Two-Way Antigenic Cross-Reactivity between Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Group 1 Animal CoVs Is Mediated through an Antigenic Site in the N-Terminal Region of the SARS-CoV Nucleoprotein (original) (raw)
Abstract
In 2002, severe acute respiratory syndrome-associated coronavirus (SARS-CoV) emerged in humans, causing a global epidemic. By phylogenetic analysis, SARS-CoV is distinct from known CoVs and most closely related to group 2 CoVs. However, no antigenic cross-reactivity between SARS-CoV and known CoVs was conclusively and consistently demonstrated except for group 1 animal CoVs. We analyzed this cross-reactivity by an enzyme-linked immunosorbent assay (ELISA) and Western blot analysis using specific antisera to animal CoVs and SARS-CoV and SARS patient convalescent-phase or negative sera. Moderate two-way cross-reactivity between SARS-CoV and porcine CoVs (transmissible gastroenteritis CoV [TGEV] and porcine respiratory CoV [PRCV]) was mediated through the N but not the spike protein, whereas weaker cross-reactivity occurred with feline (feline infectious peritonitis virus) and canine CoVs. Using _Escherichia coli_-expressed recombinant SARS-CoV N protein and fragments, the cross-reactive region was localized between amino acids (aa) 120 to 208. The N-protein fragments comprising aa 360 to 412 and aa 1 to 213 reacted specifically with SARS convalescent-phase sera but not with negative human sera in ELISA; the fragment comprising aa 1 to 213 cross-reacted with antisera to animal CoVs, whereas the fragment comprising aa 360 to 412 did not cross-react and could be a potential candidate for SARS diagnosis. Particularly noteworthy, a single substitution at aa 120 of PRCV N protein diminished the cross-reactivity. We also demonstrated that the cross-reactivity is not universal for all group 1 CoVs, because HCoV-NL63 did not cross-react with SARS-CoV. One-way cross-reactivity of HCoV-NL63 with group 1 CoVs was localized to aa 1 to 39 and at least one other antigenic site in the N-protein C terminus, differing from the cross-reactive region identified in SARS-CoV N protein. The observed cross-reactivity is not a consequence of a higher level of amino acid identity between SARS-CoV and porcine CoV nucleoproteins, because sequence comparisons indicated that SARS-CoV N protein has amino acid identity similar to that of infectious bronchitis virus N protein and shares a higher level of identity with bovine CoV N protein within the cross-reactive region. The TGEV and SARS-CoV N proteins are RNA chaperons with long disordered regions. We speculate that during natural infection, antibodies target similar short antigenic sites within the N proteins of SARS-CoV and porcine group 1 CoVs that are exposed to an immune response. Identification of the cross-reactive and non-cross-reactive N-protein regions allows development of SARS-CoV-specific antibody assays for screening animal and human sera.
Severe acute respiratory syndrome-associated coronavirus (SARS-CoV), which caused a global epidemic in 2002-2003 was identified as a new group 2b member of the Coronavirus genus (6, 21, 34), most closely related to new animal CoVs from civet cats (11, 50, 54) and bats (23, 26). Extensive characterization of the SARS-CoV indicated that it shares common features of virion structure and genome organization with the other members of the CoV genus but also that it possesses some unique traits and is distinct from known human CoVs (HCoVs). Isolation of SARS-like CoVs from Himalayan palm civets, raccoon dogs (11, 50, 54), and recently bats (23, 26) supports the idea that SARS-CoV is a recently emerged zoonosis (39, 40). Advances in understanding the molecular mechanisms of SARS-CoV replication (27), the efficient experimental transmission of the virus to a number of animals (33, 35, 37, 38, 43, 53), and the recognized propensity of CoVs for mutation/recombination with an extended host range have provided further support for the proposed animal origin of SARS-CoV (39). Phylogenetic analysis revealed that SARS-CoV does not belong to any of the three previously known phylogenetic groups of CoVs and forms a new group (2b) in the Coronavirus genus (20). Based on its distinct antigenic characteristics, SARS-CoV could also not be classified within any of the existing CoV antigenic groups. Although many studies of the genetic characterization of SARS-CoV revealed that SARS-CoV was most closely related to group 2 CoVs (9, 20, 41, 59), others reported that SARS-CoV is more closely related to group 1 or 3 CoVs (32, 55) or represents a mosaic genome structure (36, 42). In addition, researchers demonstrated antigenic cross-reactivity between SARS-CoV and animal group 1 but not group 2a CoVs (21, 44). However, the basis for this cross-reactivity at the level of the N protein was not examined. Additionally, some contradictory data on SARS-CoV and HCoV OC43 and 229E one-way cross-reactivity have been published (1a, 3), but the data were not consistent for all patients, nor was it clear which proteins and antigenic determinants were involved, although it was suggested that this cross-reactivity was not due to the nucleocapsid proteins but to other viral components (3). Based on these discrepancies, the uncertain origin and the segregated position of SARS-CoV among other CoVs, additional information is needed to comprehensively assess both the antigenic and genetic relationship between SARS-CoV and other CoVs and thus to understand SARS-CoV origin and evolution.
In this study, we performed a comprehensive analysis of the antigenic cross-reactivity between SARS-CoV and other animal CoVs (groups 1, 2, and 3) and a group 1 HCoV (NL63). Because cross-reactivity between SARS-CoV and polyclonal antisera to antigenic group 1 animal CoVs has been previously attributed to the nucleocapsid protein (N protein) by others (44) and in our preliminary studies (H. S. Nagesha, M. G. Han, L. J. Saif, T. G. Ksiazek, L. J. Anderson, and L. Haynes, presented at the 23rd annual meeting of the American Society for Virology, McGill University, Montreal, Quebec, Canada, 2004), our focus was to delineate the nature of this cross-reactivity, to define the region of the N protein involved, and to map the boundaries of the cross-reactive antigenic sites. We further demonstrated that the observed two-way antigenic cross-reactivity is mediated only by the N protein and not the spike (S) protein, and we localized the potential cross-reactive antigenic sites in the N protein. The analyses performed allowed us to ascertain the range of group 1 CoVs (animal and human) cross-reactive with SARS-CoV and to demonstrate that the observed cross-reactivity is not universal among group 1 CoVs, because HCoV-NL63 did not cross-react with SARS-CoV in our study. Full-length SARS-CoV N-protein-based serologic assays were reported to produce false positive results with sera from healthy humans (29, 30, 51). Therefore, identification and characterization of non-cross-reactive fragments of the SARS-CoV N protein will allow development of serologic tests for screening of SARS-CoV-specific antibodies (Abs) in human patients and in animal reservoirs to exclude false positives due to the cross-reactivity with group 1 human or animal CoVs.
MATERIALS AND METHODS
Reference viruses, antisera, and human sera used in the experiments.
The following panel of animal CoVs, maintained in L. J. Saif's laboratory, was used in the present study: transmissible gastroenteritis viruses (TGEVs; virulent Miller-M6 and attenuated Purdue-P115 strains), a porcine respiratory CoV (PRCV; strain ISU1) that represents an S-gene deletion mutant of TGEV, canine CoV (CCoV; strain UCD1), feline infectious peritonitis virus (FIPV; type 2, strain 79-1146), bovine enteric CoVs (BCoVs; Mebus and DB2 strains), and bovine respiratory CoV (strain 440). The infectious bronchitis virus (IBV; strain Massachusetts) and turkey CoV (TCoV; strain IN) were provided by Y. M. Saif. The group 1 HCoV-NL63 strain was kindly provided by Lia van der Hoek and Ben Berkhout from the Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands (48). The detergent-extracted, inactivated SARS-CoV-Urbani strain (SPR786) and uninfected Vero E6 antigen (SPR527) were provided by Thomas Ksiazek (Centers for Disease Control and Prevention [CDC], Atlanta, GA). The following cell lines used for growing these viruses were mock infected and used as negative controls: ST, CrFK, A72, LLCK12, and HRT-18. Allantoic fluids from uninfected, specific-pathogen-free chicken embryos or uninfected turkey gut suspension were used as negative controls for the group 3 CoVs.
Hyperimmune antisera against animal and avian CoVs were produced in gnotobiotic pigs and guinea pigs and used in the present study. The anti-TGEV-Miller (strain M6) serum (M2), anti-TGEV-Purdue (strain P115) serum (MM973), and anti-PRCV (strain ISU1) serum (PP12) were produced in gnotobiotic pigs as combined postinfection (oral/intranasal [i.n.] exposure to live CoV) and hyperimmune (four parenteral injections of inactivated CoV) antisera. The anti-CCoV (strain UCD1), anti-FIPV (strain 79-1146), anti-BCoV (enteric strain Mebus), anti-HCoV-NL63, and anti-IBV (strain Massachusetts) sera were produced by parenteral hyperimmunization of guinea pigs with the inactivated CoVs. The last antiserum was provided by Y. M. Saif. An IBV-Massachusetts antiserum was also produced in chickens (by intratracheal inoculation with 105 50% egg infective doses of IBV-Massachusetts, followed by intravenous inoculation with at least 105 egg infective doses of IBV-Massachusetts) and kindly provided by M. Jackwood (University of Georgia). Also included was antiserum to enteric BCoV (BO12722), which was produced in a gnotobiotic calf by oral/i.n. exposure to a live, virulent BCoV-DB2 strain, followed by four subsequent intraperitoneal injections with inactivated cell culture-adapted BCoV-Mebus. Sera from mock-inoculated gnotobiotic pigs, gnotobiotic calves, chickens, and guinea pigs were used as negative controls in all assays. Mouse polyclonal SARS-CoV-Urbani antiserum was produced in BALB/c mice immunized with inactivated SARS-CoV lysate (Urbani strain) in Titermax adjuvant, followed by a boost with 18 μg of recombinant SARS-CoV nucleocapsid protein in phosphate-buffered saline (PBS), and provided by Lia M. Haynes (CDC, Atlanta, GA). Normal mouse serum (Sigma-Aldrich, St. Louis, MO) was used as a negative control serum. The hyperimmune rabbit antiserum to the SARS-CoV S protein (generated by four DNA immunizations by a gene gun with codon-optimized DNA vaccine expressing the full-length S protein of the SARS-CoV-Urbani strain) and negative rabbit sera were provided by Shan Lu (Medical School, University of Massachusetts, Worcester, MA).
