGGQYGYT-J1.2 (1/32) | Vα5-CAESTGYQKVT-J13 (1/22) | |
| PRLS 22 | Vβ19-CASTGGS YGYT-J1.2 (18/35) | ND |
| Vβ19-CASSGTSYGYT-J1.2 (15/35) | ||
| Vβ29.1-CSARLRDRGYEQY-J2.7 (2/35) | ||
| HLA-B*5703 | ||
| L8227 | Vβ20.1-CSARPQGQTVALH-J1.6 (5/29) | Vα14-CAMRESISSGSARQLT-J22 (15/22) |
| Vβ20.1-CSARPGQGTVALH-J1.6 (6/29) | Vα14-CAMRESTSSGSARQLT-J22 (1/22) | |
| Vβ20.1-CSARAGTKTYEQY-J2.7 (1/29) | Vα14-CAMRESVSGANNLF-J36 (4/22) | |
| Vβ7.9-CASSPDRARDGYT-J1.2 (5/29) | Vα14-CAMRESVSGVNNLF-FGTGTSLTVI (2/22) | |
| Vβ7.9-CASSSDRARDGYT-J1.2 (1/29) | ||
| Vβ7.6-CASSQLGQSTDTQYF-J1.2 (9/29) | ||
| Vβ5.4-CASSFLLDGYT-J1.2 (2/29) | ||
| 020018 | Vβ7.6-CASSSWEGLDTQY-J2.3 (19/27) | Vα14-CAMRESVSSGSARQLT-J22 (25/25) |
| Vβ5.8-CASSLVSNYGYT-J1.2 (1/27) | ||
| Vβ29.1-CSVEDPDRPSYGYT-J1.2 (5/27) | ||
| Vβ7.9-CASSGLGGYT-J1.2 (1/27) | ||
| Vβ29.1-CSVKGQGDTELF-J2.2 (1/27) | ||
| 120024 | Vβ24.1-CATSDLRGGKF-J2.1 (13/31) | Vα14-CAMRDPRSKII-J3 (11/20) |
| Vβ7.9-CASEDSSDGANYGYT-J1.2 (6/31) | Vα14-CAMRESVSGANNLF-J36 (4/20) | |
| Vβ7.9-CVSEDSSDGANYGYT-J1.2 (1/31) | Vα14-CAMRESVSGNNRLA-J7 (4/20) | |
| Vβ7.9-CASEAGNTIY-J1.3 (2/31) | Vα14-CAMREGVNQGGKLI-J23 (1/20) | |
| Vβ7.9-CASSWSGGQRGYT-J1.2 (9/31) | ||
| 013196G | Vβ28-CASSRQGFT-J1.2 (23/36) | Vα14-CAMRESISSGSARQLT-J22 (26/26) |
| Vβ14-CASSPRDSKETQY-J2.5 (6/36) | ||
| Vβ5.6-CASSLAGGQETQY-J2.5 (7/36) | ||
| CR0464T | Vβ6.6-CASSYSDGNEAF-J1.1 (2/31) | ND |
| Vβ5.6-CASSLAGTQETQY-J2.5 (6/31) | ||
| Vβ5.6-CASSSAGTQETQY-J2.5 (1/31) | ||
| Vβ5.6-CASSLAGTHETQY-J2.5 (1/31) | ||
| Vβ5.6-CVSSLAGTQETQY-J2.5 (9/31) | ||
| Vβ7.9-CASSQRQDSSYNEQF-J2.1 (7/31) | ||
| Vβ28-CASTASYGYT-J1.2 (5/31) | ||
| 014791D | Vβ27-CASSPYAGSEQF-J2.1 (31/31) | ND |
| 2385 | Vβ7.8-CASSLGASISYEQY-J2.7 (1/27) | Vα14-CAMRESVSGYNKLI-J4 (29/29) |
| Vβ15-CATSGEGQGGPYGYT-J1.2 (26/27) | ||
| 100089 | Vβ7.9-CAGSLSNGYT-J1.2 (1/13) | Vα13.1-CAASLNSGGYQKVT-FGTGTKLQVIP (12/27) |
| Vβ9-CASSGTASFDEQF-J2.1 (2/13) | Vα14-CAMRESISSGSARQLT-J22 (9/27) | |
| Vβ9-CASSGTTSFDEQF-J2.1 (2/13) | Vα14-CAMREGVNQGGKLI-J23 (3/27) | |
| Vβ9-CASSVGEGFDIQY-J2.4 (5/13) | Vα14-CAMRDSVAAGNKLT-J17 (1/27) | |
| Vβ28-CASSSLIRGLGNQPQH-J1.5 (1/13) | Vα14-CAMRESVAGANNLF-J36 (2/27) | |
| Vβ25.1-CASSESRMGSQANYGYT-J1.2 (1/13) | ||
| Vβ25.1-CVSSESRMGSQANYGYT-J1.2 (1/13) | ||
| SKI-183-P2 | Vβ5.6- CASSLAGTQETQY-J2.5 (20/25) | ND |
| Vβ5.6-CASSLAGTQETQY-FGPGTRLPVL (1/25) | ||
| Vβ5.1-CASSLAETSADGYT-J1.2 (4/25) | ||
| PS-579-PD | Vβ28-CASSSFLSSGETQY-J2.5 (31/32) | ND |
| Vβ28-CANSSFLSSGETQY-J2.5 (1/32) |
In contrast to the diversity in the TCR β chain in HLA-B*5703-expressing persons, TCR α chains were highly conserved, although different between individuals expressing HLA-B*5701 and -B*5703. In the context of HLA-B*5701, TCR α-chain recruitment involved Vα chain 5 usage in combination with a similar or identical CDR3α amino acid sequence, although interclonotype differences clearly existed on the level of encoding nucleotides (Tables 2 and 3). The TCR α-chain repertoire used by HLA-B*5703-restricted KF11-specific CD8+ T cells was likewise highly conserved but distinct from those expressing HLA B*5701, with an almost exclusive recruitment of Vα chain 14 in combination with a similar CDR3α sequence (Tables 2 and 3).