A panel of human sera was used for assessment of antigenic cross-reactivity by Western blot analysis and an enzyme-linked immunosorbent assay (ELISA). The panel included six convalescent-phase serum samples (CoV samples 3 to 8) from the WHO-confirmed SARS-patients (collected 18 [CoV samples 3 and 4] and 50 [CoV samples 5 to 8] days after disease onset) provided by Thomas Ksiazek (CDC, Atlanta, GA) and 15 serum samples (CoV samples 10 to 24) obtained from healthy donors and provided by Matthias Niedrig (Robert Koch Institute, Berlin, Germany), which qualified as SARS negative in the first PCR in the external quality assurance test. CoV sample 9 was a human serum sample obtained from an adult in the United States that we demonstrated was negative for Abs to irradiated SARS-CoV in ELISA and the SARS-CoV N and S proteins in ELISA and Western blot analysis. All serum samples were stored at −70°C until use.
MAbs.
Two sets of monoclonal Abs (MAbs) were used to analyze cross-reactivity between SARS-CoV and group 1 CoVs: anti-TGEV-M6 N-protein MAbs (25H7, 14E3, 14G9) previously produced and characterized in L. J. Saif's laboratory (52) and anti-SARS-CoV-Urbani N-protein MAbs (SA 46-4 and SA 87-A1) kindly provided by Ying Fang (South Dakota State University, Brookings, SD) (7). The TGEV-M6 N MAbs recognized three distinct antigenic sites on the TGEV N protein: N1 (25H7), flanked by amino acids (aa) 1 to 120; N2 (14E3), imbedded between aa 255 and 383; and N3 (14G9), spanning from aa 1 to aa 205 (52). A MAb to SARS-CoV-Urbani spike, 341C (46), was kindly provided by Lia M. Haynes (CDC, Atlanta, GA). Negative mouse ascites SP2/0 was used as a negative control in assays with the different MAbs.
RNA extraction.
The RNA was extracted from a cell culture supernatant of CoVs, including TGEV-M6, TGEV-P115, PRCV-ISU1, HCoV-NL63, and the irradiation-inactivated, cell-cultured SARS-CoV-Urbani strain, using an RNeasy mini kit according to the manufacturer's instructions (Qiagen Inc., Valencia, CA).
Cloning of the N-protein genes (full-length and fragments).
Twelve oligonucleotide-containing, engineered restriction enzyme sites (EcoRI and SalI) for amplification and cloning of eight truncated and one full-length variant of the SARS-CoV N protein were designed to facilitate cloning in a prokaryotic (vector-pET23b) (Novagen, EMD Biosciences, San Diego, CA) expression system (Table 1). Using these primers, the nine fragments (including the full-length fragment) were amplified, digested with the EcoRI and SalI restriction enzymes, gel extracted, and ligated into the pET23b plasmid vector. Two more oligonucleotide pairs were designed for the full-length cloning of N proteins of group 1 CoVs, including two TGEV strains (Miller-M6 and Purdue-P115), the PRCV ISU1 strain, and the HCoV-NL63 strain (HCoV-NL63); three additional oligonucleotides were designed for HCoV-NL63 N-protein fragment cloning (Table 1). After reverse transcription-PCR, amplified fragments with these primers were digested with the EcoRI and NotI restriction enzymes, purified, and ligated into the pET23b plasmid. After transformation of the constructs into Escherichia coli strain Top Ten (Invitrogen, Carlsbad, CA), selected colonies were grown and then tested by PCR and restriction analysis. Positive clones carrying correct insertions in the pET23b plasmid vector were sequenced and used for recombinant protein expression in the E. coli host strain for expression, BL-21(DE3) (Novagen, EMD Biosciences).
TABLE 1.
Oligonucleotides used for recombinant protein expression, lengths of coding sequences, and predicted sizes of recombinant proteins
| Coronavirus | Fragment (nt) | Oligonucleotide sequencesa | Fragment length (nt) | Predicted recombinant protein mass (kDa) |
|---|---|---|---|---|
| TGEV (M6 and P115) and PRCV-ISU1 | 1-383 | F, TGACGAATTCAATGGCCAACCAGGGACAAC | 1,149 | 43 |
| R, GTCAGCGGCCGCGTTCGTTACCTCRTCAAT | ||||
| HCoV-NL63 | 1-378 | F, ATGCGAATTCGATGGCTAGTGTAAATTGG | 1,134 | 44 |
| R, GCATGCGGCCGCATGCAAAACCTCGTTG | ||||
| 1-183 | F, ATGCGAATTCGATGGCTAGTGTAAATTGG | 549 | 21 | |
| R, GCATGCGGCCGCGTTAGAATCAGAACGA | ||||
| 182-378 | F, ATGCGAATTCGTCTAACCAGTCTTCTTC | 588 | 23 | |
| R, GCATGCGGCCGCATGCAAAACCTCGTTG | ||||
| 39-183 | F, ATGCGAATTCGAAGGACCTTAAATTCAG | 432 | 16.8 | |
| R, GCATGCGGCCGCGTTAGAATCAGAACGA | ||||
| SARS-CoV | 1-72 | F, TGACGAATTCAATGTCTGATAATGGACC | 216 | 7.8 |
| R, GTGAGTCGACGCCCTGGCCTCGAGGG | ||||
| 120-208 | F, TGACGAATTCAGCTTCACTTCCCTACGG | 264 | 9.7 | |
| R, GTGAGTCGACAGGAGAATTTCCCCTAC | ||||
| 212-349 | F, TGACGAATTCAGCTAGCGGAGGTGGTGA | 411 | 15 | |
| R, GTGAGTCGACGTCTTTGAATTGTGGATC | ||||
| 360-412 | F, TGACGAATTCAGCATACAAAACATTCCC | 156 | 5.7 | |
| R, GTGAGTCGACCATGGAATTTTGAAGTTG | ||||
| 70-213 | F, TGACGAATTCAGGCCAGGGCGTTCCAATC | 429 | 15.7 | |
| R, GTGAGTCGACGCTAGCCATTCGAGCAGG | ||||
| 337-422 | F, TGACGAATTCAGCCATTAAATTGGATGA | 255 | 9.4 | |
| R, GTGAGTCGACTGCCTGAGTTGAATCAG | ||||
| 1-213 | F, TGACGAATTCAATGTCTGATAATGGACC | 639 | 23 | |
| R, GTGAGTCGACGCTAGCCATTCGAGCAGG | ||||
| 212-422 | F, TGACGAATTCAGCTAGCGGAGGTGGTGA | 630 | 23 | |
| R, GTGAGTCGACTGCCTGAGTTGAATCAG | ||||
| 1-422 | F, TGACGAATTCAATGTCTGATAATGGACC | 1,266 | 49 | |
| R, GTGAGTCGACTGCCTGAGTTGAATCAG |
Expression of the recombinant TGEV, PRCV, HCoV-NL63, and SARS-CoV N proteins in the prokaryotic pET system (Novagen, EMD Biosciences).
The recombinant pET23b plasmids were separately transformed into E. coli strain BL-21(DE3) (Novagen, EMD Biosciences) for inducible protein expression. Overnight cultures of freshly transformed E. coli BL-21(DE3) were diluted 1:25 (the final volume for each was 50 ml) and incubated with agitation at 37°C until the optical density at 600 nm reached 0.6 to 0.8 (∼3 h). The BL-21(DE3) cells produced T7 polymerase, and the expression of the target proteins was driven by a T7 RNA polymerase promoter after addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) according to the manufacturer's protocol (Novagen, EMD Biosciences). After induction, cells were incubated for 3 more hours, and then cell suspensions were centrifuged at 2,500 × g to pellet cells, washed once with cold 1× PBS, and centrifuged (2,500 × g) again. Pellets were then frozen at −20°C.
Purification of recombinant proteins.
Cloning in the vector of choice (pET23b) allowed expression of recombinant proteins fused with a six-His tag on the C-terminal end. HIS-Select nickel affinity gel (Sigma-Aldrich) was used for extraction of recombinant proteins, following the two protocols: one for native and one for denaturing conditions. All recombinant N proteins (rNs) expressed in E. coli BL21(DE3) formed mainly inclusion bodies and could be purified from the soluble fractions in limited quantities only. There was no difference in the antigenicities of the recombinant products purified by any of the conditions noted; thus, for all purifications after initial comparison, we used only denaturing conditions because they give rise to higher protein yields.
Denaturing conditions (batch purification method).
The E. coli BL-21(DE3) cell pellets were suspended in 5 ml of 6 M guanidine hydrochloride (Gu-HCl) buffer containing 5 mM dithiothreitol (DTT) and lysed by sonication with an ultrasonic processor (Vibra cells; Sonics & Materials, Inc., Newtown, CT). The resulting lysates were centrifuged at 2,500 × g for 10 min at 4°C using an Allegra X-15R benchtop centrifuge (Beckman Coulter, Fullerton, CA). After centrifugation, the clarified supernatants were applied to 1 ml (each) of HIS-Select nickel affinity gel equilibrated with 5 volumes (5 ml/ml) of equilibration buffer (0.1 M sodium phosphate buffer [Na2HPO4-NaH2PO4], 6 M Gu-HCl, 5 mM DTT) (pH 8.0). After gentle mixing for 3 min at room temperature, the suspensions were centrifuged at 250 × g for 5 min at 4°C. The supernatants were discarded, and the gel with bound proteins was washed two times with 5 ml of wash buffer (0.1 M Na2HPO4-NaH2PO4, 6 M Gu-HCl, 5 mM DTT) (pH 6.3). Then, the six-His-tagged proteins were eluted with 2 ml of elution buffer (0.1 M Na2HPO4-NaH2PO4, 6 M Gu-HCl, 5 mM DTT) (pH 4.5).
The purity of the purified proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and confirmed by Western blot analysis. Concentrations of the purified recombinant proteins were assessed using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) based on the method of Bradford (1).