TABLE 3.
Nucleotide diversity in CDR3α regions encoding identical amino acid sequencesa
| Study subject | Chain and CDR3α amino acid/nucleotide sequence | Frequency |
|---|---|---|
| Group 1 | Vα14-CAMRESISSGSARQLT-J22 | |
| 100089 | TGTGCAATGAGAGAGAGCATTTCTTCTGGTTCTGCAAGGCAACTGACC | 7/9 |
| 100089 | TGTGCAATGAGAGAGTCTATTTCTTCTGGTTCTGCAAGGCAACTGACC | 2/9 |
| L8227 | TGTGCAATGAGAGAGTCTATTTCTTCTGGTTCTGCAAGGCAACTGACC | 15/15 |
| 020018 | TGTGCAATGAGAGAGAGCGTGTCTTCTGGTTCTGCAAGGCAACTGACC | 24/25 |
| 020018 | TGTGCAATGAGAGAGAGCGTGTCTTCTAGTTCTGCAAGGCAACTGACC | 1/25 |
| Group 2 | Vα14-CAMRESVSGANNLF-J36 | |
| 120024 | TGTGCAATGAGAGAGTCCGTAAGTGGGGCAAACAACCTCTTC | 4/4 |
| L8227 | TGTGCAATGAGAGAGAGCGTCTCTGGGGCAAACAACCTCTTC | 4/4 |
| Group 3 | Vα5-CAESGGYQKVT-J13 | |
| AC-121 | TGTGCAGAGAGTGGGGGTTACCAGAAAGTTACC | 9/9 |
| AC-43 | TGTGCAGAATCTGGGGGTTACCAGAAAGTTACC | 2/2 |
| CTR28 | TGTGCAGAGTCTGGGGGTTACCAGAAAGTTACC | 3/3 |
| Group 4 | Vα5-CAGSGGYQKVT-J13 | |
| AC-43 | TGTGCAGGGTCTGGGGGTTACCAGAAAGTTACC | 7/7 |
| CTR28 | TGTGCAGGATCTGGGGGTTACCAGAAAGTTACC | 2/2 |
These data demonstrate that the two-amino-acid difference between HLA alleles presenting the identical epitope can result in completely divergent T-cell repertoires. The KF11 wild-type epitope recruits a more diversified TCR β-chain repertoire in HLA-B*5703-expressing persons, in whom there is a greater propensity for generation of escape variants, whereas the same epitope presented by HLA-B*5701 is associated with a very narrow TCR repertoire and stronger persistence of the original sequence in vivo.
Differential cross-recognition of KF11 peptide variants by CD8+ T cells restricted by HLA-B*5701 and -B*5703.
Prior data have suggested that a diverse TCR repertoire is required to prevent mutational escape (23, 30), and yet the above findings indicate that in this instance a narrow TCR β-chain repertoire was associated with greater conservation of sequence. In order to directly assess the recognition of naturally occurring variants by the recruited TCRs, we stimulated PBMCs from HLA-B*5701- and -B*5703-expressing individuals with a panel of peptides corresponding to the most frequent HLA-B*57-associated KF11 variant sequences detected during natural infection (Table 1) and analyzed responding CD8+ T cells using IFN-γ ELISPOT assays. In all the clade B- or clade C-infected individuals included in this analysis, only the wild-type KF11 sequence was detectable in the corresponding autologous virus in order to define cross-recognition in the absence of detectable circulating escape variants that arise over time. Both single and double amino acid variants of the KF11 epitope were tested, including the most frequently detected mutations that involved changes at positions 2 and 4.