Expression of the S protein of SARS-CoV.
The VRC 8304 human codon-optimized SARS-CoV S-gene expression vector was kindly provided by Gary Nabel (NIH/NIAID/VRC, Bethesda, MD) (17). Human lymphocyte 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL) supplemented with 10% fetal bovine serum. For transient transfection, approximately 106 293T cells were seeded into each well of six-well plates and incubated for 24 to 48 h at 37°C. Five to ten micrograms (per well) of purified recombinant VRC 8304 was transfected using the Cellfectin reagent (Invitrogen). Briefly, 5 to 10 μg DNA and 10 μl Cellfectin were separately mixed with 100 μl of unsupplemented DMEM each. Then, DNA-containing and Cellfectin-containing solutions were combined and incubated at room temperature for 30 min. Afterward, the final volume of the mixture was adjusted to 2 ml with DMEM. The 293T cells were washed before transfection with unsupplemented DMEM, and then the DNA-Cellfectin mixture was overlaid onto the cells. At 48 h after transfection, cells were fixed using 80% acetone and used for cell culture immunofluorescence (CCIF) assays as described previously (12). To obtain larger amounts of the SARS-CoV S protein, 293T cells were grown in 175-cm2 flasks and transfected with 50 μg of VRC 8304. At 48 h postinfection, cells were harvested and used for Western blot analysis and ELISA.
Western blot analysis.
The Western blot analysis was performed to verify the recombinant protein expression levels and antigenicity or to assess cross-reactivity by using the unpurified CoVs as infected cell lysates. Recombinant proteins and clarified virus- or mock-infected cell supernatants were lysed by boiling them for 5 min in 1× loading buffer (Fermentas, Hanover, MD) in the presence of 200 mM DTT. The purified or unpurified recombinant proteins (20 or 50 μg/lane, respectively) were separated by 12 to 15% SDS-PAGE and transferred to nitrocellulose membranes (trans-blot transfer medium; Bio-Rad, Hercules, CA). The membranes were subsequently blocked (overnight at 4°C) in blocking buffer (PBS, pH 7.4, and 10% nonfat dry milk [NFDM]) and incubated at room temperature for 1 h with human anti-SARS-CoV polyclonal Ab, anti-six-His tag mouse MAb (Invitrogen), or the animal CoV antisera described above. After incubation, membranes were rinsed for 20 min in PBS-0.05% Tween 20, and then the bound Abs were detected with anti-swine, anti-chicken, anti-guinea pig, anti-mouse, anti-bovine, or anti-human immunoglobulin G (IgG) conjugated with horseradish peroxidase at dilutions of 1:1,000, 1:1,000, 1:5,000, 1:1,000, 1:1,000, or 1:1,000, respectively. The immunoprecipitated bands, after rinsing for 20 min in PBS-0.05% Tween 20, were developed using the TMB membrane peroxidase substrate system (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). Developed membranes were washed with distilled water and air dried.
ELISA using crude cell lysates and purified recombinant proteins.
The 96-well ELISA plates (Nunc MaxiSorp high protein-binding capacity ELISA plates; Nunc, San Diego, CA) were coated (coating buffer [20 mM Na2CO3, 20 mM NaHCO3, pH 9.6]) overnight at 4°C with recombinant proteins (50 to 100 ng/well) or animal and human NL-63 CoV-infected cell lysates diluted 1:50 (obtained by two freeze-thaw cycles and then clarified by centrifugation) and purified SARS-CoV diluted 1:2,000. The wells were washed with PBS-0.05% Tween 20 and then blocked with 5% NFDM in PBS. Serially diluted sera were added and incubated for 1 h at 37°C. The plates were washed with PBS-0.05% Tween 20 and incubated for 1 h with anti-swine, anti-chicken, anti-guinea pig, anti-mouse, anti-bovine, or anti-human horseradish peroxidase-conjugated IgG at dilutions of 1:750, 1:1,000, 1:5,000, 1:500, 1:500, or 1:500, respectively. The plates were washed and developed using the TMB Microwell peroxidase substrate system (Kirkegaard & Perry Laboratories, Inc.). Then, the reactions were stopped with 0.1 M HCl, and results were read at an absorbance of 450 nm by using a SpectraMax 340PC384 ELISA reader (Molecular devices, Union City, CA).
All ELISAs described in the present study were performed under stringent conditions to avoid nonspecific reactions: all Abs were diluted using 5% NFDM and 1% Tween 20 in PBS; plates were washed five times at each step. Appropriate negative controls were included in all experiments: mock-infected host cell lines HRT-18, ST, Vero E6, CrFK, A72, LLCK12; mock-transfected 293T cells; vector pET23b backbone-transformed E. coli BL-21(DE3); allantoic fluids from uninfected specific-pathogen-free chicken embryos or homogenized baby turkey gut as a negative control for the group three CoVs; and CoV-negative gnotobiotic pig, guinea pig, rabbit, and mouse and SARS-CoV-negative human sera.
RESULTS
Assessment of cross-reactivity between SARS-CoV and other CoV groups (using unpurified group 1, 2, and 3 CoVs) by ELISA.
To assess the two-way antigenic cross-reactivity between SARS-CoV and animal or HCoVs, the 12 CoVs from the three groups were used in ELISA with the highly specific hyperimmune antisera. Because cross-reactivity was previously shown to exist between SARS-CoV and group 1 CoVs, we included in this study two porcine CoV TGEV strains (Purdue-P115 and Miller-M6) and PRCV-ISU1, CCoV-UCD1, FIPV-79-1146, and HCoV-NL63. BCoV-Mebus, BCoV-DB2, and BRCoV-440 were used as group 2a and IBV-Massachusetts and TCoV-IN as group 3 reference strains. First, we assessed SARS-CoV and other CoV reactivities with the CoV hyperimmune antisera, including SARS-CoV hyperimmune antiserum produced in mice (Table 2), and then we assessed CoV group 1 to 3 reactivities with the SARS-CoV convalescent-phase human sera (data not shown). Cross-reactivity was evident in ELISA between SARS-CoV antigen and group 1 CoV antisera, including TGEV-P115, TGEV-M6, PRCV-ISU1, CCoV-UCD1, and FIPV-79-1146 antisera (Table 2). Interestingly, the levels of cross-reactivity between SARS-CoV antigen and porcine CoV antisera varied: TGEV-P115 and TGEV-M6 antisera demonstrated higher levels of reactivity with SARS-CoV, whereas lower levels of reactivity were observed between SARS-CoV and PRCV-ISU1, FIPV-79-1146, and CCoV-UCD1 antisera.
TABLE 2.
ELISA results for testing intergroup cross-reactivities of CoVs by using unpurified group 1 to 3 CoVs (as CoV-infected crude cell lysates) and specific hyperimmune antisera
| Antiserum (host)b | ELISA titer (R%)a for indicated coronavirus | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group 1 | Group 2a | Group 2b | Group 3 | ||||||
| TGEV-M6c | TGEV-P115 | PRCV-ISU1 | CCoV-UCD1 | FIPV 79-1146 | HCoV-NL63 | BCoV-Mebusd | SARS-CoV Urbani | IBV-Massachusettsd | |
| Group 1 | |||||||||
| TGEV-M6 (Gn pig) | 6,400 (100)g | 6,400 (100) | 3,200 (70) | 1,600 (50) | 1,600 (35) | 1,600 | Negativee | 100 (1.1) | Negative |
| TGEV-P115 (Gn pig) | 51,200 (100) | 51,200 (100) | 25,600 (70) | 12,800 (50) | 12,800 (35) | 12,800 | Negative | 800 (1.1) | Negative |
| PRCV-ISU1 (Gn pig) | 102,400 (70) | 102,400 (70) | 102,400 (100) | 51,200 (70) | 25,600 (35) | 12,800 | Negative | 800 (0.55) | Negative |
| CCoV-UCD1 (GP) | 12,800 (50) | 12,800 (50) | 12,800 (70) | 12,800 (100) | 12,800 (100) | 6,400 | Negative | 100 | Negative |
| FIPV 79-1146 (GP) | 3,200 (35) | 3,200 (35) | 3,200 (35) | 6,400 (100) | 6,400 (100) | 3,200 | Negative | 100 | Negative |
| HCoV-NL63 (GP) | Negative | Negative | Negative | Negative | Negative | 12,800 (100) | Negative | Negative | Negative |
| Group 2a | |||||||||
| BCoV-Mebus (GP/Gn calf) | Negative | Negative | Negative | Negative | Negative | Negative | 25,600 (100) | Negative | Negative |
| Group 2b | |||||||||
| SARS-CoV-Urbani (mouse)f | 200 (1.1) | 200 (1.1) | 100 (0.55) | Negative | Negative | Negative | Negative | 25,600 (100) | Negative |
| Group 3 | |||||||||
| IBV-Massachusetts (GP/chicken) | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | 51,200,102,400 (100) |
Next, CoVs from all three groups were assessed by ELISA with the SARS convalescent-phase sera (CoV samples 3 to 8) and negative human sera (CoV samples 9 to 24). The SARS-CoV reacted with SARS convalescent-phase sera, with titers ranging from 3,200 to 25,600, but did not react with negative human sera (data not shown). All negative human sera demonstrated weak reactivities (titers of 100 to 800) with group 1 and 2a animal CoVs used in the experiment, suggesting the presence of Abs against group 1 and 2a HCoVs in these sera, whereas the reactivities of these CoVs with SARS convalescent-phase sera were slightly (1.4- to 2.5-fold) but not significantly higher, with titers ranging from 200 to 1,600 (data not shown). All human sera (SARS convalescent phase and negative) reacted strongly with the HCoV-NL63 without significant differences in titers, likely due to the presence in all sera of HCoV-NL63-specific Abs or cross-reactive group 1 HCoV-229E Abs. No reactivity was observed between the group 3 CoVs, IBV-Massachusetts and TCoV-IN, and any human sera (data not shown).
We could not confirm cross-reactivity by using only SARS convalescent-phase human sera, because the results would be compromised by the presence of Abs to other HCoVs. Therefore, SARS-CoV mouse hyperimmune antiserum was used to confirm the presence of the two-way cross-reactivity between SARS-CoV and group 1 CoVs. The results clearly demonstrated the presence of a cross-reactive antigenic site(s) for TGEV-P115 and TGEV-M6, whereas PRCV-ISU1 cross-reacted more weakly, based on the relatedness (R%) values as shown in Table 2. Although unpurified CoVs were used in this experiment at the same infectious titers, antigens still could be present in different concentrations and responsible for these variations. However, no reactivity was shown for FIPV, CCoV, HCoV-NL63, and group 2a or 3 CoVs by using the SARS-CoV mouse hyperimmune antiserum.
To demonstrate which SARS-CoV major structural proteins were responsible for the observed cross-reactivity, SARS-CoV S and N proteins and TGEV (M6 and P115), PRCV-ISU1, and HCoV-NL63 N proteins were expressed and assessed by ELISA and Western blot assays.
Cross-reactivity assessment using SARS-CoV S protein expressed in 293T cells.
To test whether any cross-reactivity was mediated through SARS-CoV S protein, the VRC 8304 human codon-optimized SARS-CoV S gene expression vector was used for SARS-CoV S-protein transient expression in 293T cells. Successful expression of the S protein was confirmed by Western blot analysis, CCIF, and ELISA with MAbs or polyclonal Abs to the SARS-CoV S protein. Transfected and subsequently fixed 293T cells were assessed by CCIF. S protein-containing crude cell lysates were used in ELISA and Western blot analysis with the panel of SARS convalescent-phase (CoV samples 3 to 8) and negative human (CoV samples 9 to 24) sera. The recombinant S protein demonstrated strong immunoreactivity with all SARS convalescent-phase sera but did not react with any SARS-negative sera (Table 3). Mouse hyperimmune SARS-CoV antiserum also reacted with S protein at a high titer (1:3,200). In addition, recombinant S protein reacted only with homologous SARS-CoV antisera and no other CoVs or recombinant CoV proteins tested reacted with the SARS-CoV monoclonal or polyclonal Abs to S protein (Table 3). Thus, no cross-reactivity between SARS-CoV and other CoVs mediated by the S protein was demonstrated in any assays.
TABLE 3.
Assessment by ELISA of homologous reactivity and cross-reactivity of the purified recombinant N proteins produced in E. coli (SARS-CoV N-protein fragments, HCoV-NL63 N protein, and TGEV-M6, TGEV-P115, and PRCV N proteins) and unpurified SARS-CoV S protein produced in human 293T cells
| Antiserum (host)c,d | ELISA titer for indicated protein fragment | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV N proteina | SARS-CoV S proteinb | Recombinant N proteins fora: | ||||||||||||
| TGEV-M6 | TGEV-P115 | PRCV-ISU1 | HCoV-NL63 | |||||||||||
| aa 1-72 | aa 70-213 | aa 120-208 | aa 1-213e | aa 212-349 | aa 337-422 | aa 360-412e | aa 212-422 | aa 1-422 | aa 1-1190 | aa 1-383 | aa 1-383 | aa 1-383 | aa 1-378 | |
| Group 1 | ||||||||||||||
| TGEV-M6 (Gn pig) | Negativef | 400 | 400 | 800 | Negative | Negative | Negative | Negative | 200 | Negative | 3,200 | 3,200 | 3,200 | 800 |
| TGEV-P115 (Gn pig) | Negative | 1,600 | 1,600 | 3,200 | Negative | Negative | Negative | Negative | 800 | Negative | 12,800 | 12,800 | 12,800 | 3,200 |
| PRCV-ISU1 (Gn pig) | Negative | 1,600 | 800 | 3,200 | Negative | Negative | Negative | Negative | 200 | Negative | 25,600 | 25,600 | 51,200 | 25,600 |
| CCoV-UCD1 (GP) | Negative | Negative | Negative | 400 | Negative | Negative | Negative | Negative | Negative | Negative | 6,400 | 6,400 | 3,200 | 3,200 |
| FIPV 79-1146 (GP) | Negative | Negative | Negative | 200 | Negative | Negative | Negative | Negative | Negative | Negative | 3,200 | 3,200 | 3,200 | 1,600 |
| HCoV-NL63 (GP) | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | 25,600 |
| Group 2a | ||||||||||||||
| BCoV-Mebus (GP/Gn calf) | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative |
| Group 2b | ||||||||||||||
| SARS-CoV-Urbani (mouse) | Negative | 6,400 | 6,400 | 12,800 | Negative | Negative | Negative | 800 | 3,200 | 3,200 | 400 | 400 | 100 | Negative |
| Group 3 | ||||||||||||||
| IBV-Massachusetts (GP/chicken) | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative |
| Human SARS convalescent-phase sera | ||||||||||||||
| CoV sample 3 | Negative | 1,600 | 3,200 | 51,200 | 1,600 | 3,200 | 12,800 | 3,200 | 1,600 | 1,600 | 200 | 200 | 100 | 12,800 |
| CoV sample 4 | Negative | 1,600 | 1,600 | 51,200 | 800 | 1,600 | 12,800 | 1,600 | 1,600 | 1,600 | 200 | 400 | Negative | 12,800 |
| CoV sample 5 | 100 | 800 | 1,600 | 12,800 | 800 | 1,600 | 1,600 | 1,600 | 800 | 800 | 100 | 100 | 100 | 800 |
| CoV sample 6 | 100 | 1,600 | 3,200 | 25,600 | 1,600 | 3,200 | 6,400 | 3,200 | 1,600 | 1,600 | 400 | 400 | 200 | 12,800 |
| CoV sample 7 | Negative | 800 | 1,600 | 12,800 | 800 | 800 | 3,200 | 800 | 800 | 800 | Negative | Negative | Negative | 1,600 |
| CoV sample 8 | Negative | 1,600 | 1,600 | 25,600 | 800 | 800 | 3,200 | 1,600 | 800 | 1,600 | 100 | 100 | Negative | 6,400 |
| Human negative sera | ||||||||||||||
| CoV samples 9-19 | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | 400-6,400 |
| CoV sample 20 | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | 3,200 |
| CoV sample 21 | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | 6,400 |
| CoV samples 22-24 | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | Negative | 800-6,400 |
Assessment of cross-reactivity by use of full-length recombinant CoV N proteins expressed in E. coli BL-21(DE3).
To confirm N-protein involvement in cross-reactivity, three oligonucleotide pairs containing engineered restriction enzyme sites were designed (Table 1) and used for full-length cloning of N proteins of SARS-CoV-Urbani, two TGEV strains (Miller-M6 and Purdue-P115), the PRCV-ISU1 strain, and HCoV-NL63. After reverse transcription-PCR, N genes were subsequently cloned in the pET23b plasmid and inducible expression was performed. Western blot analysis using specific antisera (to SARS-CoV, HCoV-NL63, TGEV, and PRCV) or anti-histidine MAb revealed high expression levels of the rNs with predicted molecular masses, which were determined to be as follows: 49 kDa for SARS-CoV and 43 to 44 kDa for TGEV, PRCV, and HCoV-NL63. For use in ELISA, recombinant proteins were purified using HIS-Select nickel affinity gel (Sigma-Aldrich).
The reactivities of all rNs were analyzed by ELISA with the panel of animal antisera to the CoVs (Table 3). The results confirmed previous findings for porcine CoV antisera: the strongest cross-reactivity was demonstrated between SARS-CoV rN and TGEV-P115 and TGEV-M6 antisera, with weaker cross-reactivity with PRCV-ISU1 antiserum. Anti-CCoV and anti-FIPV sera reacted broadly with all group 1 rNs, including HCoV-NL63 rN, but with the SARS-CoV rN, the level of cross-reactivity was below the detection threshold in ELISA, although it reacted with these sera in Western blot analysis (data not shown). No cross-reactivity was observed between the group 1 CoVs or SARS-CoV rNs and group 2a BCoV-Mebus and group 3 IBV-Massachusetts antisera. Although all group 1 CoV antisera recognized HCoV-NL63 rN, HCoV-NL63 antiserum did not cross-react with any but the homologous rN.
The ELISA results obtained using the CoV rNs and SARS convalescent-phase and negative human sera were similar to those obtained with crude CoV-infected cell lysates (Table 3), suggesting the presence of SARS-CoV N-protein specific Ab as well as cross-reactive Abs to group 1 CoV N proteins in these sera. The SARS-CoV rN discriminated positive and negative human sera. The TGEV-M6 and TGEV-P115 rNs reacted with most SARS convalescent-phase sera at low titers, but unlike for the unpurified CoVs, they did not react with the negative human sera. The level of PRCV rN reactivity with SARS convalescent-phase antisera was lower; PRCV rN reacted with low titers with half of the tested SARS convalescent-phase sera but was also undetectable with the negative human sera. The HCoV-NL63 rN reacted at high titers with both convalescent-phase and negative human sera, but with some SARS convalescent-phase sera (CoV samples 3, 4, and 6), the titers were twice as high as for negatives (Table 3). In addition, rNs from TGEV and PRCV reacted specifically with mouse-derived SARS-CoV hyperimmune antiserum at low titers (100 to 400), whereas for the SARS-CoV rN, the titer was 3,200 (Table 3).
The results from this experiment prove that cross-reactivity between SARS and animal group 1 CoVs appears to be two way. However, no cross-reactivity between SARS and porcine CoVs was shown in ELISA or Western blot analysis using SARS-CoV or TGEV-M6 anti-N MAbs. The SARS-CoV anti- N MAbs reacted only with SARS-CoV rN, and TGEV-M6 anti-N MAbs reacted with rNs of group 1 CoVs, TGEV- M6, TGEV-P115, PRCV-ISU1, and HCoV-NL63 (data not shown).
Identification of the cross-reactive region by use of recombinant fragments of SARS-CoV N protein expressed in E. coli BL-21(DE3).