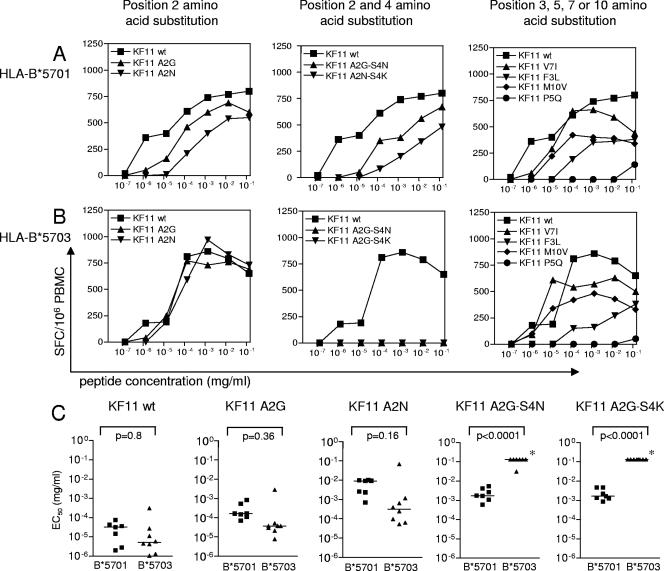
We first examined single amino acid mutations in KF11, using peptides containing the mutation most frequently encountered in vivo, according to the Los Alamos HIV sequence database (http://hiv-web.lanl.gov). Single amino acid substitutions at position 3, 7, and 10 were still recognized in the context of both alleles, although the apparent avidity of recognition decreased between 0.4 log10 and 2.1 log10 (Fig. 2A and B). Position 5 mutations, which almost never occur during natural infection, resulted in complete loss of recognition in the context of both alleles. Amino acid changes at position 2 tended to be better recognized in HLA-B*5703-expressing persons compared to HLA-B*5701-expressing persons; however, this difference failed to reach statistical significance (Fig. 2). There was, however, a marked allele-specific difference in the recognition of KF11 peptide variants with combined amino acid changes at positions 2 and 4, which represent a very frequent form of KF11 epitope variation in circulating viral clade C strains. In the context of HLA B*5701 expression, all of the double mutants were still recognized, albeit the apparent avidity of recognition decreased by an average of 1.9 log10. This cross-reactive pattern of KF11 variant recognition was remarkably consistent in all B*5701-expressing individuals tested (n = 7) (Fig. 2C) and was also observed when a single KF11-specific CD8+ T-cell clone restricted by HLA-B*5701 and expressing the previously described conserved TCR (Vβ19-CASTGTYGYT-J1.2; Vα5-CAASGGYQKVTFGTGTKLQVIP) was tested instead of PBMCs (data not shown). In contrast, there was no recognition of KF11 A2G-S4(N/K) variants in the vast majority of persons expressing HLA B*5703, even at the highest peptide concentration used (Fig. 2C). Importantly, the failure to cross-recognize these variants was not only observed in HLA-B*5703-expressing persons with a mono- or oligoclonal KF11-specific TCR β-chain architecture but also occurred in persons in whom as many as five different TCR clonotypes were recruited (Table 2). Overall, these results indicate that KF11 A2G-S4(N/K) variants can entirely abrogate KF11 recognition in many individuals expressing HLA-B*5703 who recruit a more diverse TCR β-chain repertoire, while recognition of these peptide variants is still possible in HLA-B*5701-expressing persons with a strikingly homogeneous or “public” TCR β-chain recruitment.
FIG. 2.
Cross-recognition of KF11 peptide variants in HLA-B*5701- or -B*5703-expressing individuals. (A and B) KF11 variants with amino acid substitutions at position 2, positions 2 and 4, or positions 3, 5, 7, or 10 were tested for recognition in a representative person expressing HLA-B*5701 (A) or HLA-B*5703 (B). (C) Cross-recognition of KF11 variants with single position 2 or combined position 2 and 4 mutations in individuals expressing HLA-B*5701 (seven of the patients described in Table 2 for whom sufficient PBMC samples were available) or HLA-B*5703 (eight of the patients described in Table 2 for whom sufficient samples were available). *, no IFN-γ secretion was observed at the highest peptide concentration tested.
Presentation of KF11 epitope variants by HLA-B*5701 or -B*5703.
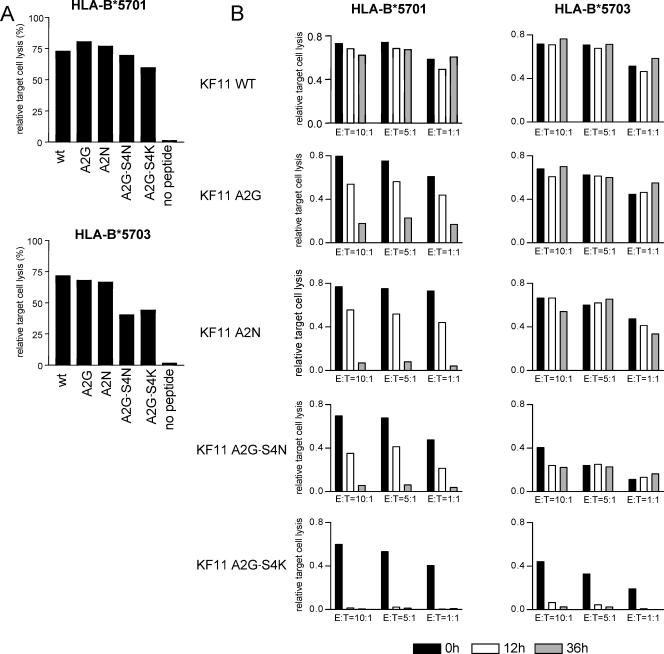
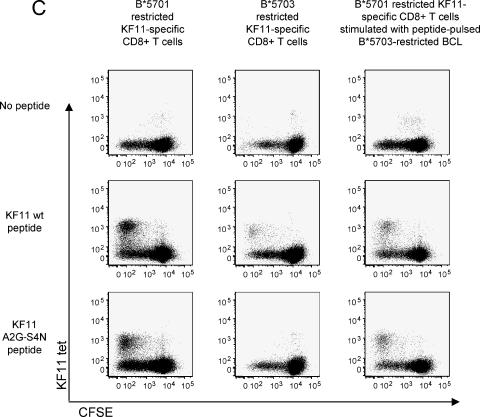
The lack of recognition of doubly mutated KF11 A2G-S4(N/K) peptides in individuals expressing HLA-B*5703 could either be related to an inability of HLA-B*5703 cells to present these peptide variants or to an inability of KF11-specific CD8+ T cells restricted by HLA-B*5703 to recognize these mutated peptides due to limitations in their TCR repertoire. To analyze KF11 peptide variant presentation by HLA-B*5701 or -B*5703 molecules, we pulsed HLA-B*5701- or HLA-B*5703-expressing B-LCL with the KF11 wild-type and variant peptides and subsequently performed chromium-release assays to test whether these peptide variants