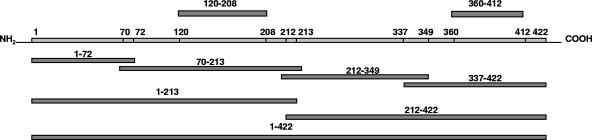
In addition to the full-length gene, eight truncated fragments of the N-protein gene of SARS-CoV (Fig. 1) were amplified with the designed primer pairs. These fragments were predicted (by Pepscan) to carry immunodominant antigenic sites and were shown to carry immunogenic sites in previous studies (15) or were used in commercial SARS-CoV Ab detection systems (Genesis Biotech Inc., Taiwan). One of the N-terminal short fragments, aa 120 to 208, was designed to exclude the possibility that cross-reactivity is mediated through the highly conserved motif FYYLGTGP (aa 111 to 118), which is present in the N proteins of all CoVs. The shortest C-terminal fragment (aa 360 to 412) was included in the study as the most specific region of the SARS-CoV N protein, with a unique lysine-rich amino acid stretch, KTFPPTEPKKDKKKKTDEAQ (aa 362 to 381).
FIG. 1.
Schematic diagram of the eight SARS-CoV N-protein fragments (covering the whole N-protein sequence) and the full-length N protein.
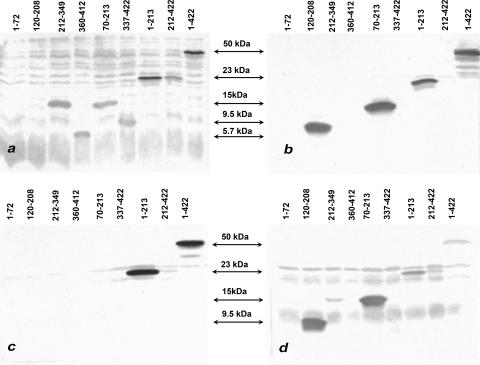
Western blot analysis with anti-histidine MAbs and crude cell lysates of SARS-CoV rN fragments revealed the presence of the eight products of predicted molecular weights. Staining with SARS convalescent-phase antisera (CoV samples 3 to 8) and two SARS-CoV N MAbs (SA 46-4 and SA 87-A1) demonstrated different levels of antigenicity for all fragments. The lowest level of binding activity was shown for the aa-1-to-72 fragment of the SARS-CoV N protein, which did not cross-react with any sera or MAbs, and the highest level of binding activity was shown for aa 1 to 213 (Fig. 2). All SARS convalescent-phase sera demonstrated the same pattern of reactivity with fragments in Western blot analysis, but the levels of reactivity with fragments in serum binding varied according to the different Ab titers. Interestingly, the fragment comprising aa 120 to 208 did not react in Western blot analysis with SARS convalescent-phase serum (Fig. 2a), but it reacted with SARS-CoV N MAb SA87-A1 (Fig. 2b) and subsequently with porcine hyperimmune TGEV/PRCV antisera (Fig. 2d). Also, low-level reactivity was observed in Western blot analysis with some negative human sera for the fragments comprising aa 1 to 213 and 212 to 422 and the full-length (aa 1 to 422) N protein.
FIG. 2.
Cross-reactivities of the _E. coli_-derived unpurified SARS-CoV N-protein fragments as assessed by Western blot analysis with SARS convalescent-phase serum (a), SARS-CoV N MAbs (b, c), or gnotobiotic pig anti-TGEV serum (d).
In the next step, the purified rN fragments of SARS-CoV were used in ELISA (50 to 100 ng per well) with the SARS convalescent-phase (CoV samples 3 to 8) and negative human (CoV samples 9 to 24) sera (Table 3). The rN fragments did not react with negative human sera. The fragment comprising aa 1 to 72 did not react with any negative human sera and reacted at low levels (titers of 100) only with CoV SARS convalescent-phase serum samples 5 and 6. Low to moderate levels of reactivity with SARS-CoV convalescent-phase sera were demonstrated for the fragments comprising aa 120 to 208, 212 to 349, 70 to 213, 337 to 422, 212 to 422, and 1 to 422, whereas the highest level of reactivity was observed for the fragments comprising aa 1 to 213 and 360 to 412 (Table 3). However, the patterns of reactivity were different for the latter two fragments (Table 3). Mouse hyperimmune antiserum to SARS-CoV was also used to assess the immunoreactivities of the N-protein fragments. Surprisingly, this antiserum failed to recognize any short fragments from the C-terminal part (aa 212 to 349, 360 to 412, and 337 to 422), reacted weakly with aa 212 to 422, and demonstrated strong immunoreactivity with all fragments from the N-terminal part of the N protein (except for aa 1 to 72) (Table 3). Thus, using our panel of SARS convalescent-phase and negative human sera and two SARS-CoV anti-N MAbs, we assessed the antigenicities of all expressed fragments and demonstrated that aa 1 to 213 and 360 to 412 were the most antigenic fragments; however, both MAbs targeted antigenic sites in the N-terminal part of the N protein (Fig. 2b and c).
We then assessed the cross-reactivities of all SARS-CoV N-protein fragments in Western blot analysis and ELISA using the group 1 hyperimmune anti-TGEV (anti-P115 [MM973] [Fig. 2d] and anti-Miller M6 [M2]) and anti-PRCV (PP12) sera produced in gnotobiotic pigs and hyperimmune antisera to FIPV-79-1146, CCoV-UCD1, HCoV-NL63 (group 1), BCoV-Mebus (group 2), and IBV-Massachusetts (group 3), all produced in guinea pigs, and to IBV-Massachusetts produced in chickens. Strong reactivity was detected between the fragments comprising aa 120 to 208, 70 to 213 (both strongest), and 1 to 213 and the porcine CoV antisera in Western blot analysis (Fig. 2d). For the full-length N protein (1 to 422), staining was weaker but detectable (Fig. 2d) with all porcine antisera. In addition, low-level reactivity was detected between the fragment comprising aa 1 to 213 and FIPV and CCoV antisera. The ELISA results with purified SARS-CoV N-protein fragments and animal group 1 antisera were mostly consistent with Western blot analysis; however, the fragment comprising aa 1 to 213 demonstrated the strongest cross-reactivity with all animal group 1 CoV antisera and no cross-reactivity was observed for the fragments from the C-terminal part of the N protein (Table 3). No reactivity was evident for any fragments with HCoV-NL63, BCoV-Mebus, or IBV-Massachusetts antisera in either Western blot analysis or ELISA.
Assessment of the antigenicity of the putative cross-reactive region by use of recombinant fragments of HCoV-NL63 N protein expressed in E. coli BL-21(DE3).
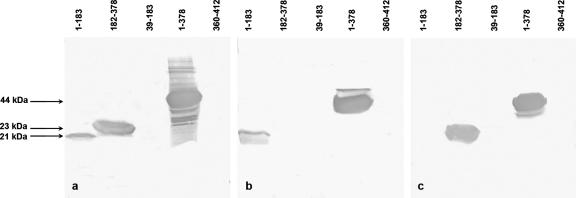
To define which HCoV-NL63 N-protein regions contribute to cross-reactivity with other group 1 CoVs and to test whether the NL63 N-protein region comprising aa 39 to 183, corresponding to the SARS-CoV N-protein cross-reactive region (based on amino acid alignment), possesses high antigenicity, three HCoV-NL63 N-protein fragments were expressed in E. coli: the N-terminal half (aa 1 to 183), the C-terminal half (aa 182 to 378), and the region comprising aa 39 to 183, which corresponds to the longer SARS-CoV cross-reactive N-protein fragment (aa 70 to 213). After successful expression was confirmed, the antigenicities of the recombinant proteins were assessed with the set of hyperimmune and SARS convalescent-phase sera in Western blot analysis and ELISA. Western blot analysis with hyperimmune guinea pig antiserum to HCoV-NL63 demonstrated the presence of a strong antigenic site in the C-terminal part of the N protein (aa 182 to 378), whereas for the N-terminal part (aa 1 to 183), a much lower antigenicity was shown and no reactivity was observed for the aa-39-to-183 fragment (Fig. 3). Hyperimmune antisera to TGEV-P115 and -M6 and TGEV-M6 anti-N MAbs 14G9 and 25H7 reacted with the N-terminal half of the HCoV-NL63 N protein but not with the aa-39-to-183 or aa-182-to-378 fragment, whereas TGEV-M6 anti-N MAb 14E3 and hyperimmune PRCV-ISU1 antiserum reacted with the aa-182-to-378 fragment (Fig. 3). Interestingly, MAbs 25H7 and 14E3 recognized distinct antigenic sites on the TGEV N protein, N1 (aa 1 to 120) and N2 (aa 255 to 383), respectively, whereas MAb 14G9 recognized a third distinct antigenic site, N3 (aa 1 to 205) (52). Human SARS convalescent-phase sera reacted strongly with the N-terminal half; weaker reactivity was observed for the C-terminal half, and no reactivity was shown for the aa-39-to-183 fragment (data not shown). The ELISA results for purified HCoV-NL63 N-protein fragments (50 to 100 ng/well) (Table 4) confirmed those of Western blot analysis by demonstrating the presence of a strong antigenic site in the C-terminal part of the N protein when tested with homologous NL63 antiserum and no reactivity for the putative cross-reactive region (aa 39 to 183) with any heterologous Abs. However, in contrast to results of Western blot analysis, antiserum to HCoV-NL63 reacted with the aa-39-to-183 fragment in ELISA at a low titer (Table 4). Although similar cross-reactivities with the N-terminal part were observed for all three antisera to porcine CoVs, the reactivities with the C-terminal half of the N protein differed dramatically between the three antisera, with the highest level detected for the PRCV-ISU1 antiserum (Table 4). In addition, no reactivity was observed between mouse hyperimmune antiserum to SARS-CoV and any of the three fragments tested in ELISA or Western blot analysis.
FIG. 3.
Cross-reactivities of the _E. coli_-derived, purified HCoV-NL63 N-protein fragments as assessed by Western blot analysis with guinea pig HCoV-NL63 hyperimmune antiserum (a), gnotobiotic pig PRCV-ISU1 antiserum (b), or gnotobiotic pig TGEV-P115 antiserum (c).
TABLE 4.
Assessment by ELISA of homologous reactivity and cross-reactivity of the HCoV-NL63 recombinant N-protein fragments
| Antiserum or TGEV anti-N MAb (host)b | ELISA titer for indicated HCoV-NL63 N-protein fragmenta | |||
|---|---|---|---|---|
| aa 1-183 | aa 182-378 | aa 39-183 | aa 1-378 | |
| Group 1 | ||||
| TGEV-M6 (Gn pig) | 400 | Negativec | Negative | 800 |
| TGEV-P115 (Gn pig) | 400 | 100 | Negative | 3,200 |
| PRCV-ISU1 (Gn pig) | 200 | 12,800 | Negative | 25,600 |
| CCoV-UCD1 (GP) | 100 | 800 | Negative | 3,200 |
| FIPV 79-1146 (GP) | 100 | 800 | Negative | 1,600 |
| HCoV-NL63 (GP) | 6,400 | 51,200 | 800 | 25,600 |
| 25H7 | 800 | Negative | Negative | 1,600 |
| 14E3 | Negative | 6,400 | Negative | 1,600 |
| 14G9 | 1,600 | Negative | Negative | 3,200 |
| Group 2a | ||||
| BCoV-Mebus (GP/Gn calf) | Negative | Negative | Negative | Negative |
| Group 2b | ||||
| SARS-CoV-Urbani (mouse) | Negative | Negative | Negative | Negative |
| Group 3 | ||||
| IBV-Massachusetts (GP/chicken) | Negative | Negative | Negative | Negative |
Analysis of the polymorphic amino acids and SARS-CoV N-protein identity levels with other CoVs N proteins.
To substantiate the different levels of the cross-reactivity observed between SARS-CoV and the porcine CoVs, TGEV (P115 and M6) and PRCV-ISU1, detailed analysis of the four N-protein sequences was performed. Twelve polymorphic positions were detected between the three porcine CoV N proteins (58), and only one of them, alanine (A) at position 120, was common between SARS-CoV and TGEV (P115 and M6), whereas valine (V) was detected at this position for PRCV-ISU1, which might contribute to its decreased cross-reactivity with SARS-CoV N proteins, although this needs to be confirmed by mutagenesis analysis. The calculated amino acid sequence identities of the SARS-CoV and the three porcine CoV N proteins were 25.8% (109 aa), 25.8% (109 aa), and 25.1% (106 aa) for TGEV-P115, TGEV-M6, and PRCV-ISU1, respectively (Table 5). Additionally, the HCoV-NL63, CCoV, FIPV, BCoV, and IBV N-protein sequences were analyzed and their amino acid identities with SARS-CoV N protein are shown in Table 5. The HCoV-NL63 strain was the only group 1 CoV tested that did not cross-react with SARS-CoV. Nevertheless, we failed to demonstrate lower sequence identity (compared to those of other group 1 CoVs) with SARS-CoV N protein or to identify critical point mutations that might diminish cross-reactivity. In total, we identified 10 polymorphic positions within the cross-reactive region common for SARS-CoV and HCoV-NL63 rNs that differed in TGEV and PRCV rNs, but we also identified 10 polymorphic positions common for SARS-CoV and porcine group 1 CoVs that differed in HCoV-NL63 rN. Any of the latter positions might influence the cross-reactive antigenic site.
TABLE 5.
Amino acid identities between SARS-CoV-Urbani N protein and N proteins of the other CoVs
| Sequence (GenBank accession no.) | % SARS-CoV N-protein aa identity | Cross-reactivity | |
|---|---|---|---|
| Total | Within cross-reactive region (aa 70-213) | ||
| TGEV-M6 (DQ811785) | 25.8 | 36 | Yes |
| TGEV-P115 (DQ811788) | 25.8 | 36 | Yes |
| PRCV-ISU1 (DQ811787) | 25.1 | 35.6 | Yes |
| CCoV-BGF10 (AY342160) | 27 | 40.5 | Yes |
| FIPV-WSU 79/1146 (NC_007025) | 26 | 35.6 | Yes |
| HCoV-NL63 (DQ846901) | 27 | 35.6 | No |
| BCoV-Mebus (U00735) | 34.6 | 46.8 | No |
| IBV-Massachusetts 41 (AY851295) | 26 | 35.6 | No |
DISCUSSION
In 2002, a new CoV emerged in Guandong province, the People's Republic of China, and within months, it spread globally and caused over 800 human deaths. The infection was associated with serious respiratory failure, and the pathogen was designated SARS-CoV (6, 21, 22, 34). The SARS-CoV is now recognized to be of zoonotic origin, with bat species as the likely wild-animal reservoir (23, 26) and Himalayan palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides) as immediate sources of infection (11, 19, 50). Previously reported interspecies transmission and wild-animal reservoirs for the different animal CoVs (18, 31, 39, 40, 47), recent progress in understanding the SARS-CoV molecular mechanisms of replication, and successful experiments for transmission of SARS-CoV to a number of animals (27, 33, 35, 37, 38, 43, 53) support the possibility of SARS-CoV emergence from wild animals. In addition, SARS-CoV transmission from humans to pigs has been reported (4). Contradictive findings regarding SARS-CoV relatedness with other CoVs (9, 20, 32, 36, 41, 42, 55, 59), along with its segregated position in the CoV genus, suggest that it has undergone more complicated evolutionary events than just an early split-off from CoV group 2a or that convergent evolutionary events occurred, leading to the outcomes observed.
Antigenic cross-reactivity between SARS-CoV and animal group 1 CoVs was reported by several research groups (21, 44), including our preliminary studies (H. S. Nagesha et al., presented at the 23rd annual meeting of the American Society for Virology, McGill University, Montreal, Quebec, Canada, 2004), and it was attributed to the N protein (44; H. S. Nagesha et al., presented at the 23rd annual meeting of the American Society for Virology, McGill University, Montreal, Quebec, Canada, 2004). To our knowledge, no antigenic cross-reactivity has been reported between SARS-CoV and animal group 2a or 3 CoVs, which is consistent with the findings in this report based on ELISA and Western blot analysis.
Also consistent with previously published results, we confirmed that the TGEV-P115, TGEV-M6, and PRCV-ISU1 antisera (group 1) from gnotobiotic pigs cross-reacts with SARS-CoV in ELISA and Western blot analysis. Gnotobiotic pigs are free from extraneous microbes and maternal or preexisting Abs to CoVs. They are the original host species for TGEV and PRCV, reflecting the full spectrum of postinfection and posthyperimmunization-induced Abs (all pigs received oral/i.n. exposure to the CoVs and were recovered from infection prior to parenteral hyperimmunization) that may influence the Ab specificities and results. The cross-reactivity of the guinea pig antisera produced against inactivated CCoV and FIPV with SARS-CoV was lower than that for TGEV antisera, possibly because the Ab spectra induced in heterologous hosts by parenteral injection of inactivated CoVs are not equivalent to those that develop in the natural host postinfection. The SARS-CoV hyperimmune mouse antiserum allowed us to confirm the two-way cross-reactivity between SARS-CoV and group 1 CoVs, with the highest level of reactivity between this antiserum and TGEV-P115 and -M6. The broad reactivity of the group 1 HCoV-NL63 with all sera of human origin suggested the widespread presence of group 1 CoV Abs to HCoV-NL63 or the cross-reactive 229E strain in the adult sera. Because we did not observe cross-reactivity between SARS-CoV and HCoV-NL63 (although the latter reacted in one-way cross-reactivity assays with all antisera to group 1 CoVs), we concluded that the cross-reactivity observed was not a common feature for all group 1 CoVs and SARS-CoV. Of interest, NL63 is in a phylogenetically separate branch relative to the animal group 1 CoVs that we tested and is grouped with HCoV-229E and the porcine epidemic diarrhea CoV (5, 10). We were unable to test porcine epidemic diarrhea CoV due to U.S. import restrictions on animal pathogens.
The two major structural proteins and immunogens, the nucleocapsid and spike proteins, were important for assessment in regard to cross-reactivity. Both proteins were previously demonstrated to be highly immunogenic, with the S protein bearing the antigenic sites for neutralizing Abs (13, 14, 16, 57), whereas the N protein was the most abundant viral protein, with its earliest appearance during SARS-CoV infection (2, 24, 25, 45, 49). Because of the absence of cross-reactivity mediated through the SARS-CoV S protein, we focused our remaining work on investigation of the N-protein contribution to cross-reactivity. Whether other SARS-CoV structural proteins (E, M) can also contribute to cross-reactivity has not been determined.
The moderate to weak cross-reactivity between TGEV-P115, TGEV-M6, and PRCV rNs and SARS convalescent-phase sera did not provide conclusive results, due to the likely presence of Abs to group 1 CoVs in these sera, as suggested by the extensive cross-reactivities of both SARS-CoV Ab-positive and -negative human sera with the group 1 HCoV-NL63 and HCoV-NL63 rNs. However, unlike the crude CoV-infected cell lysates, the rNs did not react with negative human sera, probably because the cross-reactivities with unpurified CoVs were mediated through Abs to several antigens of group 1 CoVs and not only to the N protein. Consistent with previous findings (30, 51), SARS-CoV rN in Western blot analysis produced few positive results with several of the confirmed negative human sera; however, it allowed reliable discrimination between SARS convalescent-phase and negative human sera in ELISA. The reactivity pattern of the antisera to porcine CoVs with SARS-CoV rN was similar to those obtained with the unpurified CoVs, so we concluded that the N protein is responsible for the cross-reactivity between SARS-CoV and porcine group 1 CoVs, which is consistent with previously published results (44). However, no such conclusion could be drawn for the HCoV-NL63 rN, which reacted with both SARS convalescent-phase and negative human sera but not with mouse hyperimmune antiserum to SARS-CoV. Considering the absence of cross-reactivity mediated by the SARS-CoV S protein and the fact that immunological responses in SARS patients are predominantly directed to the N protein (2, 8, 45), we hypothesized that some cross-reactive antigenic site(s) in the SARS-CoV N protein may be responsible for the observed cross-reactivity and also that variations in their primary sequences can influence the structures of the antigenic sites (56), leading to the different levels of cross-reactivity with the group 1 animal CoVs.