sensitize targets to lysis by an HLA-B*5701-restricted KF11-specific CD8+ T-cell clone which expressed the previously described typical TCR (Vβ19-CASTGTYGYT-J1.2; Vα5-CAASGGYQKVTFGTGTKLQVIP). As expected, this T-cell clone was able to recognize and lyse HLA-B*5701-expressing B-LCL pulsed with either the KF11 wild-type peptide or KF11 peptide variants with amino acid substitutions at positions 2 and 4 (Fig. 3A). Importantly, despite the fact that KF11 A2G-S4(N/K) variants were frequently completely unrecognized using ELISPOT assays in HLA-B*5703-expressing individuals, we observed that the HLA-B*5701-restricted KF11 CD8+ T-cell clone also killed KF11 A2G-S4(N/K)-labeled HLA-B*5703-restricted B-LCL, although the killing efficiency was lower compared to B*5701-restricted target cells (Fig. 3A). Moreover, HLA-B*5703-expressing B-LCL pulsed with the KF11 A2G-S4(N/K) peptide could still be recognized by this T-cell clone after 12 or 36 h of resting, indicating that the binding affinity of HLA-B*5703 to this peptide is sufficiently strong to prevent fast peptide drop-off (Fig. 3B). A lack of recognition of the KF11 A2G-S4(N/K) variant by HLA-B*5703-restricted KF11-specific CD8+ T cells was also observed when epitope recognition was tested using proliferation assays, while HLA-B*5701-restricted KF11-specific CD8+ T cells were clearly able to proliferate in response to stimulation with this peptide variant (Fig. 3C). Importantly, HLA-B*5701-restricted KF11-specific CD8+ T cells were also able to proliferate in response to stimulation with the A2G-S4(N/K) peptide presented by B*5703-expressing B-LCL, indicating that this variant peptide can be effectively presented by HLA-B*5703 (Fig. 3C, right column). Finally, a KF11-specific CD8+ T-cell clone restricted by HLA-B*5701 was also able to recognize KF11 A2G-S4(N/K) variants that were endogenously processed in HLA-B*5703-expressing B-LCL following infection with Gag-containing vaccinia viruses, although the intensity of antigen-specific IFN-γ secretion was again lower compared to HL-B*5701-expressing B-LCL (Fig. 4). Overall, these data indicate that despite the failure of HLA-B*5703-restricted KF11-specific CD8+ T cells to recognize KF11 variants with combined mutations at positions 2 and 4, both HLA-B*5701 and HLA-B*5703 molecules are able to effectively present these KF11 peptide variants. Moreover, the extremely conserved TCR used by HLA-B*5701-restricted KF11-specific CD8+ T cells can recognize the KF11 peptide and its A2G-S4(N/K) variants in context of both HLA-B*5701 and -B*5703, although it is recruited only in HLA-B*5701-expressing individuals. Therefore, the differential frequency of KF11 A2G-S4(N/K) mutations in HIV-1-infected carriers of HLA-B*5701 and -B*5703 appears to be related to a divergent induction of TCRs and not to the inability of HLA-B*5703 to present the KF11 A2G-S4(N/K) variant.
FIG. 3.
Presentation of KF11 peptide variants on HLA-B*5701- and -B*5703-expressing antigen-presenting cells. (A) Target cell lysis of HLA-B*5701- or -B*5703-expressing B-LCL pulsed with KF11 wild type or variant peptides following exposure to a KF11-specific CD8+ T-cell clone restricted to HLA-B*5701 (Vβ19-CASTGTYGYT-J1.2; Vα5-CAASGGYQKVTFGTGTKLQVIP). (B) Drop-off rate of KF11 peptide variants from HLA-B*5701- or -B*5703-expressing B-LCL. Target cell lysis of HLA-B*5701- or -B*5703-expressing B-LCL by an HLA-B*5701-restricted CD8+ T-cell clone was measured either immediately after peptide pulsing (0 h) or after 12 or 36 h of resting. One representative experiment out of three is shown. (C) Proliferative responses of B*5701- or B*5703-restricted KF11-specific CD8+ T cells to stimulation with KF11 wt or KF11 A2G-S4(N/K) peptides, as measured by CFSE dilution assays. The right column indicates proliferative responses of HLA-B*5701-restricted KF11-specific CD8+ T cells stimulated with KF11 wt peptide or KF11 A2G-S4(N/K) peptide presented by B*5703-restricted B-LCL.
FIG. 4.
Presentation of naturally processed KF11 peptide variants on HLA-B*5701- and -B*5703-expressing B-LCL. IFN-γ secretion by the same HLA-B*5701-restricted KF11-specific CD8+ T-cell clone was analyzed after stimulation with HLA-B*5701- or -B*5703-expressing B-LCL infected with vaccinia virus constructs encoding different KF11 variant sequences. In each case, one representative example with an effector:target ratio of 10:1 is shown.
DISCUSSION
HIV escape mutations associated with CD8+ T-cell-mediated immune pressure are frequently observed in primary and chronic infection; however, some HIV-1 CD8+ T-cell epitopes preserve their original sequence despite strong and long-lasting CD8+ T-cell targeting. An example is the immunodominant and highly conserved KF11 epitope, which shows little tendency for amino acid substitutions in HLA-B*5701-expressing persons, despite specific sequence alterations in KF11 being clearly allowable among circulating viral strains (8, 13). Our data indicate that the extreme sequence conservation of this epitope in the context of HLA-B*5701 is associated with a highly conserved TCR recruitment pattern of CD8+ T cells, which can mediate cross-recognition of the KF11 peptide variants with combined mutations at positions 2 and 4 [A2G-S4(N/K)]. In contrast, in HLA-B*5703-expressing individuals, the identical KF11 peptide is recognized by inter-individually diverse TCR β-chain repertoires that fail to cross-recognize the KF11 A2G-S4(N/K) variant and are associated with a substantially higher frequency of KF11 escape in vivo. Thus, these data suggest that (i) a two-amino-acid difference between restricting HLA-B*57 allele subtypes that present the identical epitope can result in differential and mutually exclusive TCR recruitment patterns, and (ii) TCR usage can be a dominant factor in modulating the evolution of immune escape variants.
The most likely reason for the different TCR recruitment patterns in HLA-B*5701- or HLA-B*5703-restricted KF11-specific CD8+ T cells is that structural features of KF11 epitope presentation substantially depend on the subtype of the restricting HLA-B*57 allele, as recently described for an Epstein-Barr virus-derived epitope in the context of two different HLA-B*35 subtypes (38). Notably, the allelic difference between HLA-B*5701 and -B*5703 consists of as aspartic acid-to-asparagine substitution at amino acid position 114 and a serine-to-tyrosine change at position 116 in the alpha 2 helix. Though both of these amino acids are located in structural proximity of the F pocket, they do not appear to significantly impact the structure of the this anchor pocket; and, indeed, both allele subtypes share similar binding motifs for antigenic peptides, with a preference for F or W as dominant F-pocket anchors residues (34). Thus, instead of impacting predominantly on the selection of antigenic peptides and their HLA binding strength, the amino acid difference between HLA-B*5701 and -B*5703 is more likely to affect the three-dimensional conformation of the central parts of the KF11 epitope, and thereby interactions with the TCR. In the context of HLA-B*5703, crystallographic studies have shown that the KF11 epitope exhibits a prominent hairpin structure with a large solvent-accessible surface area, which appears to be stabilized by a network of direct and indirect polar interactions with a platform of hydrophobic tyrosine residues in the central region of the peptide binding groove (“tyrosine bed”) (33). The three-dimensional structure of KF11 in the context of HLA B*5701 remains to be defined but may be substantially altered by the tyrosine-to-serine change at amino acid 116 in the tyrosine bed. Instead of structural differences between the KF11 epitope presentation in the context of either HLA-B*5701 or -B*5703, the differential TCR recruitment might also be due to a potential unavailability of the conserved Vβ19/Vα5 TCR in individuals expressing HLA-B*5703; however, we clearly observed Vβ19 usage in T cells of other specificities in HLA-B*5703+ persons (data not shown), making such a “hole” in the TCR repertoire of HLA-B*5703+ persons unlikely. Finally, we cannot exclude the possibility that a difference between the surface expression of HLA-B*5701 and -B*5703 contributed to the differential TCR recruitment of B*5701- and B*5703-restricted KF11-specific CD8+ T cells; however, we failed to detect a B*57-subtype-specific difference in HLA surface expression in our study person by flow cytometry (data not shown).