Identification of the cross-reactive (with TGEV-P115, TGEV-M6, and PRCV-ISU1 antisera) fragments, aa 70 to 213, 120 to 208, and 1 to 213, confirmed the results obtained with unpurified CoVs and full-length rNs, demonstrating the strongest binding efficiency for anti-P115 and -M6 sera and lower levels of reactivity for the fragment comprising aa 1 to 213 with hyperimmune anti-CCoV and hyperimmune anti-FIPV sera. Because the observed cross-reactivity was highest for the fragment comprising aa 1 to 213 but was not greatly decreased for the shortest fragment, aa 120 to 208, we speculate that most of the cross-reactive antigenic site was imbedded within the aa-120-to-208 fragment, probably spanning beyond its borders, and that the highly conserved motif FYYLGTGP (aa 111 to 118) is not likely the source of the observed cross-reactivity, because its exclusion in the fragment comprising aa 120 to 208 did not affect cross-reactivity compared with that in the fragment comprising aa 70 to 213.
Although we did not have an opportunity to use a sufficient panel of human sera, from the results that we obtained, the SARS-CoV N-protein fragments comprising aa 1 to 213 and 360 to 412 appeared to be valuable components for a SARS-CoV Ab detection ELISA. For these fragments, 100% sensitivity and specificity were shown (using the available set of sera), and moreover, these fragments were capable of recognizing all SARS convalescent-phase sera with higher efficiency than systems based on full-length S or N protein, with the detection efficiency equal to that of the SARS-CoV based ELISA. This finding suggests that these fragments carry highly antigenic linear sites, which are the main targets for immune responses to the whole N protein and, possibly, to the intact SARS-CoV. Although the aa-360-to-412 fragment failed to react with SARS-CoV hyperimmune antiserum produced in a mouse, this does not diminish but may in fact enhance its value for SARS diagnosis in the infected human population. This failure might be related to differences in the antigen presentation or immunogenicity of these sites between humans and mice (16, 28) or the fact that the human sera represented postinfection convalescent-phase sera and the mouse hyperimmune antiserum was produced using inactivated SARS-CoV given parenterally, followed by a boost with recombinant SARS-CoV N protein. The fragment comprising aa 1 to 213 also contained immunodominant sites, and although our studies demonstrated that it was responsible for the cross-reactivity with animal group 1 CoVs, surprisingly it did not seem to contain epitopes cross-reactive with another group 1 HCoV, HCoV-NL63. This finding suggests that SARS-CoV evolution was isolated from that of HCoV-NL63 and probably was more closely related to that of the animal CoVs. However, further studies including other human and animal CoVs are required to reach a more definitive conclusion.
The observed low-level reactivity of the aa-1-to-213, aa-212-to-422, and aa-1-to-422 fragments with some human negative sera in Western blot analysis did not appear to be related to sites that shared higher sequence identity between SARS- and other HCoVs, because staining was equally weak for all three fragments and could be due to other reasons, including possible N-protein sequence identity with some human tissues (51), cross-reactivity with some undetected human pathogen, or a nonspecific affinity of IgG to the recombinant nonglycosylated N protein (29, 30).
Alignment of the amino acid sequences of the three porcine CoV and SARS-CoV N proteins did not reveal major differences potentially responsible for the observed variations in cross-reactivity levels for the animal group 1 CoVs (TGEV-P115, TGEV-M6, PRCV), because the N-protein sequence is highly conserved within groups and all three porcine group 1 CoV N proteins possessed 97% amino acid identities between one another, with infrequent point mutations (58). Only 1 out of 12 polymorphic positions for the TGEV-M6, TGEV-P115, and PRCV-ISU1 N proteins appeared to be critical: a valine (V) at aa position 120 in the PRCV-ISU1 N protein (within the cross-reactive region) might have diminished its cross-reactivity with SARS-CoV, because both TGEV (M6 and p115) and SARS-CoV contain alanine (A) at this position (58). Considering that the cross-reactivity observed may not be a direct consequence of the amino acid sequence identity, one common feature of N proteins from TGEV, PRCV, and SARS-CoV should be noted: the presence of a strong immunodominant site in the N-terminal (cross-reactive) region. This was previously reported for the SARS-CoV (15), and we found similar results for TGEV and PRCV (unpublished data). The TGEV and SARS-CoV N proteins were shown to be RNA chaperons with long disordered regions (60). Similar, very short antigenic sites (as opposed to conformational sites that would have been lost during protein purification under the denaturing conditions that we used) are probably exposed during natural infection in the host species. This, together with the overall low identity of the primary sequences of SARS-CoV and porcine CoVs, suggests that one point mutation within a cross-reactive region can be critical for the cross-reactive site structure and responsible for the observed variations in the levels of cross-reactivity with porcine group 1 CoVs. Interestingly, the most antigenic site of the HCoV-NL63 rN, as reflected by the hyperimmune antiserum, was located in the C terminus, whereas cross-reactivity with group 1 CoVs was mediated through at least two antigenic sites in the N (presumably aa 1 to 39) and C (aa 182 to 378) termini of the HCoV-NL63 N protein, whereas the putative cross-reactive (aa 39 to 183) N-protein fragment of HCoV-NL63 failed to react with any heterologous Abs. Therefore, this fragment of the HCoV-NL63 N protein differed in its antigenicity and antigenic cross-reactivity from the SARS-CoV and animal group 1 CoVs. This could explain the absence of cross-reactivity with SARS-CoV, but as noted for animal group 1 CoVs, the determinants of antigenicity could not be identified at the primary amino acid sequence level.
In conclusion, we confirmed the two-way antigenic cross-reactivity between SARS-CoV and porcine group 1 CoVs (TGEVs [M6 and P115] and PRCV-ISU1) and demonstrated that it was attributable to the SARS-CoV and group 1 CoV N proteins and not the S protein. We identified the cross-reactive region and one point mutation within this region likely responsible for the observed variations in the levels of cross-reactivity with porcine group 1 CoVs. In addition, our findings suggest that the fragments comprising aa 1 to 213 and 360 to 412 of the SARS-CoV N protein when used together can be valuable discriminatory diagnostic reagents, with the latter reflecting only SARS-CoV specificity. Use of such paired reagents could overcome the problems of nonspecificity demonstrated for the full-length N protein for testing both human and animal sera.
Acknowledgments
We thank M. Azevedo, P. Lewis, G. Myers, R. McCormick, J. McCormick, and Y. Tang for technical assistance. We also thank T. Ksiazek and J. A. Comer (CDC, Atlanta, GA) and Gary Nabel (NIH/NIAID/VRC, Bethesda, MD) for the reagents for SARS-CoV diagnosis provided and Y. M. Saif and M. Jackwood for the group 3 CoVs and specific antisera provided.
This work was supported by grant R21 AI062763 from the NIAID, NIH. Salaries and research support were provided by state and federal funds to the Ohio Agricultural Research and Development Center, The Ohio State University.
Footnotes
▿
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72**:**248-254. [DOI] [PubMed] [Google Scholar]
- 1a.Chan, K. H., V. C. Cheng, P. C. Woo, S. K. Lau, L. L. Poon, Y. Guan, W. H. Seto, K. Y. Yuen, and J. S. Peiris. 2005. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 12**:**1317-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Che, X. Y., W. Hao, Y. Wang, B. Di, K. Yin, Y. C. Xu, C. S. Feng, Z. Y. Wan, V. C. Cheng, and K. Y. Yuen. 2004. Nucleocapsid protein as early diagnostic marker for SARS. Emerg. Infect. Dis. 10**:**1947-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Che, X. Y., L. W. Qiu, Z. Y. Liao, Y. D. Wang, K. Wen, Y. X. Pan, W. Hao, Y. B. Mei, V. C. Cheng, and K. Y. Yuen. 2005. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 191**:**2033-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W., M. Yan, L. Yang, B. Ding, B. He, Y. Wang, X. Liu, C. Liu, H. Zhu, B. You, S. Huang, J. Zhang, F. Mu, Z. Xiang, X. Feng, J. Wen, J. Fang, J. Yu, H. Yang, and J. Wang. 2005. SARS-associated coronavirus transmitted from human to pig. Emerg. Infect. Dis. 11**:**446-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkman, R., M. F. Jebbink, B. Wilbrink, K. Pyrc, H. L. Zaaijer, P. D. Minor, S. Franklin, B. Berkhout, V. Thiel, and L. van der Hoek. 2006. Human coronavirus 229E encodes a single ORF4 protein between the spike and the envelope genes. Virol. J. 3**:**106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten, C., W. Preiser, S. Gunther, H. Schmitz, and H. W. Doerr. 2003. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol. Med. 9**:**325-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, Y., A. Pekosz, L. Haynes, E. A. Nelson, and R. R. Rowland. 2006. Production and characterization of monoclonal antibodies against the nucleocapsid protein of SARS-CoV. Adv. Exp. Med. Biol. 581**:**153-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flego, M., P. Di Bonito, A. Ascione, S. Zamboni, A. Carattoli, F. Grasso, A. Cassone, and M. Cianfriglia. 2005. Generation of human antibody fragments recognizing distinct epitopes of the nucleocapsid (N) SARS-CoV protein using a phage display approach. BMC Infect. Dis. 5**:**73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbs, A. J., M. J. Gibbs, and J. S. Armstrong. 2004. The phylogeny of SARS coronavirus. Arch. Virol. 149**:**621-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E., E. J. Snijder, and W. J. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78**:**7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302**:**276-278. [DOI] [PubMed] [Google Scholar]
- 12.Hasoksuz, M., S. L. Lathrop, K. L. Gadfield, and L. J. Saif. 1999. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 60**:**1227-1233. [PubMed] [Google Scholar]