An unexpected finding here is that the more restricted TCR recruitment associated with HLA-B*5701 compared to B*5703 is associated with an enhanced ability to recognize sequence variants that arise in vivo. However, it is important to mention that the less cross-reactive TCRs recruited in the context of HLA-B*5703 were highly variable in terms of their TCR β-chain recruitment but consistently used TCR α chains with a conserved CDR3 binding motif; thus, their inability to cross-recognize epitope variants might be the result of the restricted TCR α-chain recruitment. A restricted TCR pattern has been described for a number of other viral CD8+ T-cell epitopes (3, 17, 27, 35), but the structural basis of the selection of such a conserved TCR repertoire is still unclear. Recently it has been shown that highly conserved or public TCRs are typically recruited for peptide-MHC class I complexes that do not exhibit structurally dominant peptide characteristics with prominent amino acid side chains but, instead, present a rather “featureless” surface area with which the residues of the TCR CDR loops can rarely form antigen-specific interactions (36). In contrast, the other extreme of highly bulged, “overprominent” peptide-MHC (pMHC) structures, which are typically seen with longer (>10 mer) antigenic peptides (7, 37), can similarly be associated with highly restricted TCR usage, most likely due to structural constraints that make such a pMHC structure similarly hard to recognize by the vast majority of available TCRs (26). Yet, in some of the B*5701-expressing individuals, the KF11-specific TCR pattern consisted only partially of public TCRs, while other “private” TCRs were also recruited, suggesting that the dominance of the Vβ19/Vα5 TCR in the context of HLA-B*5701 does not entirely rely on the absence of other suitable TCR clonotypes in the repertoire of available TCRs but that this conserved TCR rather represents a best-fit solution (4). It is noteworthy that HLA-B*5703 is able to present the KF11 wild-type peptide and all the variants to the cross-reactive HLA-B*5701 CD8+ T cells expressing the Vβ19/Vα5 TCR, and yet these same TCRs are not recruited by HLA-B*5703-restricted KF11-specific CD8+ T cells. Moreover, the conserved Vβ19/Vα5 TCR recruited in the context of HLA-B*5701 was able to cross-recognize the KF11 wt peptide and its A2G-S4(N/K) variants not only in the context of HLA-B*5701 but also in the context of HLA-B*5703, although the KF11 A2G-S4(N/K) variant presented by HLA-B*5703 is likely to exhibit a different three-dimensional conformation compared to the KF11 wild-type peptide. Clearly, the structural characteristics that enable the Vβ19/Vα5 TCR to recognize KF11 variants in the context of both HLA-B*57 allele subtypes need to be determined in detail, but it is very possible that this TCR might have an increased flexibility to adapt to altered pMHC ligands by subtle conformational changes of their CDR3 regions (31), while this appears not to be possible for the TCRs recruited in the context of HLA-B*5703 during natural infection. Thus, our data suggest that a highly conserved TCR repertoire is not necessarily easier to render ineffective through viral escape than a more broadly diversified TCR architecture. This observation contrasts with the recently described “public” TCR repertoire of simian immunodeficiency virus TL8 (Tat)-specific CD8+ T cells, for which a single amino acid substitution in the target viral epitope was sufficient to completely abrogate recognition by the entire TCR repertoire, while a broader TCR repertoire recognizing the simian immunodeficiency virus Gag CM9 epitope effectively prevented amino acid mutations in the TCR contact residues of the corresponding cognate peptide (30).
While our data provide an explanation for the higher frequency for KF11 variants with combined amino acid changes at positions 2 and 4 in HLA-B*5703-expressing persons, we also observed a higher frequency of KF11 mutations with single amino acid substitutions at position 2 in these individuals. Position 2 mutations in KF11 have recently been suggested to decrease proper intracellular processing (20), and although the KF11 A2G-S4(N/K) variant could be endogenously processed and presented on the cell surface of both HLA-B*5701- and -B*5703-expressing cells, it is possible that processing of KF11 A2(G/S/N) variants is more prominently inhibited in the context of HLA-B*5703. Moreover, the higher frequency of mutations at position 2 in the context of HLA-B*5703 makes it tempting to speculate that these amino acid changes facilitate a subsequent amino acid change at position 4, which might otherwise be more difficult to tolerate for the virus. Position 4 amino acid changes in KF11 are, indeed, rarely observed during natural infection in the absence of simultaneous amino acid substitutions at position 2. In addition, KF11 spans the boundary of structurally important regions of the p24 capsid (12) and might therefore be particularly susceptible to fitness costs associated with amino acid changes. Finally, we observed that KF11 escape mutations were substantially more frequent in clade C-infected individuals compared to clade B-infected persons, in particular with regard to the mutations involving combined amino acid substitutions at positions 2 and 4. Therefore, it appears that clade C viruses have a better capacity to structurally tolerate these KF11 mutations, potentially as a result from their circulation in areas with higher HLA-B*5703 allele frequencies (11).
In summary, our data show that minor differences between the amino acid sequence of the restricting HLA-B*5701 or -B*5703 allele subtypes resulted in an entirely different and mutually exclusive TCR repertoire of KF11-specific CD8+ T cells. Interestingly, the highly restricted KF11-specific TCR pattern observed in the context of HLA-B*5701 showed a better capacity for cross-recognition of epitope variants that arise in vivo and was associated with a lower frequency of these escape mutations in vivo than the more diversified TCR patterns observed in the context of HLA-B*5703. These data contribute to the understanding of how TCR recognition motifs can impact the selection of viral escape variants and are important considerations for the design of vaccines eliciting HIV-1-specific CD8+ T-cell responses with broadly cross-reactive TCRs.
Acknowledgments
This study was supported by grants from the National Institutes of Health (to X.G.Y., P.A.G., P.J.R.G., and B.D.W.), the Harvard Center for AIDS Research (to X.G.Y.), the Doris Duke Charitable Foundation (to X.G.Y., P.J.R.G., M.A., and B.D.W.), and the Howard Hughes Medical Institute (to B.D.W.). M.X.L. acknowledges the Laboratory Directed Research and Development Program at Los Alamos National Laboratory under the UC/Department of Energy Contract No. W-7405-ENG-36. ATN 026 was a study supported by the Adolescent Medicine Trials Network for HIV/AIDS Interventions (ATN) (www.atnonline.org) which is funded by U01 HD40533 with primary support from the NICHD.
ATN sites contributing to this effort were Children's Hospital of Los Angeles (M. Belzer), Mt. Sinai Medical Center (L. Levin), University of Maryland (L. Peralta), University of Miami (L. Friedman), and Tulane Medical Center (S. E. Abdalian).
Footnotes
▿
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78**:**7069-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407**:**386-390. [DOI] [PubMed] [Google Scholar]
- 3.Argaet, V. P., C. W. Schmidt, S. R. Burrows, S. L. Silins, M. G. Kurilla, D. L. Doolan, A. Suhrbier, D. J. Moss, E. Kieff, T. B. Sculley, and I. S. Misko. 1994. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 180**:**2335-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankovich, A. J., and K. C. Garcia. 2003. Not just any T cell receptor will do. Immunity 18**:**7-11. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415**:**335-339. [DOI] [PubMed] [Google Scholar]
- 6.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3**:**205-211. [DOI] [PubMed] [Google Scholar]
- 7.Burrows, S. R., J. Rossjohn, and J. McCluskey. 2006. Have we cut ourselves too short in mapping CTL epitopes? Trends Immunol. 27**:**11-16. [DOI] [PubMed] [Google Scholar]
- 8.Currier, J. R., M. E. Harris, J. H. Cox, F. E. McCutchan, D. L. Birx, S. Maayan, and G. Ferrari. 2005. Immunodominance and cross-reactivity of B5703-restricted CD8 T lymphocytes from HIV type 1 subtype C-infected Ethiopians. AIDS Res. Hum. Retrovir. 21**:**239-245. [DOI] [PubMed] [Google Scholar]
- 9.Draenert, R., S. Le Gall, K. J. Pfafferott, A. J. Leslie, P. Chetty, C. Brander, E. C. Holmes, S. C. Chang, M. E. Feeney, M. M. Addo, L. Ruiz, D. Ramduth, P. Jeena, M. Altfeld, S. Thomas, Y. Tang, C. L. Verrill, C. Dixon, J. G. Prado, P. Kiepiela, J. Martinez-Picado, B. D. Walker, and P. J. Goulder. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199**:**905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellexson-Turner, M. E., D. A. Sidebottom, S. Turner, T. Bennett, and W. H. Hildebrand. 2001. Precision genotyping of HLA-A, -B, and -C via direct DNA sequencing, p. 129-142. In Joyce C. Solheim (ed.), Antigen processing and presentation protocols. Humana Press, Totowa, NJ.
- 11.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, M. E. Feeney, M. M. Addo, M. Lichterfeld, M. P. Lahaie, E. Pae, A. G. Wurcel, T. Roach, M. A. St John, M. Altfeld, F. M. Marincola, C. Moore, S. Mallal, M. Carrington, D. Heckerman, T. M. Allen, J. I. Mullins, B. T. Korber, P. J. Goulder, B. D. Walker, and C. Brander. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7**:**173-178. [DOI] [PubMed] [Google Scholar]
- 12.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87**:**1285-1294. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie, G. M., R. Kaul, T. Dong, H. B. Yang, T. Rostron, J. J. Bwayo, P. Kiama, T. Peto, F. A. Plummer, A. J. McMichael, and S. L. Rowland-Jones. 2002. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16**:**961-972. [DOI] [PubMed] [Google Scholar]
- 14.Goulder, P. J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retrovir. 12**:**1691-1698. [DOI] [PubMed] [Google Scholar]
- 15.Goulder, P. J., Y. Tang, S. I. Pelton, and B. D. Walker. 2000. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J. Virol. 74**:**5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, B. H. Hahn, M. A. Nowak, G. M. Shaw, and P. Borrow. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200**:**1243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedzierska, K., S. J. Turner, and P. C. Doherty. 2004. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc. Natl. Acad. Sci. USA 101**:**4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432**:**769-775. [DOI] [PubMed] [Google Scholar]
- 19.Lefranc, M. P., C. Pommie, M. Ruiz, V. Giudicelli, E. Foulquier, L. Truong, V. Thouvenin-Contet, and G. Lefranc. 2003. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 27**:**55-77. [DOI] [PubMed] [Google Scholar]