- 13.He, Y., Y. Zhou, S. Liu, Z. Kou, W. Li, M. Farzan, and S. Jiang. 2004. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 324**:**773-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, Y., Y. Zhou, P. Siddiqui, and S. Jiang. 2004. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 325**:**445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, Y., Y. Zhou, H. Wu, Z. Kou, S. Liu, and S. Jiang. 2004. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42**:**5309-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, Y., Y. Zhou, H. Wu, B. Luo, J. Chen, W. Li, and S. Jiang. 2004. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 173**:**4050-4057. [DOI] [PubMed] [Google Scholar]
- 17.Huang, Y., Z. Y. Yang, W. P. Kong, and G. J. Nabel. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78**:**12557-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ismail, M. M., K. O. Cho, L. A. Ward, L. J. Saif, and Y. M. Saif. 2001. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. 45**:**157-163. [PubMed] [Google Scholar]
- 19.Kan, B., M. Wang, H. Jing, H. Xu, X. Jiang, M. Yan, W. Liang, H. Zheng, K. Wan, Q. Liu, B. Cui, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, X. Qi, K. Chen, L. Du, K. Gao, Y. T. Zhao, X. Z. Zou, Y. J. Feng, Y. F. Gao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 79**:11892-11900. (Erratum, 80:**7786.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, O. J., D. H. Lee, and C. H. Lee. 2006. Close relationship between SARS-coronavirus and group 2 coronavirus. J. Microbiol. 44**:**83-91. [PubMed] [Google Scholar]
- 21.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and S. W. Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348**:**1953-1966. [DOI] [PubMed] [Google Scholar]
- 22.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362**:**263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102**:**14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung, D. T., F. C. Tam, C. H. Ma, P. K. Chan, J. L. Cheung, H. Niu, J. S. Tam, and P. L. Lim. 2004. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 190**:**379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, S., L. Lin, H. Wang, J. Yin, Y. Ren, Z. Zhao, J. Wen, C. Zhou, X. Zhang, X. Li, J. Wang, Z. Zhou, J. Liu, J. Shao, T. Lei, J. Fang, N. Xu, and S. Liu. 2003. The epitope study on the SARS-CoV nucleocapsid protein. Genomics Proteomics Bioinformatics 1**:**198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310**:**676-679. [DOI] [PubMed] [Google Scholar]
- 27.Li, W., S. K. Wong, F. Li, J. H. Kuhn, I. C. Huang, H. Choe, and M. Farzan. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 80**:**4211-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S. J., C. H. Leng, S. P. Lien, H. Y. Chi, C. Y. Huang, C. L. Lin, W. C. Lian, C. J. Chen, S. L. Hsieh, and P. Chong. 2006. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24**:**3100-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, W., X. D. Wu, M. D. Shi, R. F. Yang, Y. Y. He, C. Bian, T. L. Shi, S. Yang, X. L. Zhu, W. H. Jiang, Y. X. Li, L. C. Yan, Y. Y. Ji, Y. Lin, G. M. Lin, L. Tian, J. Wang, H. X. Wang, Y. H. Xie, G. Pei, J. R. Wu, and B. Sun. 2005. Synthetic peptides derived from SARS coronavirus S protein with diagnostic and therapeutic potential. FEBS Lett. 579**:**2130-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maache, M., F. Komurian-Pradel, A. Rajoharison, M. Perret, J. L. Berland, S. Pouzol, A. Bagnaud, B. Duverger, J. Xu, A. Osuna, and G. Paranhos-Baccala. 2006. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid-based western blot assay were rectified by the use of two subunits (S1 and S2) of spike for detection of antibody to SARS-CoV. Clin. Vaccine Immunol. 13**:**409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majhdi, F., H. C. Minocha, and S. Kapil. 1997. Isolation and characterization of a coronavirus from elk calves with diarrhea. J. Clin. Microbiol. 35**:**2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300**:**1399-1404. [DOI] [PubMed] [Google Scholar]
- 33.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425**:**915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peiris, J. S., Y. Guan, and K. Y. Yuen. 2004. Severe acute respiratory syndrome. Nat. Med. 10**:**S88-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin, E., H. Shi, L. Tang, C. Wang, G. Chang, Z. Ding, K. Zhao, J. Wang, Z. Chen, M. Yu, B. Si, J. Liu, D. Wu, X. Cheng, B. Yang, W. Peng, Q. Meng, B. Liu, W. Han, X. Yin, H. Duan, D. Zhan, L. Tian, S. Li, J. Wu, G. Tan, Y. Li, Y. Li, Y. Liu, H. Liu, F. Lv, Y. Zhang, X. Kong, B. Fan, T. Jiang, S. Xu, X. Wang, C. Li, X. Wu, Y. Deng, M. Zhao, and Q. Zhu. 2006. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine 24**:**1028-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rest, J. S., and D. P. Mindell. 2003. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 3**:**219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, A., L. Vogel, J. Guarner, N. Hayes, B. Murphy, S. Zaki, and K. Subbarao. 2005. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 79**:**503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowe, T., G. Gao, R. J. Hogan, R. G. Crystal, T. G. Voss, R. L. Grant, P. Bell, G. P. Kobinger, N. A. Wivel, and J. M. Wilson. 2004. Macaque model for severe acute respiratory syndrome. J. Virol. 78**:**11401-11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saif, L. J. 2004. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 23**:**643-660. [DOI] [PubMed] [Google Scholar]
- 40.Saif, L. J. 2005. Comparative biology of animal coronaviruses: lessons for SARS, p. 84-89. In M. Peiris, Anderson, L. J., Osterhaus, A. D. M. E., Stohr, K. and Yuen, K. Y. (ed.), Severe acute respiratory syndrome. Blackwell Publishing, Oxford, United Kingdom.
- 41.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331**:**991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stavrinides, J., and D. S. Guttman. 2004. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 78**:**76-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W. J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78**:**3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, Z. F., and X. J. Meng. 2004. Antigenic cross-reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group I animal coronaviruses: implication for SARS diagnosis. J. Clin. Microbiol. 42**:**2351-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan, Y. J., P. Y. Goh, B. C. Fielding, S. Shen, C. F. Chou, J. L. Fu, H. N. Leong, Y. S. Leo, E. E. Ooi, A. E. Ling, S. G. Lim, and W. Hong. 2004. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 11**:**362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripp, R. A., L. M. Haynes, D. Moore, B. Anderson, A. Tamin, B. H. Harcourt, L. P. Jones, M. Yilla, G. J. Babcock, T. Greenough, D. M. Ambrosino, R. Alvarez, J. Callaway, S. Cavitt, K. Kamrud, H. Alterson, J. Smith, J. L. Harcourt, C. Miao, R. Razdan, J. A. Comer, P. E. Rollin, T. G. Ksiazek, A. Sanchez, P. A. Rota, W. J. Bellini, and L. J. Anderson. 2005. Monoclonal antibodies to SARS-associated coronavirus (SARS-CoV): identification of neutralizing and antibodies reactive to S, N, M and E viral proteins. J. Virol. Methods 128**:**21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsunemitsu, H., Z. R. el-Kanawati, D. R. Smith, H. H. Reed, and L. J. Saif. 1995. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J. Clin. Microbiol. 33**:**3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10**:**368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J., J. Wen, J. Li, J. Yin, Q. Zhu, H. Wang, Y. Yang, E. Qin, B. You, W. Li, X. Li, S. Huang, R. Yang, X. Zhang, L. Yang, T. Zhang, Y. Yin, X. Cui, X. Tang, L. Wang, B. He, L. Ma, T. Lei, C. Zeng, J. Fang, J. Yu, J. Wang, H. Yang, M. B. West, A. Bhatnagar, Y. Lu, N. Xu, and S. Liu. 2003. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin. Chem. 49**:**1989-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, M., M. Yan, H. Xu, W. Liang, B. Kan, B. Zheng, H. Chen, H. Zheng, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, Y. F. Liu, R. T. Guo, X. N. Liu, L. H. Zhan, D. H. Zhou, A. Zhao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 11**:**1860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Y., S. Sun, H. Shen, L. Jiang, M. Zhang, D. Xiao, Y. Liu, X. Ma, Y. Zhang, N. Guo, and T. Jia. 2004. Cross-reaction of SARS-CoV antigen with autoantibodies in autoimmune diseases. Cell. Mol. Immunol. 1**:**304-307. [PubMed] [Google Scholar]
- 52.Welch, S. K., and L. J. Saif. 1988. Monoclonal antibodies to a virulent strain of transmissible gastroenteritis virus: comparison of reactivity with virulent and attenuated virus. Arch. Virol. 101**:**221-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wentworth, D. E., L. Gillim-Ross, N. Espina, and K. A. Bernard. 2004. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 10**:**1293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, D., C. Tu, C. Xin, H. Xuan, Q. Meng, Y. Liu, Y. Yu, Y. Guan, Y. Jiang, X. Yin, G. Crameri, M. Wang, C. Li, S. Liu, M. Liao, L. Feng, H. Xiang, J. Sun, J. Chen, Y. Sun, S. Gu, N. Liu, D. Fu, B. T. Eaton, L. F. Wang, and X. Kong. 2005. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 79**:**2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yap, Y. L., X. W. Zhang, and A. Danchin. 2003. Relationship of SARS-CoV to other pathogenic RNA viruses explored by tetranucleotide usage profiling. BMC Bioinformatics 4**:**43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoo, D., and D. Deregt. 2001. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin. Diagn. Lab. Immunol. 8**:**297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, H., G. Wang, J. Li, Y. Nie, X. Shi, G. Lian, W. Wang, X. Yin, Y. Zhao, X. Qu, M. Ding, and H. Deng. 2004. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 78**:**6938-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, X., M. Hasoksuz, D. Spiro, R. Halpin, S. Wang, S. Stollar, D. Janies, N. Hadya, Y. Tang, E. Ghedin, and L. Saif. 2007. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology 358**:**424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, G., and H. W. Chen. 2004. Monophyletic relationship between severe acute respiratory syndrome coronavirus and group 2 coronaviruses. J. Infect. Dis. 189**:**1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuniga, S., I. Sola, J. L. Moreno, P. Sabella, J. Plana-Duran, and L. Enjuanes. 2007. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 357**:**215-227. [DOI] [PMC free article] [PubMed] [Google Scholar]