- 20.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, S. Le Gall, G. Luzzi, A. Edwards, C. Brander, A. K. Sewell, S. Moore, J. Mullins, C. Moore, S. Mallal, N. Bhardwaj, K. Yusim, R. Phillips, P. Klenerman, B. Korber, P. Kiepiela, B. Walker, and P. Goulder. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201**:**891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichterfeld, M., D. E. Kaufmann, X. G. Yu, S. K. Mui, M. M. Addo, M. N. Johnston, D. Cohen, G. K. Robbins, E. Pae, G. Alter, A. Wurcel, D. Stone, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2004. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 200**:**701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichterfeld, M., X. G. Yu, D. Cohen, M. M. Addo, J. Malenfant, B. Perkins, E. Pae, M. N. Johnston, D. Strick, T. M. Allen, E. S. Rosenberg, B. Korber, B. D. Walker, and M. Altfeld. 2004. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. AIDS 18**:**1383-1392. [DOI] [PubMed] [Google Scholar]
- 23.Meyer-Olson, D., N. H. Shoukry, K. W. Brady, H. Kim, D. P. Olson, K. Hartman, A. K. Shintani, C. M. Walker, and S. A. Kalams. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J. Exp. Med. 200**:**307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migueles, S. A., A. C. Laborico, H. Imamichi, W. L. Shupert, C. Royce, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, C. W. Hallahan, and M. Connors. 2003. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J. Virol. 77**:**6889-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97**:**2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miles, J. J., D. Elhassen, N. A. Borg, S. L. Silins, F. E. Tynan, J. M. Burrows, A. W. Purcell, L. Kjer-Nielsen, J. Rossjohn, S. R. Burrows, and J. McCluskey. 2005. CTL recognition of a bulged viral peptide involves biased TCR selection. J. Immunol. 175**:**3826-3834. [DOI] [PubMed] [Google Scholar]
- 27.Moss, P. A., R. J. Moots, W. M. Rosenberg, S. J. Rowland-Jones, H. C. Bodmer, A. J. McMichael, and J. I. Bell. 1991. Extensive conservation of alpha and beta chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc. Natl. Acad. Sci. USA 88**:**8987-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8**:**493-499. [DOI] [PubMed] [Google Scholar]
- 29.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94**:**1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price, D. A., S. M. West, M. R. Betts, L. E. Ruff, J. M. Brenchley, D. R. Ambrozak, Y. Edghill-Smith, M. J. Kuroda, D. Bogdan, K. Kunstman, N. L. Letvin, G. Franchini, S. M. Wolinsky, R. A. Koup, and D. C. Douek. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21**:**793-803. [DOI] [PubMed] [Google Scholar]
- 31.Rudolph, M. G., and I. A. Wilson. 2002. The specificity of TCR/pMHC interaction. Curr. Opin. Immunol. 14**:**52-65. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, J. J., C. Y. Lee, S. Ibrahim, P. Watts, M. Shlomchik, M. Weigert, and S. Litwin. 1997. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol. Immunol. 34**:**1067-1082. [DOI] [PubMed] [Google Scholar]
- 33.Stewart-Jones, G. B., G. Gillespie, I. M. Overton, R. Kaul, P. Roche, A. J. McMichael, S. Rowland-Jones, and E. Y. Jones. 2005. Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression. J. Immunol. 175**:**2459-2468. [DOI] [PubMed] [Google Scholar]
- 34.Stewart-Jones, G. B., A. J. McMichael, J. I. Bell, D. I. Stuart, and E. Y. Jones. 2003. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 4**:**657-663. [DOI] [PubMed] [Google Scholar]
- 35.Trautmann, L., M. Rimbert, K. Echasserieau, X. Saulquin, B. Neveu, J. Dechanet, V. Cerundolo, and M. Bonneville. 2005. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 175**:**6123-6132. [DOI] [PubMed] [Google Scholar]
- 36.Turner, S. J., K. Kedzierska, H. Komodromou, N. L. La Gruta, M. A. Dunstone, A. I. Webb, R. Webby, H. Walden, W. Xie, J. McCluskey, A. W. Purcell, J. Rossjohn, and P. C. Doherty. 2005. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat. Immunol. 6**:**382-389. [DOI] [PubMed] [Google Scholar]
- 37.Tynan, F. E., S. R. Burrows, A. M. Buckle, C. S. Clements, N. A. Borg, J. J. Miles, T. Beddoe, J. C. Whisstock, M. C. Wilce, S. L. Silins, J. M. Burrows, L. Kjer-Nielsen, L. Kostenko, A. W. Purcell, J. McCluskey, and J. Rossjohn. 2005. T cell receptor recognition of a “super-bulged” major histocompatibility complex class I-bound peptide. Nat. Immunol. 6**:**1114-1122. [DOI] [PubMed] [Google Scholar]
- 38.Tynan, F. E., D. Elhassen, A. W. Purcell, J. M. Burrows, N. A. Borg, J. J. Miles, N. A. Williamson, K. J. Green, J. Tellam, L. Kjer-Nielsen, J. McCluskey, J. Rossjohn, and S. R. Burrows. 2005. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J. Exp. Med. 202**:**1249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, X. G., H. Shang, M. M. Addo, R. L. Eldridge, M. N. Phillips, M. E. Feeney, D. Strick, C. Brander, P. J. Goulder, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2002. Important contribution of p15 Gag-specific responses to the total Gag-specific CTL responses. AIDS 16**:**321-328. [DOI] [PubMed] [Google Scholar]